A Comprehensive Investigation of Active Learning Strategies for Conducting Anti-Cancer Drug Screening
Abstract
Simple Summary
Abstract
1. Introduction
| Reference | Approach | Application |
|---|---|---|
| [41] | Monitors the normalized average loss and normalized average predictive entropy of every sample. Eliminates noisy samples and selects the most informative samples for annotation. | Histopathology image analysis |
| [44] | Queries unlabeled samples that maximize the average distance to training set samples. | Medical image analysis |
| [45] | Uncertainty sampling identifies the next set of sentences to be annotated. | Natural language processing |
| [42] | Diversity-based active learning to annotate the most informative frames and objects. | Autonomous navigation and object detection |
| [46] | Utilizes Bayesian global optimization (BGO) to select an experiment by maximizing a utility function. | Material science |
| [47] | Selects samples from the unlabeled set using uncertainty computed by a discrete information entropy measure. | Industrial fault detection |
| [48] | Uses diversity-based sampling and loss-prediction sampling to select unlabeled lung CT image samples for annotation. | Disease diagnosis (COVID-19) |
| [49] | Reduces annotations at both image-level and pixel-level using uncertainty-based active learning. Uncertainty is estimated by computing entropy at the image and pixel levels. | Semantic segmentation |
| [33] | Used uncertainty, greedy, and random active learning workflows for predicting drug responses. | Drug response prediction |
2. Materials and Methods
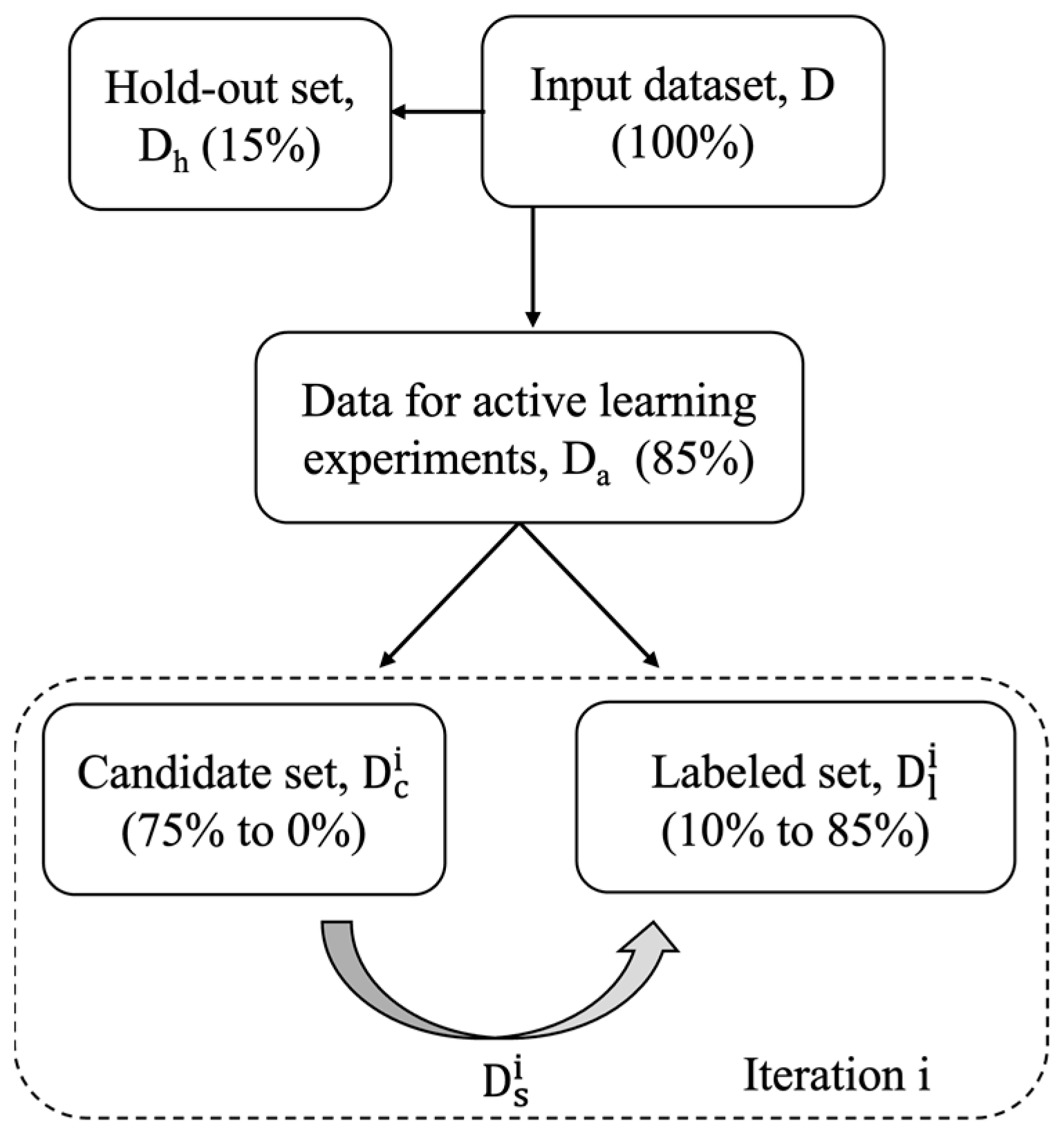
2.1. Data Sources and Data Splitting
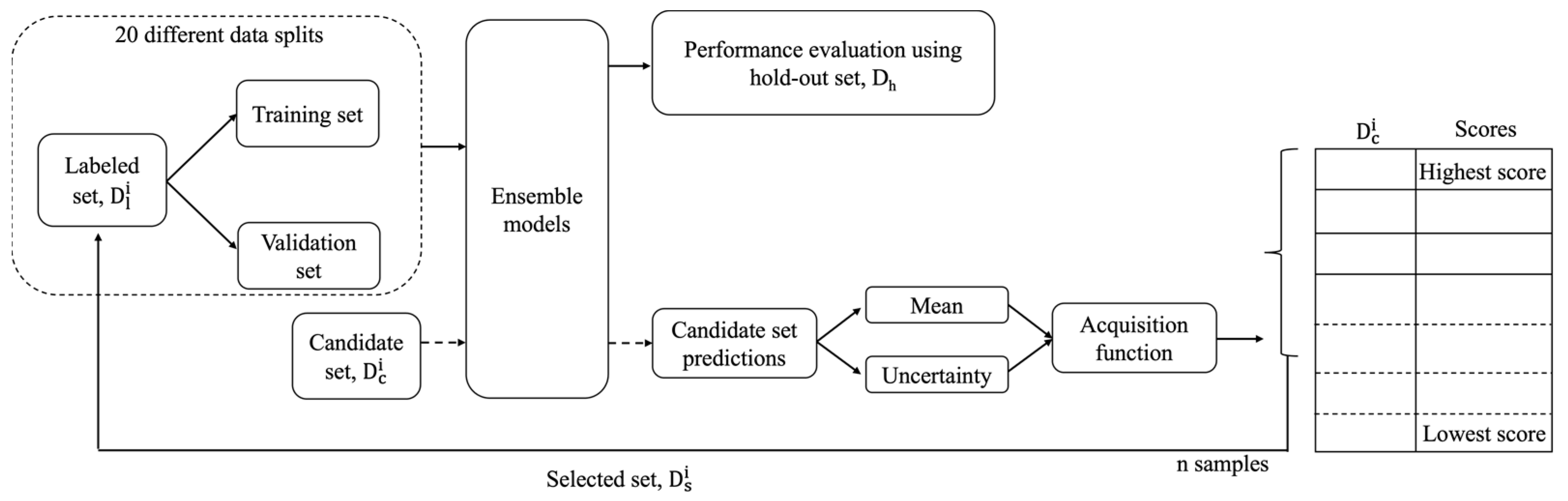
2.2. Active Learning Approaches and Workflow
- Greedy [30,33]: This approach uses an acquisition function , which considers only the mean of the prediction values generated by the 20 different models for a candidate sample. The negative sign allows the acquisition function to give a large value to a candidate sample with low AUC predictions, as a low AUC value indicates a responsive treatment.
- GU combined [30]: This approach uses an acquisition function , which is a combination of greedy and uncertainty.
- Diversity [42,48]: This approach does not consider predictions on the candidate set . It is based on the diversity of samples in . K-means clustering is performed on with the cluster number equal to ‘n’. Then in every cluster, the sample closest to the cluster centroid is chosen and added to the labelled set .
- Hybrid sampling: In every iteration of the analysis, the ‘ps’ percentage of samples is selected using random sampling. The remaining samples are selected using the GU combined acquisition function. The values of ‘ps’ are 20%, 30%, 40%, and 50%.
- Hybrid iteration: In this approach, random sampling is used in the initial ‘pi’ percentage of iterations, while the remaining iterations use the GU combined acquisition function. The values of ‘pi’ are 20%, 30%, 40%, and 50%.
2.3. Prediction Model
2.4. The Definitions and Demonstrations of Active Learning Performances
3. Results
3.1. Comprehensive Hits Analysis
3.2. Comprehensive Analysis on Drug Response Modeling Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wang, D.C.; Cheng, Y.; Qian, M.; Zhang, M.; Shen, Q.; Wang, X. Roles of Tumor Heterogeneity in the Development of Drug Resistance: A Call for Precision Therapy. Semin. Cancer Biol. 2017, 42, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Brettin, T.; Evrard, Y.A.; Partin, A.; Xia, F.; Shukla, M.; Yoo, H.; Doroshow, J.H.; Stevens, R.L. Ensemble Transfer Learning for the Prediction of Anti-Cancer Drug Response. Sci. Rep. 2020, 10, 18040. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular Profiling for Precision Cancer Therapies. Genome Med. 2020, 12, 1–19. [Google Scholar] [CrossRef]
- Chiu, Y.-C.; Zheng, S.; Wang, L.-J.; Iskra, B.S.; Rao, M.K.; Houghton, P.J.; Huang, Y.; Chen, Y. Predicting and Characterizing a Cancer Dependency Map of Tumors with Deep Learning. Sci. Adv. 2021, 7, eabh1275. [Google Scholar] [CrossRef]
- Partin, A.; Brettin, T.S.; Zhu, Y.; Narykov, O.; Clyde, A.; Overbeek, J.; Stevens, R.L. Deep Learning Methods for Drug Response Prediction in Cancer: Predominant and Emerging Trends. Front. Med. 2023, 10, 1086097. [Google Scholar] [CrossRef]
- Firoozbakht, F.; Yousefi, B.; Schwikowski, B. An Overview of Machine Learning Methods for Monotherapy Drug Response Prediction. Brief Bioinform. 2022, 23, bbab408. [Google Scholar] [CrossRef]
- Ammad-Ud-Din, M.; Khan, S.A.; Wennerberg, K.; Aittokallio, T. Systematic Identification of Feature Combinations for Predicting Drug Response with Bayesian Multi-View Multi-Task Linear Regression. Bioinformatics 2017, 33, i359–i368. [Google Scholar] [CrossRef]
- Huang, C.; Clayton, E.A.; Matyunina, L.V.; McDonald, L.D.E.; Benigno, B.B.; Vannberg, F.; McDonald, J.F. Machine Learning Predicts Individual Cancer Patient Responses to Therapeutic Drugs with High Accuracy. Sci. Rep. 2018, 8, 16444. [Google Scholar] [CrossRef]
- Parca, L.; Pepe, G.; Pietrosanto, M.; Galvan, G.; Galli, L.; Palmeri, A.; Sciandrone, M.; Ferrè, F.; Ausiello, G.; Helmer-Citterich, M. Modeling Cancer Drug Response through Drug-Specific Informative Genes. Sci. Rep. 2019, 9, 15222. [Google Scholar] [CrossRef]
- Lind, A.P.; Anderson, P.C. Predicting Drug Activity against Cancer Cells by Random Forest Models Based on Minimal Genomic Information and Chemical Properties. PLoS ONE 2019, 14, e0219774. [Google Scholar] [CrossRef]
- Su, R.; Liu, X.; Wei, L.; Zou, Q. Deep-Resp-Forest: A Deep Forest Model to Predict Anti-Cancer Drug Response. Methods 2019, 166, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Turki, T.; Wang, J.T.L. Clinical Intelligence: New Machine Learning Techniques for Predicting Clinical Drug Response. Comput. Biol. Med. 2019, 107, 302–322. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, M.; Qin, Y. Drug-Induced Cell Viability Prediction from LINCS-L1000 through WRFEN-XGBoost Algorithm. BMC Bioinform. 2021, 22, 13. [Google Scholar] [CrossRef]
- Sharifi-Noghabi, H.; Zolotareva, O.; Collins, C.C.; Ester, M. MOLI: Multi-Omics Late Integration with Deep Neural Networks for Drug Response Prediction. Bioinformatics 2019, 35, i501–i509. [Google Scholar] [CrossRef]
- Jiang, Y.; Rensi, S.; Wang, S.; Altman, R.B. DrugOrchestra: Jointly Predicting Drug Response, Targets, and Side Effects via Deep Multi-Task Learning. bioRxiv 2020, 11, 385757. [Google Scholar] [CrossRef]
- Tang, Y.C.; Gottlieb, A. Explainable Drug Sensitivity Prediction through Cancer Pathway Enrichment. Sci. Rep. 2021, 11, 3128. [Google Scholar] [CrossRef] [PubMed]
- Narykov, O.; Zhu, Y.; Brettin, T.; Evrard, Y.A.; Partin, A.; Shukla, M.; Xia, F.; Clyde, A.; Vasanthakumari, P.; Doroshow, J.H.; et al. Integration of Computational Docking into Anti-Cancer Drug Response Prediction Models. Cancers 2023, 16, 50. [Google Scholar] [CrossRef]
- Nguyen, T.; Nguyen, G.T.T.; Nguyen, T.; Le, D.H. Graph Convolutional Networks for Drug Response Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 146–154. [Google Scholar] [CrossRef]
- Liu, P.; Li, H.; Li, S.; Leung, K.S. Improving Prediction of Phenotypic Drug Response on Cancer Cell Lines Using Deep Convolutional Network. BMC Bioinform. 2019, 20, 408. [Google Scholar] [CrossRef]
- Liu, Q.; Hu, Z.; Jiang, R.; Zhou, M. DeepCDR: A Hybrid Graph Convolutional Network for Predicting Cancer Drug Response. Bioinformatics 2020, 36, I911–I918. [Google Scholar] [CrossRef]
- Manica, M.; Oskooei, A.; Born, J.; Subramanian, V.; Saéz-Rodríguez, J.; Rodríguez Martínez, M. Toward Explainable Anticancer Compound Sensitivity Prediction via Multimodal Attention-Based Convolutional Encoders. Mol. Pharm. 2019, 16, 4797–4806. [Google Scholar] [CrossRef]
- Jiang, L.; Jiang, C.; Yu, X.; Fu, R.; Jin, S.; Liu, X. DeepTTA: A Transformer-Based Model for Predicting Cancer Drug Response. Brief Bioinform. 2022, 23, bbac100. [Google Scholar] [CrossRef] [PubMed]
- Jin, I.; Nam, H. HiDRA: Hierarchical Network for Drug Response Prediction with Attention. J. Chem. Inf. Model 2021, 61, 3858–3867. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.; Nguyen, T. Graph Transformer for Drug Response Prediction. IEEE/ACM Transact. Comput. Biol. Bioinform. 2022, 20, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Ren, S.; Ding, M.Q.; Schwartz, R.; Lu, X.; Tao, Y.; Ren, S.; Ding, M.Q.; Schwartz, R.; Lu, X. Predicting Drug Sensitivity of Cancer Cell Lines via Collaborative Filtering with Contextual Attention. In Proceedings of the Machine Learning for Healthcare Conference; Carnegie Mellon University: Pittsburgh, PN, USA, 2020; pp. 660–680. [Google Scholar]
- Van Norman, G.A. Drugs, Devices, and the FDA: Part 1: An Overview of Approval Processes for Drugs. JACC Basic Transl. Sci. 2016, 1, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Olivier, T.; Haslam, A.; Prasad, V. Anticancer Drugs Approved by the US Food and Drug Administration from 2009 to 2020 According to Their Mechanism of Action. JAMA Netw. Open 2021, 4, e2138793. [Google Scholar] [CrossRef] [PubMed]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R.; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e16. [Google Scholar] [CrossRef]
- Bertin, P.; Rector-Brooks, J.; Sharma, D.; Gaudelet, T.; Anighoro, A.; Gross, T.; Martinez-Pena, F.; Tang, E.L.; Regep, C.; Hayter, J.; et al. RECOVER: Sequential Model Optimization Platform for Combination Drug Repurposing Identifies Novel Synergistic Compounds in Vitro. arXiv 2022, arXiv:2202.04202. [Google Scholar]
- Naik, A.W.; Kangas, J.D.; Langmead, C.J.; Murphy, R.F. Efficient Modeling and Active Learning Discovery of Biological Responses. PLoS ONE 2013, 8, e83996. [Google Scholar] [CrossRef]
- Naik, A.W.; Kangas, J.D.; Sullivan, D.P.; Murphy, R.F. Active Machine Learning-Driven Experimentation to Determine Compound Effects on Protein Patterns. Elife 2016, 5, e10047. [Google Scholar] [CrossRef]
- Kangas, J.D.; Naik, A.W.; Murphy, R.F. Efficient Discovery of Responses of Proteins to Compounds Using Active Learning. BMC Bioinform. 2014, 15, 1–11. [Google Scholar] [CrossRef]
- Murphy, R.F. An Active Role for Machine Learning in Drug Development. Nat. Chem. Biol. 2011, 7, 327–330. [Google Scholar] [CrossRef]
- Temerinac-Ott, M.; Naik, A.W.; Murphy, R.F. Deciding When to Stop: Efficient Experimentation to Learn to Predict Drug-Target Interactions. BMC Bioinform. 2015, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pinsler, R. Advances in Active Learning and Sequential Decision Making; University of Cambridge: Cambridge, UK, 2022. [Google Scholar]
- Wu, M.; Li, C.; Yao, Z. Deep Active Learning for Computer Vision Tasks: Methodologies, Applications, and Challenges. Appl. Sci. 2022, 12, 8103. [Google Scholar] [CrossRef]
- Ren, P.; Xiao, Y.; Chang, X.; Huang, P.Y.; Li, Z.; Gupta, B.B.; Chen, X.; Wang, X. A Survey of Deep Active Learning. ACM Comput. Surv. 2022, 54, 1–40. [Google Scholar] [CrossRef]
- Smailagic, A.; Costa, P.; Gaudio, A.; Khandelwal, K.; Mirshekari, M.; Fagert, J.; Walawalkar, D.; Xu, S.; Galdran, A.; Zhang, P.; et al. O-MedAL: Online Active Deep Learning for Medical Image Analysis. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2020, 10, e1353. [Google Scholar] [CrossRef]
- Budd, S.; Robinson, E.C.; Kainz, B. A Survey on Active Learning and Human-in-the-Loop Deep Learning for Medical Image Analysis. Med. Image Anal. 2021, 71, 102062. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, J.; Wang, Z.; Polson, J.; Sisk, A.E.; Sajed, D.P.; Speier, W.; Arnold, C.W. PathAL: An Active Learning Framework for Histopathology Image Analysis. IEEE Trans. Med. Imaging 2022, 41, 1176–1187. [Google Scholar] [CrossRef]
- Liang, Z.; Xu, X.; Deng, S.; Cai, L.; Jiang, T.; Jia, K. Exploring Diversity-Based Active Learning for 3D Object Detection in Autonomous Driving. arXiv 2022, arXiv:2205.07708. [Google Scholar]
- Haussmann, E.; Fenzi, M.; Chitta, K.; Ivanecky, J.; Xu, H.; Roy, D.; Mittel, A.; Koumchatzky, N.; Farabet, C.; Alvarez, J.M. Scalable Active Learning for Object Detection. In Proceedings of the 2020 IEEE Intelligent Vehicles Symposium (IV), Las Vegas, NV, USA, 19 October 2020–13 November 2020; pp. 1430–1435. [Google Scholar]
- Smailagic, A.; Costa, P.; Young Noh, H.; Walawalkar, D.; Khandelwal, K.; Galdran, A.; Mirshekari, M.; Fagert, J.; Xu, S.; Zhang, P.; et al. MedAL: Accurate and Robust Deep Active Learning for Medical Image Analysis. In Proceedings of the 17th IEEE International Conference on Machine Learning and Applications, ICMLA, Orlando, FL, USA, 17–20 December 2018; pp. 481–488. [Google Scholar]
- Shen, Y.; Yun, H.; Lipton, Z.C.; Kronrod, Y.; Anandkumar, A. Deep Active Learning for Named Entity Recognition. arXiv 2017, arXiv:1707.05928. [Google Scholar]
- Lookman, T.; Balachandran, P.V.; Xue, D.; Yuan, R. Active Learning in Materials Science with Emphasis on Adaptive Sampling Using Uncertainties for Targeted Design. NPJ Comput. Mater. 2019, 5, 21. [Google Scholar] [CrossRef]
- Jian, C.; Yang, K.; Ao, Y. Industrial Fault Diagnosis Based on Active Learning and Semi-Supervised Learning Using Small Training Set. Eng. Appl. Artif. Intell. 2021, 104, 104365. [Google Scholar] [CrossRef]
- Wu, X.; Chen, C.; Zhong, M.; Wang, J.; Shi, J. COVID-AL: The Diagnosis of COVID-19 with Deep Active Learning. Med. Image Anal. 2021, 68, 101913. [Google Scholar] [CrossRef]
- Kasarla, T.; Nagendar, G.; Hegde, G.M.; Balasubramanian, V.; Jawahar, C.V. Region-Based Active Learning for Efficient Labeling in Semantic Segmentation. In Proceedings of the 2019 IEEE Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 7–11 January 2019; pp. 1109–1117. [Google Scholar]
- Basu, A.; Bodycombe, N.E.; Cheah, J.H.; Price, E.V.; Liu, K.; Schaefer, G.I.; Ebright, R.Y.; Stewart, M.L.; Ito, D.; Wang, S.; et al. An Interactive Resource to Identify Cancer Genetic and Lineage Dependencies Targeted by Small Molecules. Cell 2013, 154, 1151–1161. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Settles, B.; Craven, M. An Analysis of Active Learning Strategies for Sequence Labeling Tasks. In Proceedings of the 2008 Conference on Empirical Methods in Natural Language Processing, Honolulu, HI, USA, 25–27 October 2008; pp. 1070–1079. [Google Scholar]
- Tian, Y.; Yuan, R.; Xue, D.; Zhou, Y.; Ding, X.; Sun, J.; Lookman, T. Role of Uncertainty Estimation in Accelerating Materials Development via Active Learning. J. Appl. Phys. 2020, 128, 014103. [Google Scholar] [CrossRef]
- Mohamadi, S.; Amindavar, H. Deep Bayesian Active Learning, A Brief Survey on Recent Advances. arXiv 2020, arXiv:2012.08044. [Google Scholar]
- Desai, S.; Ghose, D. Active Learning for Improved Semi-Supervised Semantic Segmentation in Satellite Images. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 4–8 January 2022; pp. 553–563. [Google Scholar]
- Duran-Sierra, E.; Cheng, S.; Cuenca, R.; Ahmed, B.; Ji, J.; Yakovlev, V.V.; Martinez, M.; Al-Khalil, M.; Al-Enazi, H.; Lisa Cheng, Y.S.; et al. Machine-Learning Assisted Discrimination of Precancerous and Cancerous from Healthy Oral Tissue Based on Multispectral Autofluorescence Lifetime Imaging Endoscopy. Cancers 2021, 13, 4751. [Google Scholar] [CrossRef] [PubMed]
- Vasanthakumari, P.; Romano, R.A.; Rosa, R.G.T.; Salvio, A.G.; Yakovlev, V.; Kurachi, C.; Hirshburg, J.M.; Jo, J.A. Discrimination of Cancerous from Benign Pigmented Skin Lesions Based on Multispectral Autofluorescence Lifetime Imaging Dermoscopy and Machine Learning. J. Biomed. Opt. 2022, 27, 066002. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Zhang, J. PalmHash Code vs. PalmPhasor Code. Neurocomputing 2013, 108, 1–12. [Google Scholar] [CrossRef]
- Leng, L.; Li, M.; Kim, C.; Bi, X. Dual-Source Discrimination Power Analysis for Multi-Instance Contactless Palmprint Recognition. Multimed. Tools. Appl. 2017, 76, 333–354. [Google Scholar] [CrossRef]
- Ogunleye, A.Z.; Piyawajanusorn, C.; Gonçalves, A.; Ghislat, G.; Ballester, P.J. Interpretable Machine Learning Models to Predict the Resistance of Breast Cancer Patients to Doxorubicin from Their MicroRNA Profiles. Adv. Sci. 2022, 9, 2201501. [Google Scholar] [CrossRef]
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A Survey on Ensemble Learning. Front. Comput. Sci. 2020, 14, 241–258. [Google Scholar] [CrossRef]
- Ding, Z.; Zu, S.; Gu, J. Evaluating the Molecule-Based Prediction of Clinical Drug Responses in Cancer. Bioinformatics 2016, 32, 2891–2895. [Google Scholar] [CrossRef]
- Bomane, A.; Gonçalves, A.; Ballester, P.J. Paclitaxel Response Can Be Predicted with Interpretable Multi-Variate Classifiers Exploiting DNA-Methylation and MiRNA Data. Front. Genet. 2019, 10, 1041. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Sander, C.; Stuart, J.M.; Chang, K.; Creighton, C.J.; et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]




| Analysis ‘X’ | p-Value with ‘Greedy’ | (AUChit)X − (AUChit)Greedy | p-Value with ‘Random’ | (AUChit)X − (AUChit)Random |
|---|---|---|---|---|
| Greedy | - | - | 1.02 × 10−39 | 0.24 |
| Random | 1.02 × 10−39 | −0.24 | - | - |
| Uncertainty | 0.55 | 0.01 | 9.13 × 10−24 | 0.26 |
| GU combined | 0.015 | 0.04 | 1.22 × 10−40 | 0.29 |
| Diversity | 1.19 × 10−25 | −0.19 | 8.54 × 10−8 | 0.05 |
| Hybrid sampling—0.2 | 0.15 | 0.03 | 6.44 × 10−38 | 0.27 |
| Hybrid sampling—0.3 | 0.43 | 0.01 | 2.85 × 10−36 | 0.26 |
| Hybrid sampling—0.4 | 0.99 | 2.0 × 10−4 | 1.71 × 10−34 | 0.24 |
| Hybrid sampling—0.5 | 0.33 | −0.02 | 1.89 × 10−32 | 0.23 |
| Hybrid iteration—0.2 | 1.00 × 10−4 | −0.06 | 9.95 × 10−42 | 0.18 |
| Hybrid iteration—0.3 | 1.00 × 10−10 | −0.10 | 1.51 × 10−40 | 0.15 |
| Hybrid iteration—0.4 | 1.06 × 10−18 | −0.13 | 4.17 × 10−41 | 0.11 |
| Hybrid iteration—0.5 | 8.16 × 10−26 | −0.16 | 1.82 × 10−40 | 0.07 |
| Analysis ‘X’ | p-Value with ‘Greedy’ | (AUChit Rank)Greedy − (AUChit Rank)X | p-Value with ‘Random’ | (AUChit rank)Random − (AUChit Rank)X |
|---|---|---|---|---|
| Greedy | - | - | 4.36 × 10−11 | 8.79 |
| Random | 4.36 × 10−11 | −8.79 | - | - |
| Uncertainty | 0.35 | −0.72 | 4.97 × 10−11 | 8.07 |
| GU combined | 8.30 × 10−9 | 2.45 | 1.91 × 10−11 | 11.24 |
| Diversity | 4.47 × 10−11 | −7.79 | 6.16 × 10−8 | 1.00 |
| Hybrid sampling—0.2 | 7.00 × 10−3 | 1.12 | 3.85 × 10−12 | 9.91 |
| Hybrid sampling—0.3 | 0.79 | −0.09 | 5.31 × 10−12 | 8.70 |
| Hybrid sampling—0.4 | 5.00 × 10−4 | −1.17 | 7.12 × 10−12 | 7.61 |
| Hybrid sampling—0.5 | 4.26 × 10−7 | −2.52 | 1.20 × 10−11 | 6.26 |
| Hybrid iteration—0.2 | 1.10 × 10−9 | −3.35 | 1.66 × 10−11 | 5.44 |
| Hybrid iteration—0.3 | 1.00 × 10−10 | −4.77 | 4.64 × 10−12 | 4.02 |
| Hybrid iteration—0.4 | 4.80 × 10−11 | −5.89 | 5.45 × 10−12 | 2.89 |
| Hybrid iteration—0.5 | 4.52 × 10−11 | −7.02 | 6.06 × 10−12 | 1.77 |
| Analysis ‘X’ | p-Value with ‘Greedy’ | (AUCper)X − (AUCper)Greedy | p-Value with ‘Random’ | (AUCper)X − (AUCper)Random |
|---|---|---|---|---|
| Greedy | - | - | 2.90 × 10−11 | −0.13 |
| Random | 2.90 ×10−11 | 0.13 | - | - |
| Uncertainty | 3.00 ×10−3 | 0.07 | 2.5 × 10−4 | −0.06 |
| GU combined | 0.57 | 0.02 | 3.61 × 10−8 | −0.11 |
| Diversity | 6.00 ×10−10 | 0.12 | 0.19 | −6.40 × 10−3 |
| Hybrid sampling—0.2 | 0.28 | 0.03 | 1.54 × 10−8 | −0.10 |
| Hybrid sampling—0.3 | 0.14 | 0.03 | 2.50 × 10−9 | −0.09 |
| Hybrid sampling—0.4 | 0.12 | 0.04 | 3.00 × 10−10 | −0.09 |
| Hybrid sampling—0.5 | 0.06 | 0.04 | 1.03 × 10−11 | −0.09 |
| Hybrid iteration—0.2 | 5.50 ×10−6 | 0.09 | 3.30 × 10−5 | −0.04 |
| Hybrid iteration—0.3 | 7.81 ×10−8 | 0.11 | 2.50 × 10−4 | −0.02 |
| Hybrid iteration—0.4 | 2.40 ×10−9 | 0.11 | 2.10 × 10−3 | −0.01 |
| Hybrid iteration—0.5 | 3.00 ×10−10 | 0.12 | 0.01 | −0.006 |
| Analysis ‘X’ | p-Value with ‘Greedy’ | (AUCper Rank)Greedy − (AUCper Rank)X | p-Value with ‘Random’ | (AUCper Rank)Random − (AUCper Rank)X |
| Greedy | - | - | 8.60 × 10−9 | −7.12 |
| Random | 8.60 ×10−9 | 7.12 | - | - |
| Uncertainty | 2.00 ×10−5 | 3.94 | 6.30 × 10−6 | −3.17 |
| GU combined | 0.07 | 0.38 | 9.00 × 10−10 | −6.73 |
| Diversity | 2.00 ×10−10 | 6.54 | 0.03 | −0.57 |
| Hybrid sampling—0.2 | 0.54 | 0.50 | 1.44 × 10−8 | −6.61 |
| Hybrid sampling—0.3 | 0.02 | 1.19 | 1.18 × 10−7 | −5.93 |
| Hybrid sampling—0.4 | 0.03 | 1.05 | 2.50 × 10−8 | −6.07 |
| Hybrid sampling—0.5 | 0.04 | 1.14 | 1.72 × 10−7 | −5.98 |
| Hybrid iteration—0.2 | 3.90 ×10−9 | 4.24 | 3.2 × 10−5 | −2.87 |
| Hybrid iteration—0.3 | 4.00 ×10−9 | 5.29 | 5.00 × 10−4 | −1.82 |
| Hybrid iteration—0.4 | 2.60 ×10−9 | 6.29 | 0.02 | −0.82 |
| Hybrid iteration—0.5 | 5.40 ×10−9 | 6.77 | 0.15 | −0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasanthakumari, P.; Zhu, Y.; Brettin, T.; Partin, A.; Shukla, M.; Xia, F.; Narykov, O.; Weil, M.R.; Stevens, R.L. A Comprehensive Investigation of Active Learning Strategies for Conducting Anti-Cancer Drug Screening. Cancers 2024, 16, 530. https://doi.org/10.3390/cancers16030530
Vasanthakumari P, Zhu Y, Brettin T, Partin A, Shukla M, Xia F, Narykov O, Weil MR, Stevens RL. A Comprehensive Investigation of Active Learning Strategies for Conducting Anti-Cancer Drug Screening. Cancers. 2024; 16(3):530. https://doi.org/10.3390/cancers16030530
Chicago/Turabian StyleVasanthakumari, Priyanka, Yitan Zhu, Thomas Brettin, Alexander Partin, Maulik Shukla, Fangfang Xia, Oleksandr Narykov, Michael Ryan Weil, and Rick L. Stevens. 2024. "A Comprehensive Investigation of Active Learning Strategies for Conducting Anti-Cancer Drug Screening" Cancers 16, no. 3: 530. https://doi.org/10.3390/cancers16030530
APA StyleVasanthakumari, P., Zhu, Y., Brettin, T., Partin, A., Shukla, M., Xia, F., Narykov, O., Weil, M. R., & Stevens, R. L. (2024). A Comprehensive Investigation of Active Learning Strategies for Conducting Anti-Cancer Drug Screening. Cancers, 16(3), 530. https://doi.org/10.3390/cancers16030530





