Machine Learning as a Tool for Early Detection: A Focus on Late-Stage Colorectal Cancer across Socioeconomic Spectrums
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Machine Learning Approaches in Predicting Cancer Outcomes—A Literature Review
1.2. Role of Social Determinants of Health in Cancer Research
1.3. Study Purpose
2. Materials and Methods
2.1. Study Design and Setting
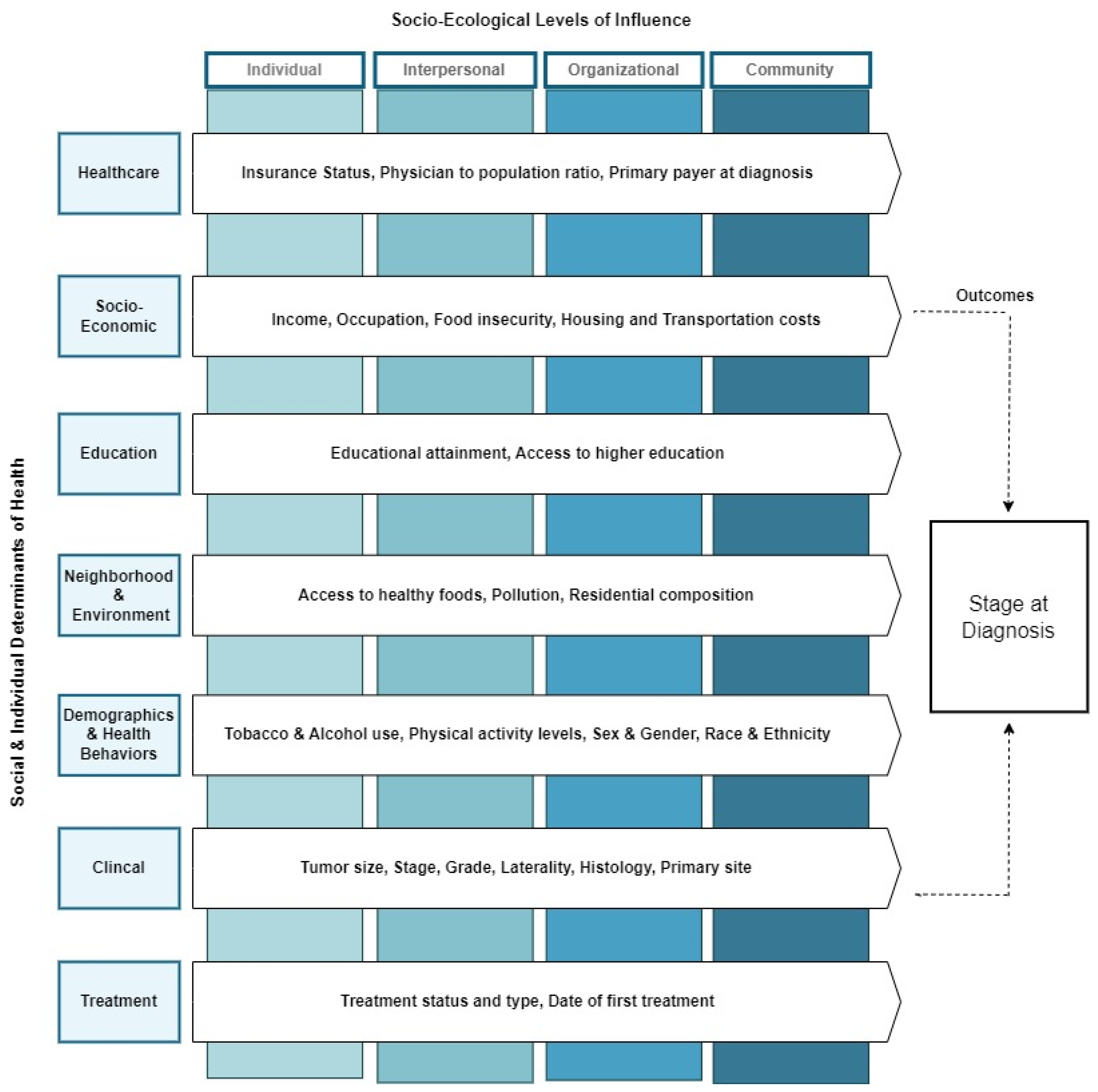
2.2. Theoretical Framework
2.3. Data Sources
2.4. Study Population
2.5. Data Collection and Variables Definitions
2.6. Data Linkage and Management
2.7. An Overview of Machine Learning Techniques in Predicting Cancer Outcomes
2.8. Statistical Analyses
3. Results
3.1. Baseline Characteristics
3.2. Census Tracts Characteristics by Stage at Diagnosis
3.3. Results of the Predictive Machine Learning Models
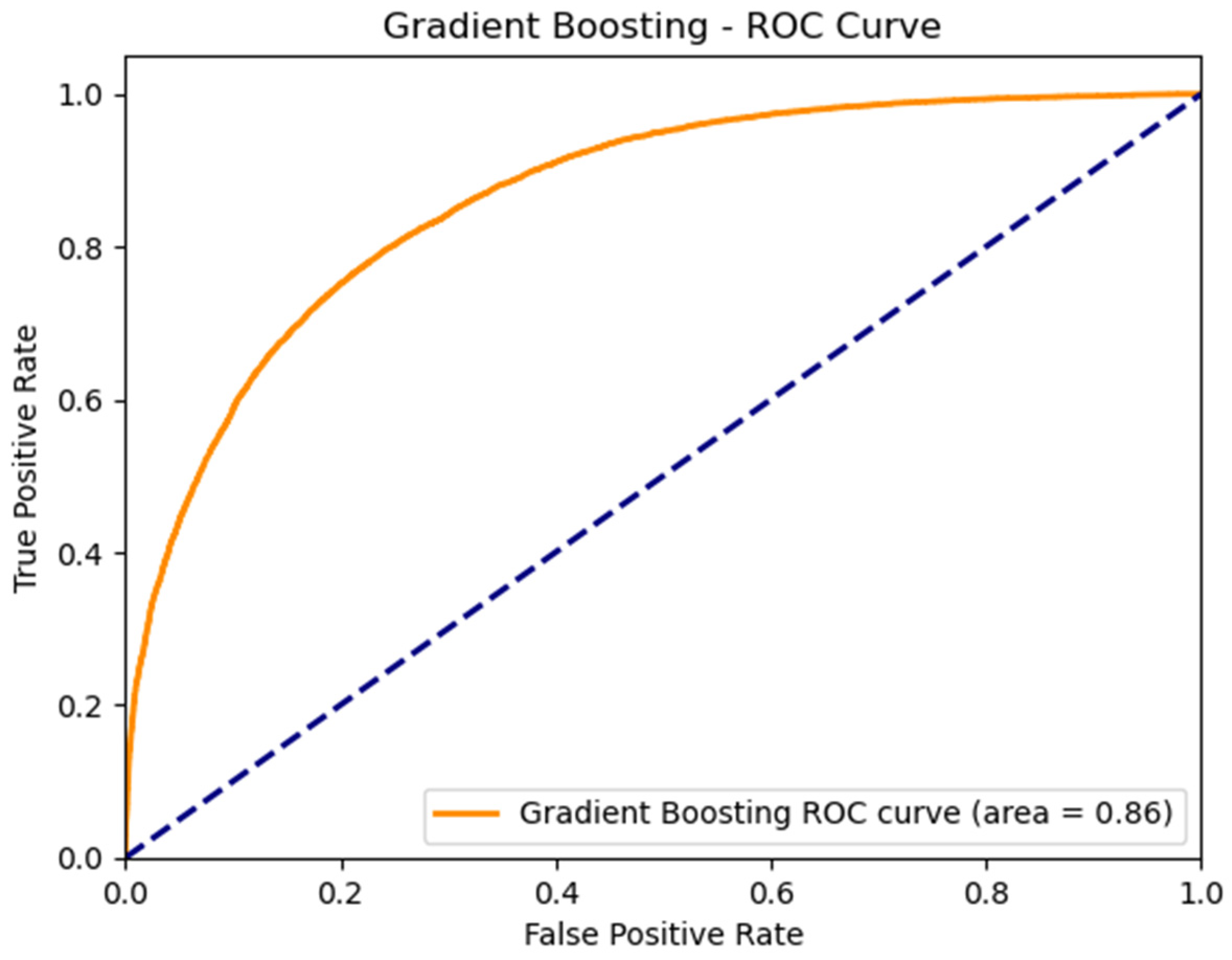
3.4. Performance of the Best ML Predictive Model
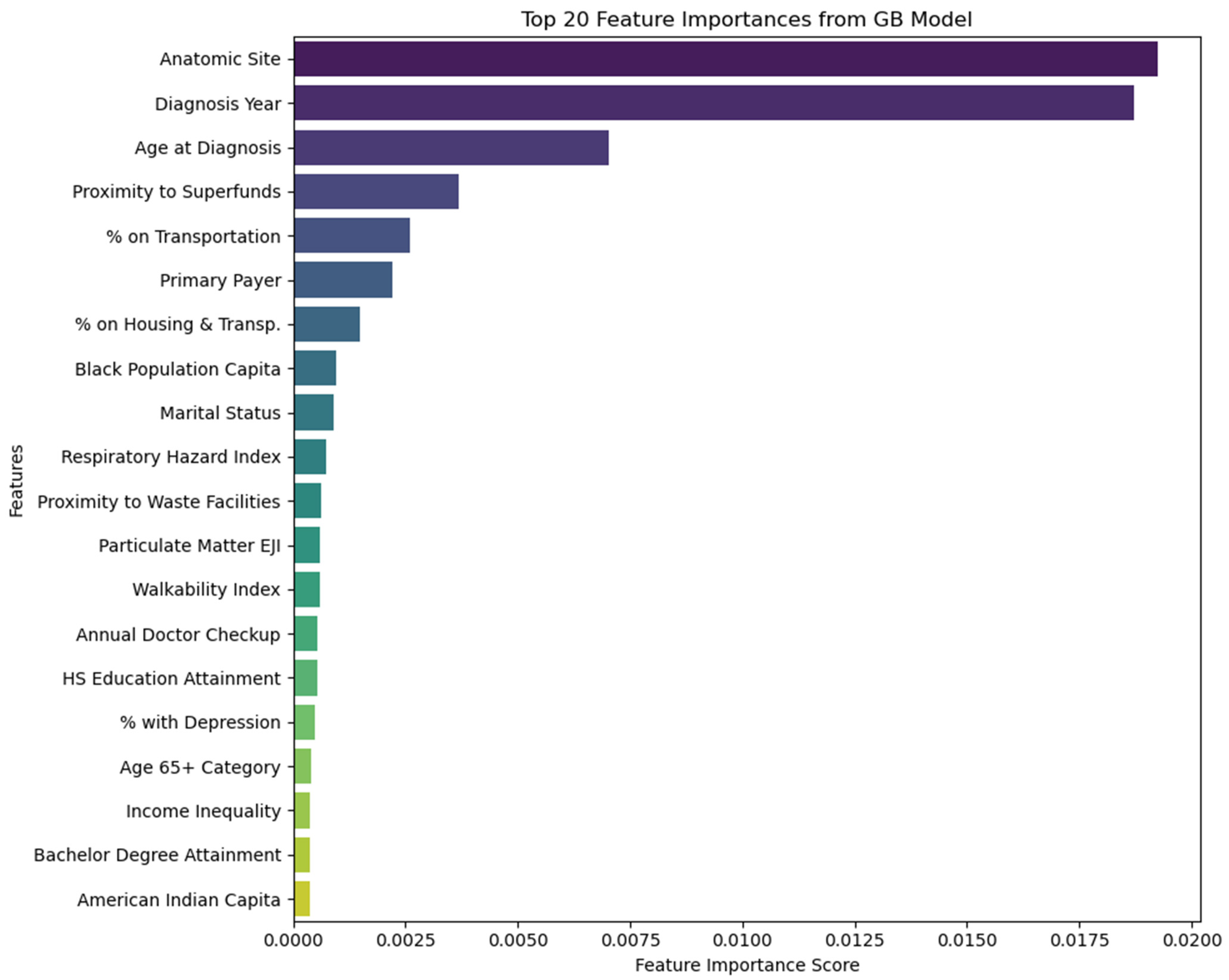
3.5. Feature Importance
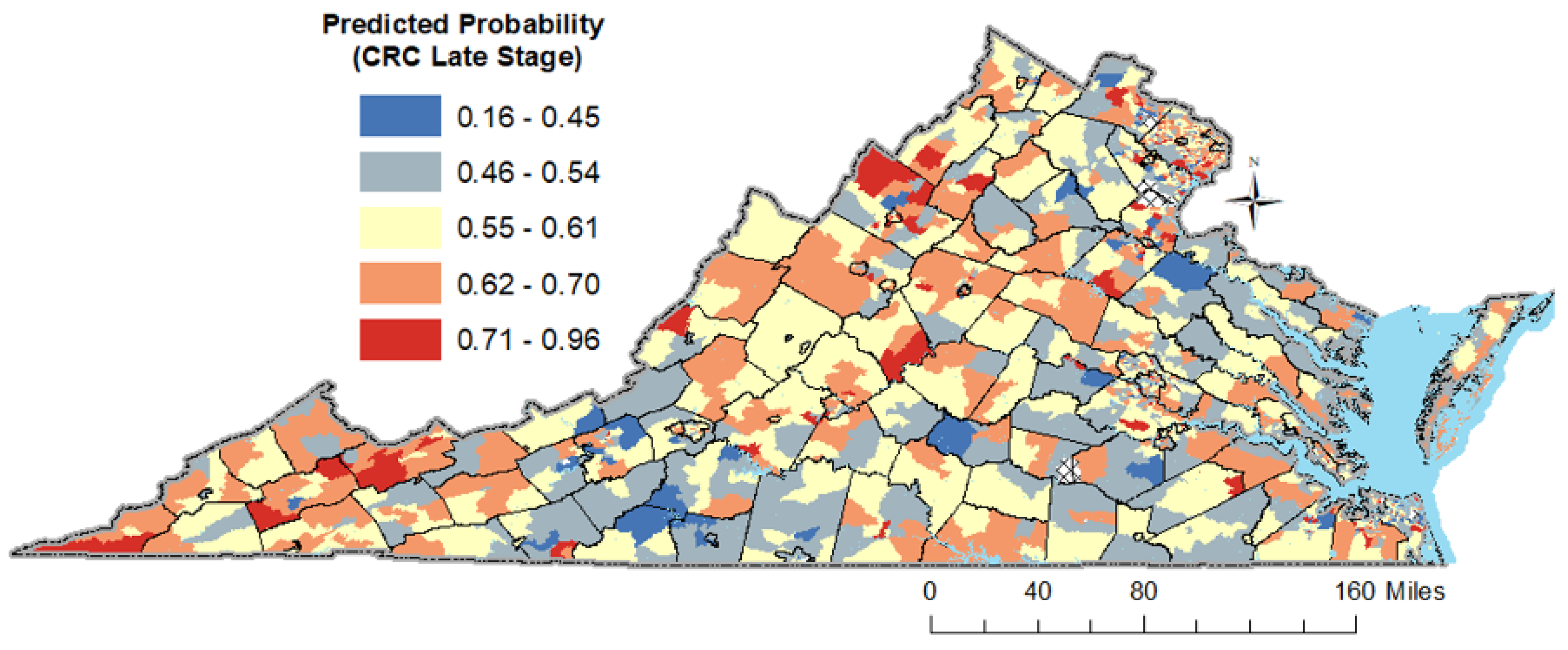
3.6. Results of the Spatio-Temporal Analysis
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Moyer, V.A.; US Preventive Services Task Force. Screening for prostate cancer: US Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2012, 157, 120–134. [Google Scholar] [CrossRef]
- Zauber, A.G. The impact of screening on colorectal cancer mortality and incidence: Has it really made a difference? Dig. Dis. Sci. 2015, 60, 681–691. [Google Scholar] [CrossRef]
- Zauber, A.G.; Winawer, S.J.; O’Brien, M.J.; Lansdorp-Vogelaar, I.; van Ballegooijen, M.; Hankey, B.F.; Shi, W.; Bond, J.H.; Schapiro, M.; Panish, J.F. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 2012, 366, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Fedewa, S.A.; Anderson, W.F.; Miller, K.D.; Ma, J.; Rosenberg, P.S.; Jemal, A. Colorectal cancer incidence patterns in the United States, 1974–2013. JNCI J. Natl. Cancer Inst. 2017, 109, djw322. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.; Lieberman, D.A.; McFarland, B.; Andrews, K.S.; Brooks, D.; Bond, J.; Dash, C.; Giardiello, F.M.; Glick, S.; Johnson, D. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: A joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology 2008, 134, 1570–1595. [Google Scholar] [CrossRef] [PubMed]
- Kushi, L.H.; Doyle, C.; McCullough, M.; Rock, C.L.; Demark-Wahnefried, W.; Bandera, E.V.; Gapstur, S.; Patel, A.V.; Andrews, K.; Gansler, T. American Cancer Society Guidelines on nutrition and physical activity for cancer prevention: Reducing the risk of cancer with healthy food choices and physical activity. CA Cancer J. Clin. 2012, 62, 30–67. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterol. Rev. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Jasperson, K.W.; Tuohy, T.M.; Neklason, D.W.; Burt, R.W. Hereditary and familial colon cancer. Gastroenterology 2010, 138, 2044–2058. [Google Scholar] [CrossRef]
- Commission on Social Determinants of Health. Closing the Gap in a Generation: Health Equity through Action on the Social Determinants of Health: Final Report of the Commission on Social Determinants of Health; World Health Organization: Geneva, Switzerland, 2008. [Google Scholar]
- Braveman, P.; Gottlieb, L. The social determinants of health: It’s time to consider the causes of the causes. Public Health Rep. 2014, 129, 19–31. [Google Scholar] [CrossRef]
- Pinheiro, L.C.; Reshetnyak, E.; Akinyemiju, T.; Phillips, E.; Safford, M.M. Social determinants of health and cancer mortality in the Reasons for Geographic and Racial Differences in Stroke (REGARDS) cohort study. Cancer 2022, 128, 122–130. [Google Scholar] [CrossRef]
- Yu, K.-H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef]
- Chen, M.; Decary, M. Artificial Intelligence in Healthcare: An Essential Guide for Health Leaders; SAGE Publications: Los Angeles, CA, USA, 2020; pp. 10–18. [Google Scholar]
- Silva, T.C.; Zhao, L. Machine Learning in Complex Networks; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: Berlin/Heidelberg, Germany, 2009; Volume 2. [Google Scholar]
- Alpaydin, E. Introduction to Machine Learning; MIT Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Fakoor, R.; Ladhak, F.; Nazi, A.; Huber, M. Using Deep Learning to Enhance Cancer Diagnosis and Classification; ACM: New York, NY, USA, 2013; pp. 3937–3949. [Google Scholar]
- Aliferis, C.F.; Tsamardinos, I.; Massion, P.P.; Statnikov, A.R.; Fananapazir, N.; Hardin, D.P. Machine Learning Models for Classification of Lung Cancer and Selection of Genomic Markers Using Array Gene Expression Data. In Proceedings of the FLAIRS Conference, St. Augustine, FL, USA, 12–14 May 2003; pp. 67–71. [Google Scholar]
- Passos, I.C.; Mwangi, B.; Kapczinski, F. Big data analytics and machine learning: 2015 and beyond. Lancet Psychiatry 2016, 3, 13–15. [Google Scholar] [CrossRef] [PubMed]
- Hornbrook, M.C.; Goshen, R.; Choman, E.; O’Keeffe-Rosetti, M.; Kinar, Y.; Liles, E.G.; Rust, K.C. Early colorectal cancer detected by machine learning model using gender, age, and complete blood count data. Dig. Dis. Sci. 2017, 62, 2719–2727. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Li, Z.Q.; Luo, Z.W.; Liao, W.J.; Liu, Z.L.; Liu, J.M. Machine learning for the prediction of bone metastasis in patients with newly diagnosed thyroid cancer. Cancer Med. 2021, 10, 2802–2811. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.J.; Cho, C.W.; Jang, J.; Kim, S.S.; Ahn, K.S.; Park, S.K.; Park, D.I. Application of deep learning to predict advanced neoplasia using big clinical data in colorectal cancer screening of asymptomatic adults. Korean J. Intern. Med. 2021, 36, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Tran, T.; Luo, W.; Phung, D.; Kennedy, R.L.; Broad, A.; Campbell, D.; Kipp, D.; Singh, M.; Khasraw, M.; et al. Machine-learning prediction of cancer survival: A retrospective study using electronic administrative records and a cancer registry. BMJ Open 2014, 4, e004007. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.; Park, J.; Hon, E.S.; Bounsanga, J.; Moazzami, S.; Ruiz-Negrón, B.; Wang, D. Artificial intelligence in dentistry: Harnessing big data to predict oral cancer survival. World J. Clin. Oncol. 2020, 11, 918–934. [Google Scholar] [CrossRef]
- Manz, C.R.; Chen, J.; Liu, M.; Chivers, C.; Regli, S.H.; Braun, J.; Draugelis, M.; Hanson, C.W.; Shulman, L.N.; Schuchter, L.M.; et al. Validation of a Machine Learning Algorithm to Predict 180-Day Mortality for Outpatients with Cancer. JAMA Oncol. 2020, 6, 1723–1730. [Google Scholar] [CrossRef]
- Lynch, C.M.; Abdollahi, B.; Fuqua, J.D.; de Carlo, A.R.; Bartholomai, J.A.; Balgemann, R.N.; van Berkel, V.H.; Frieboes, H.B. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int. J. Med. Inform. 2017, 108, 1–8. [Google Scholar] [CrossRef]
- Praiss, A.M.; Huang, Y.; St Clair, C.M.; Tergas, A.I.; Melamed, A.; Khoury-Collado, F.; Hou, J.Y.; Hu, J.; Hur, C.; Hershman, D.L.; et al. Using machine learning to create prognostic systems for endometrial cancer. Gynecol. Oncol. 2020, 159, 744–750. [Google Scholar] [CrossRef]
- Kudo, S.E.; Ichimasa, K.; Villard, B.; Mori, Y.; Misawa, M.; Saito, S.; Hotta, K.; Saito, Y.; Matsuda, T.; Yamada, K.; et al. Artificial Intelligence System to Determine Risk of T1 Colorectal Cancer Metastasis to Lymph Node. Gastroenterology 2021, 160, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- She, Y.; Jin, Z.; Wu, J.; Deng, J.; Zhang, L.; Su, H.; Jiang, G.; Liu, H.; Xie, D.; Cao, N.; et al. Development and Validation of a Deep Learning Model for Non-Small Cell Lung Cancer Survival. JAMA Netw. Open 2020, 3, e205842. [Google Scholar] [CrossRef] [PubMed]
- Bergquist, S.L.; Brooks, G.A.; Keating, N.L.; Landrum, M.B.; Rose, S. Classifying Lung Cancer Severity with Ensemble Machine Learning in Health Care Claims Data. Proc. Mach. Learn. Res. 2017, 68, 25–38. [Google Scholar] [PubMed]
- Wang, R.; Weng, Y.; Zhou, Z.; Chen, L.; Hao, H.; Wang, J. Multi-objective ensemble deep learning using electronic health records to predict outcomes after lung cancer radiotherapy. Phys. Med. Biol. 2019, 64, 245005. [Google Scholar] [CrossRef]
- Nartowt, B.J.; Hart, G.R.; Muhammad, W.; Liang, Y.; Stark, G.F.; Deng, J. Robust Machine Learning for Colorectal Cancer Risk Prediction and Stratification. Front. Big Data 2020, 3, 6. [Google Scholar] [CrossRef]
- Levitsky, A.; Pernemalm, M.; Bernhardson, B.M.; Forshed, J.; Kölbeck, K.; Olin, M.; Henriksson, R.; Lehtiö, J.; Tishelman, C.; Eriksson, L.E. Early symptoms and sensations as predictors of lung cancer: A machine learning multivariate model. Sci. Rep. 2019, 9, 16504. [Google Scholar] [CrossRef] [PubMed]
- Council, N.R. Analysis of Cancer Risks in Populations Near Nuclear Facilities: Phase 1; National Academies Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Elena, J.W.; Travis, L.B.; Simonds, N.I.; Ambrosone, C.B.; Ballard-Barbash, R.; Bhatia, S.; Cerhan, J.R.; Hartge, P.; Heist, R.S.; Kushi, L.H. Leveraging epidemiology and clinical studies of cancer outcomes: Recommendations and opportunities for translational research. J. Natl. Cancer Inst. 2013, 105, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, K.M.; Udesky, J.O.; Rudel, R.A.; Brody, J.G. Environmental chemicals and breast cancer: An updated review of epidemiological literature informed by biological mechanisms. Environ. Res. 2018, 160, 152–182. [Google Scholar] [CrossRef]
- Patel, C.J.; Kerr, J.; Thomas, D.C.; Mukherjee, B.; Ritz, B.; Chatterjee, N.; Jankowska, M.; Madan, J.; Karagas, M.R.; McAllister, K.A. Opportunities and challenges for environmental exposure assessment in population-based studies. Cancer Epidemiol. Prev. Biomark. 2017, 26, 1370–1380. [Google Scholar] [CrossRef]
- McClish, D.; Penberthy, L. Using Medicare data to estimate the number of cases missed by a cancer registry: A 3-source capture-recapture model. Med. Care 2004, 42, 1111–1116. [Google Scholar] [CrossRef]
- Izquierdo, J.N.; Schoenbach, V.J. The potential and limitations of data from population-based state cancer registries. Am. J. Public Health 2000, 90, 695. [Google Scholar] [PubMed]
- McClure, L.A.; Miller, E.A.; Tannenbaum, S.L.; Hernandez, M.N.; MacKinnon, J.A.; He, Y.; LeBlanc, W.G.; Lee, D.J. Linking the National Health Interview Survey with the Florida Cancer Data System: A Pilot Study. J. Regist. Manag. 2016, 43, 16. [Google Scholar]
- Sugarman, J.R.; Holliday, M.; Ross, A.; Castorina, J.; Hui, Y. Improving American Indian cancer data in the Washington State Cancer Registry using linkages with the Indian Health Service and tribal records. Cancer Interdiscip. Int. J. Am. Cancer Soc. 1996, 78, 1564–1568. [Google Scholar]
- Coughlin, S.S. Social determinants of breast cancer risk, stage, and survival. Breast Cancer Res. Treat. 2019, 177, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Buehler, J.W.; Castro, J.C.; Cohen, S.; Zhao, Y.; Melly, S.; Moore, K. Personal and Neighborhood Attributes Associated with Cervical and Colorectal Cancer Screening in an Urban African American Population. Prev. Chronic Dis. 2019, 16, E118. [Google Scholar] [CrossRef] [PubMed]
- Shariff-Marco, S.; Breen, N.; Stinchcomb, D.G.; Klabunde, C.N. Multilevel predictors of colorectal cancer screening use in California. Am. J. Manag. Care 2013, 19, 205–216. [Google Scholar]
- Mitchell, J.A.; Watkins, D.C.; Modlin, C.S., Jr. Social Determinants Associated with Colorectal Cancer Screening in an Urban Community Sample of African-American Men. J. Mens. Health 2013, 10, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Leech, M.M.; Weiss, J.E.; Markey, C.; Loehrer, A.P. Influence of Race, Insurance, Rurality, and Socioeconomic Status on Equity of Lung and Colorectal Cancer Care. Ann. Surg. Oncol. 2022, 29, 3630–3639. [Google Scholar] [CrossRef] [PubMed]
- Tawk, R.; Abner, A.; Ashford, A.; Brown, C.P. Differences in Colorectal Cancer Outcomes by Race and Insurance. Int. J. Environ. Res. Public Health 2015, 13, 48. [Google Scholar] [CrossRef] [PubMed]
- Balan, N.; Petrie, B.A.; Chen, K.T. Racial Disparities in Colorectal Cancer Care for Black Patients: Barriers and Solutions. Am. Surg. 2022, 88, 2823–2830. [Google Scholar] [CrossRef]
- Salem, M.E.; Puccini, A.; Trufan, S.J.; Sha, W.; Kadakia, K.C.; Hartley, M.L.; Musselwhite, L.W.; Symanowski, J.T.; Hwang, J.J.; Raghavan, D. Impact of Sociodemographic Disparities and Insurance Status on Survival of Patients with Early-Onset Colorectal Cancer. Oncologist 2021, 26, e1730–e1741. [Google Scholar] [CrossRef] [PubMed]
- Warren Andersen, S.; Blot, W.J.; Lipworth, L.; Steinwandel, M.; Murff, H.J.; Zheng, W. Association of Race and Socioeconomic Status with Colorectal Cancer Screening, Colorectal Cancer Risk, and Mortality in Southern US Adults. JAMA Netw. Open 2019, 2, e1917995. [Google Scholar] [CrossRef] [PubMed]
- Coughlin, S.S. Social determinants of colorectal cancer risk, stage, and survival: A systematic review. Int. J. Color. Dis. 2020, 35, 985–995. [Google Scholar] [CrossRef]
- Bauer, C.; Zhang, K.; Xiao, Q.; Lu, J.; Hong, Y.R.; Suk, R. County-Level Social Vulnerability and Breast, Cervical, and Colorectal Cancer Screening Rates in the US, 2018. JAMA Netw. Open 2022, 5, e2233429. [Google Scholar] [CrossRef]
- Korn, A.R.; Walsh-Bailey, C.; Pilar, M.; Sandler, B.; Bhattacharjee, P.; Moore, W.T.; Brownson, R.C.; Emmons, K.M.; Oh, A.Y. Social determinants of health and cancer screening implementation and outcomes in the USA: A systematic review protocol. Syst. Rev. 2022, 11, 117. [Google Scholar] [CrossRef]
- Siegel, R.L.; Sahar, L.; Robbins, A.; Jemal, A. Where Can Colorectal Cancer Screening Interventions Have the Most Impact? Where Can Colorectal Cancer Screening Have the Most Impact? Cancer Epidemiol. Biomark. Prev. 2015, 24, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.R.; Moore, J.X.; Qeadan, F.; Gu, L.Y.; Huntington, M.S.; Holowatyj, A.N. Examining factors underlying geographic disparities in early-onset colorectal cancer survival among men in the United States. Am. J. Cancer Res. 2020, 10, 1592–1607. [Google Scholar] [PubMed]
- Kuo, T.-M.; Meyer, A.M.; Baggett, C.D.; Olshan, A.F. Examining determinants of geographic variation in colorectal cancer mortality in North Carolina: A spatial analysis approach. Cancer Epidemiol. 2019, 59, 8–14. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. The Social-Ecological Model: A Framework for Prevention; Centers for Disease Control and Prevention: Atlanta, GE, USA, 2015. Available online: https://www.cdc.gov/violenceprevention/about/social-ecologicalmodel.html (accessed on 30 August 2023).
- US Department of Health and Human Services; Office of Disease Prevention and Health Promotion. Healthy People 2030. 2021. Available online: https://health.gov/healthypeople/objectives-and-data/social-determinants-health (accessed on 30 August 2023).
- Artiga, S.; Hinton, E. Beyond Health Care: The Role of Social Determinants in Promoting Health and Health Equity; Kaiser Family Foundation: San Francisco, CA, USA, 2018; Volume 10. [Google Scholar]
- Health, V.D.o. Virginia Cancer Registry (VCR)—Data. Available online: https://www.vdh.virginia.gov/virginia-cancer-registry/data/ (accessed on 1 May 2023).
- United States Census Bureau. 2015–2019 American Community Survey 5-Year Estimates; US Department of Commerce: Washington, DC, USA, 2019.
- mySidewalk. Data Is for the People. Available online: https://www.mysidewalk.com/ (accessed on 28 November 2023).
- Cui, W.; Finkelstein, J. Using EHR Data to Identify Social Determinants of Health Affecting Disparities in Cancer Survival. Stud. Health Technol. Inform. 2022, 290, 967–971. [Google Scholar] [CrossRef]
- Gehlert, S.; Hudson, D.; Sacks, T. A Critical Theoretical Approach to Cancer Disparities: Breast Cancer and the Social Determinants of Health. Front. Public Health 2021, 9, 674736. [Google Scholar] [CrossRef]
- Yu, Y.; Carey, M.; Pollett, W.; Green, J.; Dicks, E.; Parfrey, P.; Yilmaz, Y.E.; Savas, S. The long-term survival characteristics of a cohort of colorectal cancer patients and baseline variables associated with survival outcomes with or without time-varying effects. BMC Med. 2019, 17, 150. [Google Scholar] [CrossRef]
- He, Z.; Zhang, J.; Yuan, X.; Xi, J.; Liu, Z.; Zhang, Y. Stratification of Breast Cancer by Integrating Gene Expression Data and Clinical Variables. Molecules 2019, 24, 631. [Google Scholar] [CrossRef]
- Galadima, H.I.; Adunlin, G.; Hughes, M.S.; Cropp, C.D.; Lucero, L.; Akpinar-Elci, M. Racial disparities and treatment trends among young-onset colorectal cancer patients: An analysis of a hospital cancer registry. Cancer Epidemiol. 2021, 72, 101911. [Google Scholar] [CrossRef]
- Harvey, V.M.; Enos, C.W.; Chen, J.T.; Galadima, H.; Eschbach, K. The Role of Neighborhood Characteristics in Late Stage Melanoma Diagnosis among Hispanic Men in California, Texas, and Florida, 1996–2012. J. Cancer Epidemiol. 2017, 2017, 8418904. [Google Scholar] [CrossRef] [PubMed]
- Zahnd, W.E.; Gomez, S.L.; Steck, S.E.; Brown, M.J.; Ganai, S.; Zhang, J.; Arp Adams, S.; Berger, F.G.; Eberth, J.M. Rural-urban and racial/ethnic trends and disparities in early-onset and average-onset colorectal cancer. Cancer 2021, 127, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Abualkhair, W.H.; Zhou, M.; Ochoa, C.O.; Lacayo, L.; Murphy, C.; Wu, X.C.; Karlitz, J.J. Geographic and intra-racial disparities in early-onset colorectal cancer in the SEER 18 registries of the United States. Cancer Med. 2020, 9, 9150–9159. [Google Scholar] [CrossRef] [PubMed]
- Missouri Census Data Center. MABLE/Geocorr: Geographic Correspondence Engine. 2018. Available online: https://mcdc.missouri.edu/applications/geocorr.html (accessed on 3 May 2023).
- Hosmer, D.W., Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression; John Wiley & Sons: Hoboken, NJ, USA, 2013; Volume 398. [Google Scholar]
- Rafique, R.; Islam, S.R.; Kazi, J.U. Machine learning in the prediction of cancer therapy. Comput. Struct. Biotechnol. J. 2021, 19, 4003–4017. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bo, D.; Shi, C.; Fan, S.; Ye, Y.; Philip, S.Y. A survey on heterogeneous graph embedding: Methods, techniques, applications and sources. IEEE Trans. Big Data 2022, 9, 415–436. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Bertsimas, D.; Wiberg, H. Machine learning in oncology: Methods, applications, and challenges. JCO Clin. Cancer Inform. 2020, 4, CCI.20.00072. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Levin, T.R.; Corley, D.A.; Jensen, C.D.; Schottinger, J.E.; Quinn, V.P.; Zauber, A.G.; Lee, J.K.; Zhao, W.K.; Udaltsova, N.; Ghai, N.R. Effects of organized colorectal cancer screening on cancer incidence and mortality in a large community-based population. Gastroenterology 2018, 155, 1383–1391.e1385. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A.; Soman, A.V.; Berkowitz, Z.; Fedewa, S.A.; Sabatino, S.A.; de Moor, J.S.; Clarke, T.C.; Doria-Rose, V.P.; Breslau, E.S.; Jemal, A. Screening for colorectal cancer in the United States: Correlates and time trends by type of test. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1554–1565. [Google Scholar] [CrossRef]
- Hubers, J.; Sonnenberg, A.; Gopal, D.; Weiss, J.; Holobyn, T.; Soni, A. Trends in wait time for colorectal cancer screening and diagnosis 2013–2016. Clin. Transl. Gastroenterol. 2020, 11, e00113. [Google Scholar] [CrossRef]
- Demb, J.; Earles, A.; Martínez, M.E.; Bustamante, R.; Bryant, A.K.; Murphy, J.D.; Liu, L.; Gupta, S. Risk factors for colorectal cancer significantly vary by anatomic site. BMJ Open Gastroenterol. 2019, 6, e000313. [Google Scholar] [CrossRef]
- Parikh-Patel, A.; Bates, J.H.; Campleman, S. Colorectal cancer stage at diagnosis by socioeconomic and urban/rural status in California, 1988–2000. Cancer 2006, 107, 1189–1195. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zou, Z.; Gao, Z.; Wang, Y.; Xiao, M.; Xu, C.; Jiang, G.; Wang, H.; Jin, L.; Wang, J. Prediction of lung cancer risk in Chinese population with genetic-environment factor using extreme gradient boosting. Cancer Med. 2022, 11, 4469–4478. [Google Scholar] [CrossRef] [PubMed]
- Bibault, J.-E.; Chang, D.T.; Xing, L. Development and validation of a model to predict survival in colorectal cancer using a gradient-boosted machine. Gut 2021, 70, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jia, Z.; Mercola, D.; Xie, X. A gradient boosting algorithm for survival analysis via direct optimization of concordance index. Comput. Math. Methods Med. 2013, 2013, 873595. [Google Scholar] [CrossRef] [PubMed]
- Aryal, S.; Paudel, B.; Aryal, S.; Paudel, B. Supervised classification using gradient boosting machine: Wisconsin breast cancer dataset. Int. J. Sci. Res. Eng. Trends 2020, 6, 1887–1892. [Google Scholar]
- Vo, D.M.; Nguyen, N.-Q.; Lee, S.-W. Classification of breast cancer histology images using incremental boosting convolution networks. Inf. Sci. 2019, 482, 123–138. [Google Scholar] [CrossRef]
- Liu, B.; Zhu, L.; Zou, J.; Chen, H.-S.; Miller, K.D.; Jemal, A.; Siegel, R.L.; Feuer, E.J. Updated methodology for projecting US-and state-level cancer counts for the current calendar year: Part I: Spatio-temporal modeling for cancer incidence. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1620–1626. [Google Scholar] [CrossRef]
- Guo, Y.; Zeng, H.; Zheng, R.; Li, S.; Barnett, A.G.; Zhang, S.; Zou, X.; Huxley, R.; Chen, W.; Williams, G. The association between lung cancer incidence and ambient air pollution in China: A spatiotemporal analysis. Environ. Res. 2016, 144, 60–65. [Google Scholar] [CrossRef]
- Brady, L.A.; Tumiel-Berhalter, L.M.; Schad, L.A.; Bentham, A.; Vitale, K.; Norton, A.; Noronha, G.; Swanger, C.; Morley, C.P. Increasing Breast, Cervical, and Colorectal Cancer Screenings: A Qualitative Assessment of Barriers and Promoters in Safety-Net Practices. J. Patient Cent. Res. Rev. 2021, 8, 323–330. [Google Scholar] [CrossRef]
- Choy, A.M.; Lebwohl, B.; Krigel, A. Impact of social determinants of health on colorectal cancer screening and surveillance in the COVID reopening phase. Eur. J. Gastroenterol. Hepatol. 2022, 34, 739–743. [Google Scholar] [CrossRef]


| Patient Characteristics | Early n = 17,884 (42.74%) | Late n = 23,955 (57.26%) | |
|---|---|---|---|
| Frequency (%) or Mean (SE) | Frequency (%) or Mean (SE) | p-Value | |
| Age at Diagnosis | <0.0001 | ||
| Age in years, Mean (SE) | 66.72 (0.1) | 65.85 (0.1) | |
| Age Group | <0.0001 | ||
| 18–39 | 471 (2.63%) | 790 (3.30%) | |
| 40–49 | 1262 (7.06%) | 2288 (9.55%) | |
| 50–64 | 5678 (31.75%) | 7578 (31.63%) | |
| 65+ | 10,473 (58.56%) | 13,299 (55.52%) | |
| Race & Ethnicity | 0.005 | ||
| NH White | 12,263 (68.67%) | 16,305 (68.19%) | |
| NH Black | 3263 (18.27%) | 4279 (17.89%) | |
| Hispanic | 1786 (10.00%) | 2517 (10.53%) | |
| NH Asian | 440 (2.46%) | 702 (2.94%) | |
| NH Pacific Islander | 68 (0.38%) | 74 (0.31%) | |
| Unknown/Not Documented | 39 (0.22%) | 35 (0.15%) | |
| Marital Status at Diagnosis | <0.0001 | ||
| Married | 9578 (57.00%) | 11,935 (53.95%) | |
| Unmarried | 6523 (38.82%) | 9437 (42.66%) | |
| Unknown | 702 (4.18%) | 752 (3.40%) | |
| Primary Payer at Diagnosis | <0.0001 | ||
| Medicaid | 284 (1.70%) | 636 (2.83%) | |
| Medicare | 9267 (55.52%) | 11,762 (52.28%) | |
| Private Insurance | 6124 (36.69%) | 8295 (36.87%) | |
| Military | 601 (3.60%) | 680 (3.02%) | |
| Indian/Public Health Service | 2 (0.01%) | 1 (0.00%) | |
| Self-pay/Uninsured | 413 (2.47%) | 1122 (4.99%) | |
| Year of Diagnosis | <0.0001 | ||
| 2000–2004 | 5346 (29.89%) | 7242 (30.23%) | |
| 2005–2009 | 4836 (27.04%) | 5882 (24.55%) | |
| 2010–2014 | 3763 (21.04%) | 4917 (20.53%) | |
| 2015–2019 | 3821 (21.37%) | 5705 (23.82%) | |
| 2020 | 118 (0.66%) | 209 (0.87%) | |
| Stage of disease at diagnosis | <0.0001 | ||
| In Situ | 2594 (14.50%) | 0 (0.00%) | |
| Localized | 15,290 (85.50%) | 0 (0.00%) | |
| Regional | 0 (0.00%) | 16,005 (66.81%) | |
| Distant | 0 (0.00%) | 7950 (33.19%) | |
| Not Staged/Unknown | 0 (0.00%) | 0 (0.00%) | |
| Primary Site | <0.0001 | ||
| Colon | 12,521 (70.01%) | 17,645 (73.66%) | |
| Rectum | 5363 (29.99%) | 6310 (26.34%) | |
| Grade | <0.0001 | ||
| Grade I | 3421 (19.13%) | 2390 (9.98%) | |
| Grade II | 8589 (48.03%) | 13,067 (54.55%) | |
| Grade III | 1069 (5.98%) | 4162 (17.37%) | |
| Grade IV | 194 (1.08%) | 570 (2.38%) | |
| T-cell | 1 (0.01%) | 1 (0.00%) | |
| B-cell | 89 (0.50%) | 92 (0.38%) | |
| NK Cell | 1 (0.01%) | 0 (0.00%) | |
| Unknown Grade | 4520 (25.27%) | 3673 (15.33%) | |
| Treatment Status | <0.0001 | ||
| No treatment given | 351 (4.54%) | 711 (6.54%) | |
| Treatment given | 7256 (93.88%) | 10,044 (92.44%) | |
| Active surveillance (watchful waiting) | 17 (0.22%) | 5 (0.05%) | |
| Unknown if treatment was given | 105 (1.36%) | 106 (0.98%) | |
| Days between date of initial diagnosis and date first course of treatment | 0.2142 | ||
| Time Lag, Mean (SE) | 9.93 (1.97) | 13.67 (2.28) | |
| Surgery | <0.0001 | ||
| Yes | 16,627 (92.97%) | 19,698 (82.23%) | |
| No | 1077 (6.02%) | 4060 (16.95%) | |
| Unknown | 180 (1.01%) | 197 (0.82%) | |
| Chemotherapy | <0.0001 | ||
| Yes | 1835 (10.26%) | 13,276 (55.42%) | |
| No | 15,575 (87.09%) | 10,330 (43.12%) | |
| Unknown | 474 (2.65%) | 349 (1.46%) | |
| Radiation Therapy | <0.0001 | ||
| Yes | 1384 (7.75%) | 4063 (16.97%) | |
| No | 15,541 (86.98%) | 18,717 (78.19%) | |
| Unknown | 943 (5.28%) | 1157 (4.83%) | |
| Vital Status | <0.0001 | ||
| Dead | 7275 (40.68%) | 14,859 (62.03%) | |
| Alive | 10,609 (59.32%) | 9096 (37.97%) |
| Census Tract Characteristics | Early n = 17,884 (42.74%) | Late n = 23,955 (57.26%) | |
|---|---|---|---|
| Mean (SE) | Mean (SE) | p-Value | |
| Access to Healthcare | |||
| Percent Uninsured | 7.816 (0.04) | 7.9552 (0.04) | 0.0085 |
| Doctor Checkup in Past Year Among Adults (2020) | 76.7273 (2.98) | 76.5405 (3.03) | <0.0001 |
| Doctor Checkup in Past Year Among Adults (2020) | 76.7273 (2.98) | 76.5405 (3.03) | <0.0001 |
| Population with a Disability (2017–2021) | 13.2833 (0.05) | 13.1551 (0.04) | 0.0372 |
| Socio Economic | |||
| % Spent on Housing & Transportation | 53.5814 (0.06) | 53.2928 (0.05) | 0.0003 |
| %Spent on Housing | 26.0618 (0.01) | 26.0798 (0.01) | 0.2533 |
| % Spent on Transportation | 27.5196 (0.06) | 27.213 (0.06) | 0.0003 |
| Employment Access Index (2016) | 20,613.6 (177) | 21,241.2 (158) | 0.0082 |
| Labor Force Participation Rate (2017–2021) | 63.3817 (0.08) | 63.6564 (0.07) | 0.0093 |
| Median Household Income (2017–2021) | 82,806.2 (326.1) | 83,742.1 (285.2) | 0.0311 |
| % Pop with Access to Healthy Food | 0.7214 (0.002) | 0.7144 (0.001) | 0.0088 |
| Educational Attainment | |||
| Less than High school education | 9.8888 (0.05) | 10.027 (0.05) | 0.0446 |
| Educational Attainment—Some College No Degree (2017–2021) | 8.1371 (0.03) | 8.0137 (0.02) | 0.0005 |
| Behaviors | |||
| Binge Drinking Among Adults (2020) | 15.3011 (0.02) | 15.3652 (0.01) | 0.0017 |
| Race and Ethnicity | |||
| White per capita | 0.633 (0.002) | 0.6335 (0.002) | 0.8212 |
| Black per capita | 0.2003 (0.002) | 0.1952 (0.001) | 0.0141 |
| Asian per capita | 0.0483 (0.006) | 0.0495 (0.005) | 0.0987 |
| Hispanic per capita | 0.0789 (0) | 0.0821 (0) | 0.0008 |
| Environmental Exposures | |||
| Air Quality: Respiratory Hazard Index (2014) | 0.4039 (0.005) | 0.4051 (0.004) | 0.0707 |
| Diesel Particulate Matter Environmental Justice Index (2021) | 14.1463 (0.1) | 14.398 (0.09) | 0.0501 |
| Diesel Particulate Matter Level in Air (2021) | 0.2206 (0.008) | 0.224 (0.007) | 0.0026 |
| Underground Storage Tanks (2021) | 5.044 (0.05) | 5.2027 (0.04) | 0.0114 |
| Population Weighted Density | 3359.9 (43.17) | 3546.9 (39.87) | 0.0015 |
| Model | ROC-AUC | Overall Prediction Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Lasso (Penalized Logistic Regression) | 0.7864 | 0.7159 | 0.7405 | 0.6975 |
| Decision Tree | 0.7006 | 0.7068 | 0.6580 | 0.7432 |
| Random Forest | 0.7554 | 0.7032 | 0.5640 | 0.8072 |
| Gradient Boosting | 0.8549 | 0.7725 | 0.7263 | 0.8070 |
| SVM (SGD Classifier) | 0.7298 | 0.6760 | 0.6793 | 0.6735 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galadima, H.; Anson-Dwamena, R.; Johnson, A.; Bello, G.; Adunlin, G.; Blando, J. Machine Learning as a Tool for Early Detection: A Focus on Late-Stage Colorectal Cancer across Socioeconomic Spectrums. Cancers 2024, 16, 540. https://doi.org/10.3390/cancers16030540
Galadima H, Anson-Dwamena R, Johnson A, Bello G, Adunlin G, Blando J. Machine Learning as a Tool for Early Detection: A Focus on Late-Stage Colorectal Cancer across Socioeconomic Spectrums. Cancers. 2024; 16(3):540. https://doi.org/10.3390/cancers16030540
Chicago/Turabian StyleGaladima, Hadiza, Rexford Anson-Dwamena, Ashley Johnson, Ghalib Bello, Georges Adunlin, and James Blando. 2024. "Machine Learning as a Tool for Early Detection: A Focus on Late-Stage Colorectal Cancer across Socioeconomic Spectrums" Cancers 16, no. 3: 540. https://doi.org/10.3390/cancers16030540
APA StyleGaladima, H., Anson-Dwamena, R., Johnson, A., Bello, G., Adunlin, G., & Blando, J. (2024). Machine Learning as a Tool for Early Detection: A Focus on Late-Stage Colorectal Cancer across Socioeconomic Spectrums. Cancers, 16(3), 540. https://doi.org/10.3390/cancers16030540







