Association of HOTAIR, MIR155HG, TERC, miR-155, -196a2, and -146a Genes Polymorphisms with Papillary Thyroid Cancer Susceptibility and Prognosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Histopathological Analysis, Pathological Evaluation, and Tumor Staging
2.3. DNA Isolation and Gene Polymorphism Analysis
2.4. Statistical Analysis and Bioinformatics
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seib, C.D.; Sosa, J.A. Evolving Understanding of the Epidemiology of Thyroid Cancer. Endocrinol. Metab. Clin. N. Am. 2019, 48, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Haugen, B.R.; Alexander, E.K.; Bible, K.C.; Doherty, G.M.; Mandel, S.J.; Nikiforov, Y.E.; Pacini, F.; Randolph, G.W.; Sawka, A.M.; Schlumberger, M.; et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid 2016, 26, 1–133. [Google Scholar] [CrossRef]
- Kim, K.; Kim, J.H.; Park, I.S.; Rho, Y.S.; Kwon, G.H.; Lee, D.J. The Updated AJCC/TNM Staging System for Papillary Thyroid Cancer (8th Edition): From the Perspective of Genomic Analysis. World J. Surg. 2018, 42, 3624–3631. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, M.; Chorti, A.G.; Chatzikyriakidou, A.; Giannoulis, K.; Bakkar, S.; Papavramidis, T.S. MicroRNAs in Papillary Thyroid Cancer: What Is New in Diagnosis and Treatment. Front. Oncol. 2021, 11, 755097. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhang, K.; Ma, L.; Xu, J.; Chang, W. The Role of Long Non-Coding RNAs in Thyroid Cancer. Front. Oncol. 2020, 10, 941. [Google Scholar] [CrossRef]
- Liz, J.; Esteller, M. LncRNAs and MicroRNAs with a Role in Cancer Development. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 169–176. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A CeRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Botti, G.; Scognamiglio, G.; Aquino, G.; Liguori, G.; Cantile, M. LncRNA HOTAIR in Tumor Microenvironment: What Role? Int. J. Mol. Sci. 2019, 20, 2279. [Google Scholar] [CrossRef]
- Niu, L.; Lou, F.; Sun, Y.; Sun, L.; Cai, X.; Liu, Z.; Zhou, H.; Wang, H.; Wang, Z.; Bai, J.; et al. A Micropeptide Encoded by LncRNA MIR155HG Suppresses Autoimmune Inflammation via Modulating Antigen Presentation. Sci. Adv. 2020, 6, eaaz2059. [Google Scholar] [CrossRef]
- Kuo, F.-C.; Wang, Y.-T.; Liu, C.-H.; Li, Y.-F.; Lu, C.-H.; Su, S.-C.; Liu, J.-S.; Li, P.-F.; Huang, C.-L.; Ho, L.-J.; et al. LncRNA HOTAIR Impairs the Prognosis of Papillary Thyroid Cancer via Regulating Cellular Malignancy and Epigenetically Suppressing DLX1. Cancer Cell Int. 2022, 22, 396. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.; Li, Z.; Qiu, S.; Cao, J.; Zhao, Y.; Huang, Z.; He, J.; Luo, F.; Yang, K. Diagnostic Value of Serum LncRNA HOTAIR Combined with Galectin-3 in Benign and Papillary Thyroid Carcinoma. Cancer Manag. Res. 2021, 13, 6517–6525. [Google Scholar] [CrossRef]
- Mahmoud, A.A.; Mohamed, H.O.; Abdel Aal, A.M.; Abdelghafour, H.S.; Jabir, M.A. Long Noncoding RNA HOTAIR and Midkine as Biomarkers in Thyroid Cancer. Egypt. J. Immunol. 2023, 30, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Possieri, C.; Locantore, P.; Salis, C.; Bacci, L.; Aiello, A.; Fadda, G.; De Crea, C.; Raffaelli, M.; Bellantone, R.; Grassi, C.; et al. Combined Molecular and Mathematical Analysis of Long Noncoding RNAs Expression in Fine Needle Aspiration Biopsies as Novel Tool for Early Diagnosis of Thyroid Cancer. Endocrine 2021, 72, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Hann, S.S. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell Physiol. Biochem. 2018, 47, 893–913. [Google Scholar] [CrossRef]
- Wu, L.; Shi, Y.; Liu, B.; Zhao, M. Expression of LncRNA-HOTAIR in the Serum of Patients with Lymph Node Metastasis of Papillary Thyroid Carcinoma and Its Impact. Oncol. Lett. 2020, 20, 907–913. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Mahesh, G.; Biswas, R. MicroRNA-155: A Master Regulator of Inflammation. J. Interferon Cytokine Res. 2019, 39, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Chen, Z.; Chen, Y.; Wang, X.; Tang, N. MIR155HG Is a Prognostic Biomarker and Associated with Immune Infiltration and Immune Checkpoint Molecules Expression in Multiple Cancers. Cancer Med. 2019, 8, 7161–7173. [Google Scholar] [CrossRef]
- Cui, W.; Meng, W.; Zhao, L.; Cao, H.; Chi, W.; Wang, B. TGF-β-Induced Long Non-Coding RNA MIR155HG Promotes the Progression and EMT of Laryngeal Squamous Cell Carcinoma by Regulating the MiR-155-5p/SOX10 Axis. Int. J. Oncol. 2019, 54, 2005–2018. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Yu, T.; Nie, E.; Hu, Q.; Wu, W.; Zhi, T.; Jiang, K.; Wang, X.; Lu, X.; et al. Blocking MIR155HG/MiR-155 Axis Inhibits Mesenchymal Transition in Glioma. Neuro-Oncol. 2017, 19, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.C.; Vardinogiannis, I.; Gilmore, T.D. Identification of an NF-ΚB P50/P65-Responsive Site in the Human MIR155HG Promoter. BMC Mol. Biol. 2013, 14, 24. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, Y.; Ge, Y.; Liu, J.; Zhao, Y. TERC Promotes Cellular Inflammatory Response Independent of Telomerase. Nucleic Acids Res. 2019, 47, 8084–8095. [Google Scholar] [CrossRef] [PubMed]
- Barthel, F.P.; Wei, W.; Tang, M.; Martinez-Ledesma, E.; Hu, X.; Amin, S.B.; Akdemir, K.C.; Seth, S.; Song, X.; Wang, Q.; et al. Systematic Analysis of Telomere Length and Somatic Alterations in 31 Cancer Types. Nat. Genet. 2017, 49, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Montero-Conde, C.; Leandro-García, L.J.; Martínez-Montes, Á.M.; Martínez, P.; Moya, F.J.; Letón, R.; Gil, E.; Martínez-Puente, N.; Guadalix, S.; Currás-Freixes, M.; et al. Comprehensive Molecular Analysis of Immortalization Hallmarks in Thyroid Cancer Reveals New Prognostic Markers. Clin. Transl. Med. 2022, 12, e1001. [Google Scholar] [CrossRef]
- Daveri, E.; Vergani, E.; Shahaj, E.; Bergamaschi, L.; La Magra, S.; Dosi, M.; Castelli, C.; Rodolfo, M.; Rivoltini, L.; Vallacchi, V.; et al. MicroRNAs Shape Myeloid Cell-Mediated Resistance to Cancer Immunotherapy. Front. Immunol. 2020, 11, 1214. [Google Scholar] [CrossRef]
- Qiu, Z.; Li, H.; Wang, J.; Sun, C. MiR-146a and MiR-146b in the Diagnosis and Prognosis of Papillary Thyroid Carcinoma. Oncol. Rep. 2017, 38, 2735–2740. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-Z.; Zheng, K.; Zhang, H.-H.; Chen, L.; Wu, K.-L.; Ren, C.-H.; Wang, Z.-C.; Kong, L.-J.; Ruan, W.-H.; Chen, X.-J. Expression of microRNA-155 in papillary thyroid carcinoma and its clinical significance. Nan Fang Yi Ke Da Xue Xue Bao 2016, 36, 1364–1368. [Google Scholar]
- Fu, Y.-T.; Zhang, D.-Q.; Zhou, L.; Li, S.-J.; Sun, H.; Liu, X.-L.; Zheng, H.-B. Has-MiR-196a-2 Is up-Regulated and Acts as an Independent Unfavorable Prognostic Factor in Thyroid Carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2707–2714. [Google Scholar] [CrossRef]
- Dong, G.; Zhang, R.; Xu, J.; Guo, Y. Association between MicroRNA Polymorphisms and Papillary Thyroid Cancer Susceptibility. Int. J. Clin. Exp. Pathol. 2015, 8, 13450–13457. [Google Scholar]
- Khan, R.; Abbasi, S.A.; Mansoor, Q.; Ahmed, M.N.; Mir, K.B.; Baig, R.M. Analysis of Rare Alleles of MiRNA-146a (Rs2910164) and MiRNA-34b/c (Rs4938723) as a Prognostic Marker in Thyroid Cancer in Pakistani Population. Diagnostics 2022, 12, 2495. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, L.; Fu, G.; Sun, F.; Shi, J.; Wei, J.; Lu, C.; Zhou, C.; Yuan, Q.; Yang, M. The Identification of an ESCC Susceptibility SNP Rs920778 That Regulates the Expression of LncRNA HOTAIR via a Novel Intronic Enhancer. Carcinogenesis 2014, 35, 2062–2067. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lv, Z.; An, C.; Shi, M.; Pan, W.; Zhou, L.; Yang, W.; Yang, M. Onco-LncRNA HOTAIR and Its Functional Genetic Variants in Papillary Thyroid Carcinoma. Sci. Rep. 2016, 6, 31969. [Google Scholar] [CrossRef] [PubMed]
- Baloch, Z.W.; Asa, S.L.; Barletta, J.A.; Ghossein, R.A.; Juhlin, C.C.; Jung, C.K.; LiVolsi, V.A.; Papotti, M.G.; Sobrinho-Simões, M.; Tallini, G.; et al. Overview of the 2022 WHO Classification of Thyroid Neoplasms. Endocr. Pathol. 2022, 33, 27–63. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to Build a Bridge from a Population-Based to a More “Personalized” Approach to Cancer Staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic Mining of Putative Causal Variants, Cell Types, Regulators and Target Genes for Human Complex Traits and Disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef]
- Li, J.; Han, L.; Roebuck, P.; Diao, L.; Liu, L.; Yuan, Y.; Weinstein, J.N.; Liang, H. TANRIC: An Interactive Open Platform to Explore the Function of LncRNAs in Cancer. Cancer Res. 2015, 75, 3728–3737. [Google Scholar] [CrossRef]
- Yan, R.; Cao, J.; Song, C.; Chen, Y.; Wu, Z.; Wang, K.; Dai, L. Polymorphisms in LncRNA HOTAIR and Susceptibility to Breast Cancer in a Chinese Population. Cancer Epidemiol. 2015, 39, 978–985. [Google Scholar] [CrossRef]
- Hassanzarei, S.; Hashemi, M.; Sattarifard, H.; Hashemi, S.M.; Bahari, G.; Ghavami, S. Genetic Polymorphisms of HOTAIR Gene Are Associated with the Risk of Breast Cancer in a Sample of Southeast Iranian Population. Tumour Biol. 2017, 39, 101042831772753. [Google Scholar] [CrossRef]
- Pan, W.; Liu, L.; Wei, J.; Ge, Y.; Zhang, J.; Chen, H.; Zhou, L.; Yuan, Q.; Zhou, C.; Yang, M. A Functional LncRNA HOTAIR Genetic Variant Contributes to Gastric Cancer Susceptibility: FUNCTIONAL POLYMORPHISMS OF LncRNA HOTAIR IN GASTRIC CANCER. Mol. Carcinog. 2016, 55, 90–96. [Google Scholar] [CrossRef]
- Kim, J.O.; Jun, H.H.; Kim, E.J.; Lee, J.Y.; Park, H.S.; Ryu, C.S.; Kim, S.; Oh, D.; Kim, J.W.; Kim, N.K. Genetic Variants of HOTAIR Associated with Colorectal Cancer Susceptibility and Mortality. Front. Oncol. 2020, 10, 72. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Jiang, R.; Zhang, M.; Wang, H.; Zhang, L.; Tang, J.; Liang, C. Analyzing 37,900 Samples Shows Significant Association between Hotair Polymorphisms and Cancer Susceptibility: A Meta-Analysis. Int. J. Biol. Markers 2017, 32, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Chen, Y.; Yuan, Q.; Hua, Q.; Zhang, X.; Wang, M.; Tong, N.; Zhang, W.; Chen, J.; Zhang, Z. The HOTAIR, PRNCR1 and POLR2E Polymorphisms Are Associated with Cancer Risk: A Meta-Analysis. Oncotarget 2017, 8, 43271–43283. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qiu, H.; Wang, X.; Guo, R.; Liu, Q.; Wang, Y.; Yuan, Z.; Li, J.; Shi, H. HOTAIR Rs920778 Polymorphism Is Associated with Ovarian Cancer Susceptibility and Poor Prognosis in a Chinese Population. Future Oncol. 2017, 13, 347–355. [Google Scholar] [CrossRef]
- Bayram, S.; Sümbül, A.T.; Batmacı, C.Y.; Genç, A. Effect of HOTAIR Rs920778 Polymorphism on Breast Cancer Susceptibility and Clinicopathologic Features in a Turkish Population. Tumour Biol. 2015, 36, 3863–3870. [Google Scholar] [CrossRef]
- Lv, Z.; Kou, C.; Chen, N.; Jia, L.; Sun, X.; Gao, Y.; Bai, R.; Yang, M.; Cui, J. Single Nucleotide Polymorphisms in HOTAIR Are Related to Breast Cancer Risk and Prognosis in the Northeastern Chinese Population. Front. Oncol. 2021, 11, 706428. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Y. Identification of Differentiated Functional Modules in Papillary Thyroid Carcinoma by Analyzing Differential Networks. J. Cancer Res. Ther. 2018, 14, S969–S974. [Google Scholar] [CrossRef]
- Guo, R.; Ning, Y.; Ma, Y.; Lin, Q.; Shen, N.; Shi, P. Long Non-coding RNA HOTAIR/microRNA -761 Sponge Regulates PPME1 and Further Influences Cell Biological Functions in Thyroid Carcinoma. Laryngoscope Investig. Otolaryngol. 2021, 6, 438–445. [Google Scholar] [CrossRef]
- Xia, F.; Xia, W.; Yu, X. LncRNA HOTAIR Influences the Growth, Migration, and Invasion of Papillary Thyroid Carcinoma via Affection on the MiR-488-5p/NUP205 Axis. Technol. Cancer Res. Treat. 2020, 19, 153303382096212. [Google Scholar] [CrossRef]
- Fujisaka, Y.; Iwata, T.; Tamai, K.; Nakamura, M.; Mochizuki, M.; Shibuya, R.; Yamaguchi, K.; Shimosegawa, T.; Satoh, K. Long Non-Coding RNA HOTAIR Up-regulates Chemokine (C-C Motif) Ligand 2 and Promotes Proliferation of Macrophages and Myeloid-derived Suppressor Cells in Hepatocellular Carcinoma Cell Lines. Oncol. Lett. 2017, 15, 509–514. [Google Scholar] [CrossRef]
- Yang, L.; Peng, X.; Li, Y.; Zhang, X.; Ma, Y.; Wu, C.; Fan, Q.; Wei, S.; Li, H.; Liu, J. Long Non-Coding RNA HOTAIR Promotes Exosome Secretion by Regulating RAB35 and SNAP23 in Hepatocellular Carcinoma. Mol. Cancer 2019, 18, 78. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, N.; Zheng, Z.; Che, Y.; Suzuki, M.; Kano, S.; Lu, J.; Wang, P.; Sun, Y.; Homma, A. Exosomal LncRNA HOTAIR Induce Macrophages to M2 Polarization via PI3K/ p-AKT /AKT Pathway and Promote EMT and Metastasis in Laryngeal Squamous Cell Carcinoma. BMC Cancer 2022, 22, 1208. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Zhou, Y.; Lam, A.K. Long Non-Coding RNAs Profiling Using Microarray in Papillary Thyroid Carcinoma. Methods Mol. Biol. 2022, 2534, 135–148. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, J.; Chen, J.; Wang, J.; Xu, J. Construction and Analysis of an Aberrant LncRNA-MiRNA-MRNA Network Associated with Papillary Thyroid Cancer. Medicine 2020, 99, e22705. [Google Scholar] [CrossRef]
- Li, H.; Yang, H.; Wen, D.; Luo, Y.; Liang, C.; Pan, D.; Ma, W.; Chen, G.; He, Y.; Chen, J. Overexpression of LncRNA HOTAIR Is Associated with Poor Prognosis in Thyroid Carcinoma: A Study Based on TCGA and GEO Data. Horm. Metab. Res. 2017, 49, 388–399. [Google Scholar] [CrossRef]
- Chao, X.; Feng, X.; Wang, X.; Shi, H.; Li, H.; Wang, Y.; Wang, L.; Shen, H.; Zha, Q.; Chen, Y. MiRNA155HG Polymorphisms Influenced the Risk of Liver Cancer among the Han Chinese Population. BMC Med. Genet. 2020, 21, 134. [Google Scholar] [CrossRef]
- Zou, W.; Li, X.; Li, C.; Liu, D.; Lv, Y.; Yang, Y.; Ye, N.; Guo, D.; He, S. Analysis of the Relationship between MIR155HG Variants and Gastric Cancer Susceptibility. BMC Gastroenterol. 2020, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; He, G.; Han, H.; Xiong, W.; Song, T.; Chen, H.; Chen, X.; Wu, X.; Huang, G.; Zhang, Y.; et al. Analysis of MIR155HG Variants and Colorectal Cancer Susceptibility in Han Chinese Population. Molec Gen. Gen. Med. 2019, 7, e778. [Google Scholar] [CrossRef]
- Wu, H.R.; Zhang, J. AP-2α Expression in Papillary Thyroid Carcinoma Predicts Tumor Progression and Poor Prognosis. Cancer Manag. Res. 2018, 10, 2615–2625. [Google Scholar] [CrossRef]
- Subhi, O.; Schulten, H.-J.; Bagatian, N.; Al-Dayini, R.; Karim, S.; Bakhashab, S.; Alotibi, R.; Al-Ahmadi, A.; Ata, M.; Elaimi, A.; et al. Genetic Relationship between Hashimoto‘s Thyroiditis and Papillary Thyroid Carcinoma with Coexisting Hashimoto’s Thyroiditis. PLoS ONE 2020, 15, e0234566. [Google Scholar] [CrossRef]
- Qin, Y.; Liu, X.; Pan, L.; Zhou, R.; Zhang, X. Long Noncoding RNA MIR155HG Facilitates Pancreatic Cancer Progression through Negative Regulation of MiR-802. J. Cell. Biochem. 2019, 120, 17926–17934. [Google Scholar] [CrossRef]
- Thiele, J.-A.; Hosek, P.; Kralovcova, E.; Ostasov, P.; Liska, V.; Bruha, J.; Vycital, O.; Rosendorf, J.; Opattova, A.; Horak, J.; et al. LncRNAs in Non-Malignant Tissue Have Prognostic Value in Colorectal Cancer. Int. J. Mol. Sci. 2018, 19, 2672. [Google Scholar] [CrossRef]
- Lin, H.; Ni, R.; Li, D.; Zhao, M.; Li, Y.; Li, K.; Zhang, Q.; Huang, C.; Huang, S. LncRNA MIR155HG Overexpression Promotes Proliferation, Migration, and Chemoresistance in Gastric Cancer Cells. Int. J. Med. Sci. 2023, 20, 933–942. [Google Scholar] [CrossRef]
- Wen, A.; Luo, L.; Du, C.; Luo, X. Long Non-Coding RNA MiR155HG Silencing Restrains Ovarian Cancer Progression by Targeting the MicroRNA-155-5p/Tyrosinase-Related Protein 1 Axis. Exp. Ther. Med. 2021, 22, 1237. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.-Y.; Han, Y.-D.; Lin, Q. Long Non-Coding RNA MIR155HG Knockdown Suppresses Cell Proliferation, Migration and Invasion in NSCLC by Upregulating TP53INP1 Directly Targeted by MiR-155-3p and MiR-155-5p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4822–4835. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.R.; Liao, Y.; Cai, M.; Qiu, H.; Wen, F.; Peng, M.; Wang, S.; Liu, S.; Guo, G.; Chi, X.; et al. MIR155HG Plays a Bivalent Role in Regulating Innate Antiviral Immunity by Encoding Long Noncoding RNA-155 and MicroRNA-155-5p. mBio 2022, 13, e0251022. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Pang, G.; Yang, L.; Chen, S.; Xu, R.; Shao, W. Long Noncoding RNAs Regulate the Inflammatory Responses of Macrophages. Cells 2021, 11, 5. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; Liao, M.; Zhang, Q.; Yang, M. LncRNA MIR155HG Induces M2 Macrophage Polarization and Drug Resistance of Colorectal Cancer Cells by Regulating ANXA2. Cancer Immunol. Immunother. 2022, 71, 1075–1091. [Google Scholar] [CrossRef]
- Li, N.; Liu, Y.; Cai, J. LncRNA MIR155HG Regulates M1/M2 Macrophage Polarization in Chronic Obstructive Pulmonary Disease. Biomed. Pharmacother. 2019, 117, 109015. [Google Scholar] [CrossRef]
- Peng, L.; Pan, B.; Zhang, X.; Wang, Z.; Qiu, J.; Wang, X.; Tang, N. Lipopolysaccharide Facilitates Immune Escape of Hepatocellular Carcinoma Cells via M6A Modification of LncRNA MIR155HG to Upregulate PD-L1 Expression. Cell Biol. Toxicol. 2022, 38, 1159–1173. [Google Scholar] [CrossRef]
- Xie, K.; Ma, H.; Liang, C.; Wang, C.; Qin, N.; Shen, W.; Gu, Y.; Yan, C.; Zhang, K.; Dai, N.; et al. A Functional Variant in MiR-155 Regulation Region Contributes to Lung Cancer Risk and Survival. Oncotarget 2015, 6, 42781–42792. [Google Scholar] [CrossRef]
- Ji, J.; Xu, M.; Tu, J.; Zhao, Z.; Gao, J.; Chen, M.; Song, J.; Zhu, H.; Cheng, X.; Hui, J.; et al. MiR-155 and Its Functional Variant Rs767649 Contribute to the Susceptibility and Survival of Hepatocellular Carcinoma. Oncotarget 2016, 7, 60303–60309. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, H.; Liu, Y.; Chen, Z.; Gu, J.; Cui, D.; Yang, T. MiR-146a Rs2910164 Polymorphism and Risk of Papillary Thyroid Carcinoma: A Meta-Analysis. Genet. Test. Mol. Biomark. 2018, 22, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Conzo, G.; Docimo, G.; Ruggiero, R.; Napolitano, S.; Palazzo, A.; Gambardella, C.; Mauriello, C.; Tartaglia, E.; Cavallo, F.; Santini, L. Surgical Treatment of Papillary Thyroid Carcinoma without Lymph Nodal Involvement. G. Chir. 2012, 33, 339–342. [Google Scholar] [PubMed]
| Variables | Controls | PTC Cases | p | |||
|---|---|---|---|---|---|---|
| N = 106 | % | N = 102 | % | |||
| Sex | Male | 39 | 36.79 | 31 | 30.39 | 0.379 |
| Female | 67 | 63.21 | 71 | 69.61 | ||
| Age | 55< | 75 | 70.75 | 67 | 65.69 | 0.459 |
| ≥55 | 31 | 29.25 | 35 | 34.31 | ||
| HOTAIR rs920778 | CC | 51 | 48.11 | 40 | 39.22 | 0.016 |
| CT | 37 | 34.91 | 27 | 26.47 | ||
| TT | 18 | 16.98 | 35 | 34.31 | ||
| MIR155HG rs1893650 | TT | 53 | 50.00 | 75 | 73.53 | 0.0002 |
| TC | 46 | 43.40 | 17 | 16.67 | ||
| CC | 7 | 6.60 | 10 | 9.80 | ||
| TERC rs10936599 | CC | 65 | 61.32 | 68 | 66.67 | 0.146 |
| CT | 33 | 31.13 | 21 | 20.59 | ||
| TT | 8 | 7.55 | 13 | 12.75 | ||
| miR-155 rs767649 | TT | 95 | 89.62 | 86 | 84.31 | 0.255 |
| TA | 11 | 10.38 | 16 | 15.69 | ||
| AA | 0 | 0 | 0 | 0 | ||
| miR-196a2 rs11614913 | CC | 55 | 51.89 | 56 | 54.90 | 0.149 |
| CT | 41 | 38.68 | 29 | 28.43 | ||
| TT | 10 | 9.43 | 17 | 16.67 | ||
| miR-146a rs2910164 | GG | 72 | 67.92 | 72 | 70.59 | 0.473 |
| GC | 27 | 25.47 | 20 | 19.61 | ||
| CC | 7 | 6.60 | 10 | 9.80 | ||
| Variables | N | HOTAIR rs920778 wt/ht/mt | MIR155HG rs1893650 wt/ht/mt | TERC rs10936599 wt/ht/mt | miR-155 rs767649 wt/ht | miR-196a2 rs11614913 wt/ht/mt | miR-146a rs2910164 wt/ht/mt | |
|---|---|---|---|---|---|---|---|---|
| Age | <55 | 67 | 22/21/24 | 52/9/6 | 46/11/10 | 56/11 | 38/20/9 | 46/15/6 |
| ≥55 | 35 | 18/6/11 | 23/8/4 | 22/10/3 | 30/5 | 18/9/8 | 26/5/4 | |
| p/p * | 0.142/0.414 | 0.404/0.733 | 0.287/0.534 | 1 | 0.477/0.175 | 0.602/0.733 | ||
| Sex | Male | 31 | 12/8/11 | 22/6/3 | 19/9/3 | 26/5 | 14/10/7 | 23/4/4 |
| Female | 71 | 28/19/24 | 53/11/7 | 49/12/10 | 60/11 | 42/19/10 | 49/16/6 | |
| p/p * | 0.986/1 | 0.890/1 | 0.356/0.749 | 1 | 0.380/0.386 | 0.463/0.487 | ||
| Multifocality | Absent | 23 | 8/5/10 | 19/0/4 | 18/2/3 | 20/3 | 17/5/1 | 16/5/2 |
| Present | 79 | 32/22/25 | 56/17/6 | 50/19/10 | 66/13 | 39/24/16 | 56/15/8 | |
| p/p * | 0.587/0.621 | 0.030/0.227 | 0.267/1 | 1 | 0.077/0.110 | 0.946/1 | ||
| ETE | Absent | 84 | 33/23/28 | 61/15/6 | 52/19/13 | 69/15 | 45/23/16 | 60/16/8 |
| Present | 18 | 7/4/7 | 14/2/2 | 16/2/0 | 17/1 | 11/6/1 | 12/4/2 | |
| p/p * | 0.868/0.785 | 0.780/1 | 0.069/0.117 | 0.292 | 0.376/0.294 | 0.922/1 | ||
| Vascular invasion | Absent | 48 | 22/16/10 | 35/8/5 | 32/10/6 | 40/8 | 25/18/5 | 33/11/4 |
| Present | 54 | 18/11/25 | 40/9/5 | 36/11/7 | 46/8 | 31/11/12 | 39/9/6 | |
| p/p * | 0.024/0.012 | 0.981/1 | 0.997/1 | 0.797 | 0.087/0.110 | 0.687/0.746 | ||
| Calcifications | Absent | 72 | 31/14/27 | 52/11/9 | 46/16/10 | 64/8 | 41/21/10 | 51/15/6 |
| Present | 30 | 9/13/8 | 23/6/1 | 22/5/3 | 22/8 | 15/8/7 | 21/5/4 | |
| p/p * | 0.045/0.363 | 0.340/0.274 | 0.653/0.592 | 0.049 | 0.505/0.244 | 0.694/0.475 | ||
| ATA risk | Low | 7 | 2/2/3 | 6/0/1 | 6/1/0 | 6/1 | 3/4/0 | 5/1/1 |
| Intermediate | 81 | 34/22/25 | 58/16//7 | 51/17/13 | 68/13 | 47/17/17 | 57/16/8 | |
| High | 14 | 4/3/7 | 11/1/2 | 11/3/0 | 12/2 | 6/8/0 | 10/3/1 | |
| p/p * | 0.670/0.336 | 0.531/0.741 | 0.359/0.145 | 0.981 | 0.012/0.071 | 0.985/0.873 | ||
| T | T1 | 43 | 17/12/14 | 30/9/4 | 32/6/5 | 36/7 | 28/8/7 | 33/8/2 |
| T2 | 35 | 17/8/10 | 27/4/4 | 17/11/7 | 28/7 | 15/13/7 | 22/9/4 | |
| T3 | 24 | 6/7/11 | 18/4/2 | 19/4/1 | 21/3 | 13/8/3 | 17/3/4 | |
| p/p * | 0.464/0.371 | 0.852/0.916 | 0.071/0.193 | 0.295 | 0.305/0.747 | 0.374/0.263 | ||
| N | Absent | 45 | 23/12/10 | 30/9/6 | 32/7/6 | 41/4 | 27/11/7 | 28/10/7 |
| Present | 57 | 17/15/25 | 45/8/4 | 36/14/7 | 45/12 | 29/18/10 | 44/10/3 | |
| p/p * | 0.042/0.022 | 0.354/0.330 | 0.535/0.874 | 0.093 | 0.640/0.789 | 0.150/0.102 | ||
| Stage | I | 31 | 15/8/8 | 22/6/3 | 20/6/5 | 29/2 | 17/11/3 | 22/7/2 |
| II | 60 | 23/16/21 | 43/10/7 | 38/14/8 | 46/14 | 34/14/12 | 42/10/8 | |
| III | 11 | 2/3/6 | 10/1/0 | 10/1/0 | 11/0 | 5/4/2 | 8/3/0 | |
| p/p * | 0.420/0.223 | 0.679/0.489 | 0.443/0.378 | 0.035 | 0.580/0.452 | 0.568/0.296 | ||
| Recurrence | Absent | 88 | 38/24/26 | 66/15/7 | 58/17/13 | 75/13 | 51/22/15 | 62/16/10 |
| Present | 14 | 2/3/9 | 9/2/3 | 10/4/0 | 11/3 | 5/7/2 | 10/4/0 | |
| p/p * | 0.031/0.016 | 0.289/0.138 | 0.269/0.206 | 0.457 | 0.150/1 | 0.323/0.350 | ||
| COX Regression Analysis | Variables | Progression-Free Survival | ||
|---|---|---|---|---|
| HR | (95% CI) | p | ||
| Univariate Analysis | Age (55 years) | 1.068 | (0.357–3.188) | 0.907 |
| Sex | 0.412 | (0.144–1.175) | 0.097 | |
| Hashimoto thyroiditis | 0.466 | (0.146–1.486) | 0.197 | |
| Multifocality | 3.719 | (0.486–28.427) | 0.206 | |
| ETE | 4.099 | (1.420–11.831) | 0.009 | |
| Vascular invasion | 1.768 | (0.592–5.279) | 0.308 | |
| ATA risk | 11.994 | (4.176–34.448) | 0.000004 | |
| T | 1.482 | (0.799–2.751) | 0.212 | |
| Nodal metastases | 2.565 | (1.107–5.941) | 0.028 | |
| Stage | 2.405 | (1.206–4.798) | 0.013 | |
| Calcifications | 0.368 | (0.082–1.644) | 0.190 | |
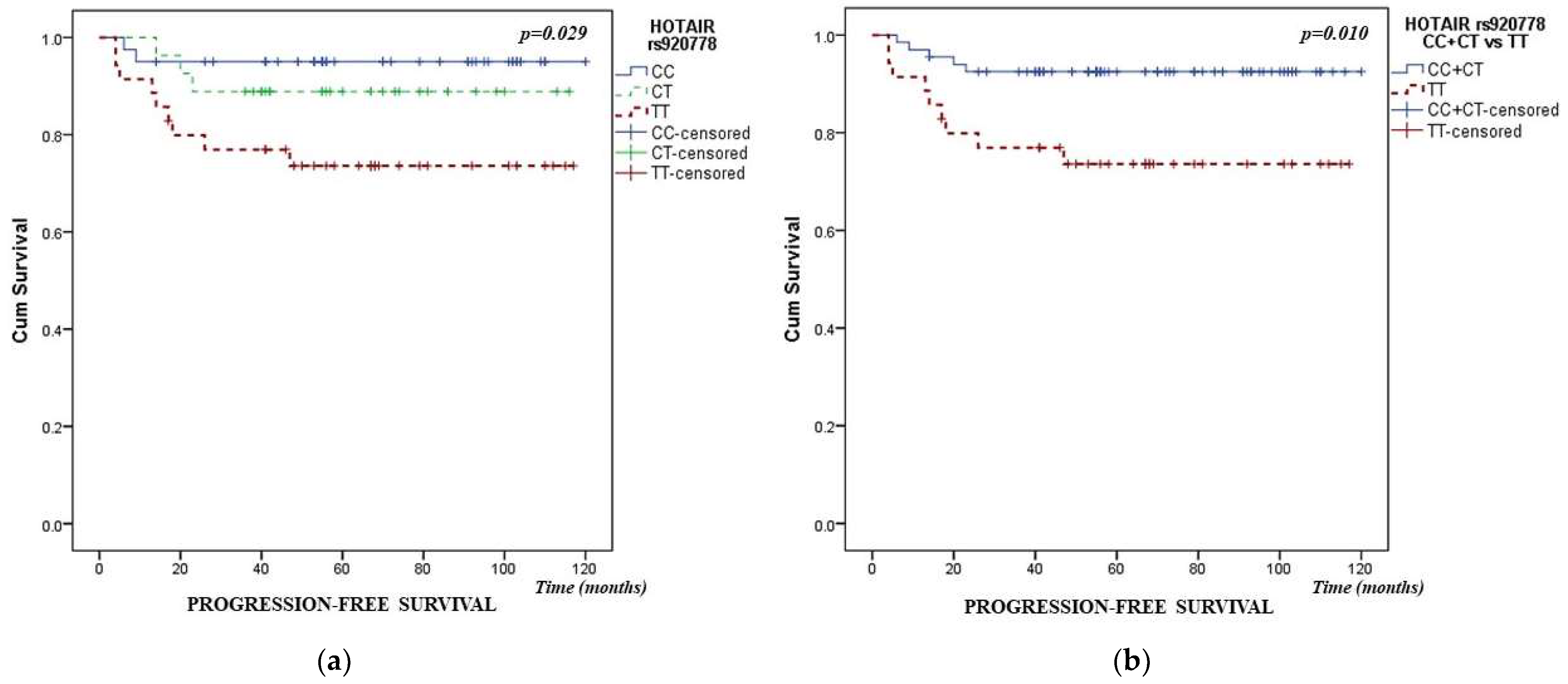
| HOTAIR rs920778 | 2.467 | (1.195–5.094) | 0.015 | |
| TERC rs10936599 | 0.649 | (0.263–1.601) | 0.348 | |
| MI155HG rs1893650 | 1.607 | (0.794–3.253) | 0.187 | |
| miR-155 rs767649 | 1.474 | (0.411–5.289) | 0.552 | |
| miR-196a2 rs11614913 | 1.286 | (0.673–2.458) | 0.446 | |
| miR-146a rs2910164 | 0.755 | (0.305–1.873) | 0.544 | |
| Multivariate Analysis | ATA risk | 14.210 | (4.589–43.999) | 0.000004 |
| HOTAIR rs920778 | 2.811 | (1.275–6.197) | 0.010 | |
| Gene/SNP | Genotype | Controls | PTC Cases | Age and Sex Adjusted OR, (95% CI) | p | ||
|---|---|---|---|---|---|---|---|
| N = 106 | % | N = 102 | % | ||||
| HOTAIR rs920778 | CC | 51 | 48.11 | 40 | 39.22 | 1 | Reference |
| CT | 37 | 34.91 | 27 | 26.47 | 0.956 (0.499–1.832) | 0.892 | |
| TT | 18 | 16.98 | 35 | 34.31 | 2.466 (1.219–4.990) | 0.012 | |
| Additive model | 1.497 (1.063–2.110) | 0.021 | |||||
| Recessive model-mt vs. wt + ht (Ref.) | 2.512 (1.306–4.829) | 0.006 | |||||
| Dominant model-wt vs. ht + mt (Ref.) | 0.684 (0.393–1.190) | 0.179 | |||||
| Over-dominant model-ht vs. wt + mt (Ref.) | 0.693 (0.380–1.261) | 0.230 | |||||
| MIR155HG rs1893650 | TT | 53 | 50.00 | 75 | 73.53 | 1 | Reference |
| TC | 46 | 43.40 | 17 | 16.67 | 0.268 (0.139–0.520) | 0.00009 | |
| CC | 7 | 6.60 | 10 | 9.80 | 1.013 (0.361–2.846) | 0.980 | |
| Additive model | 0.610 (0.392–0.951) | 0.029 | |||||
| Recessive model-mt vs. wt + ht (Ref.) | 1.841 (0.639–5.304) | 0.258 | |||||
| Dominant model-wt vs. ht + mt (Ref.) | 1.921 (0.665–5.546) | 0.228 | |||||
| Over-dominant model-ht vs. wt + mt (Ref.) | 0.268 (0.140–0.513) | 0.00007 | |||||
| TERC rs10936599 | CC | 65 | 61.32 | 68 | 66.67 | 1 | Reference |
| CT | 33 | 31.13 | 21 | 20.59 | 0.586 (0.304–1.129) | 0.110 | |
| TT | 8 | 7.55 | 13 | 12.75 | 1.666 (0.636–4.362) | 0.299 | |
| Additive model | 1.005 (0.670–1.510) | 0.979 | |||||
| Recessive model-mt vs. wt + ht (Ref.) | 1.345 (0.844–2.142) | 0.212 | |||||
| Dominant model-wt vs. ht + mt (Ref.) | 1.234 (0.693–2.197) | 0.476 | |||||
| Over-dominant model-ht vs. wt + mt (Ref.) | 0.584 (0.308–1.108) | 0.100 | |||||
| miR-155 rs767649 | TT | 95 | 89.62 | 86 | 84.31 | 1 | Reference |
| TA | 11 | 10.38 | 16 | 15.69 | 1.655 (0.718–3.813) | 0.237 | |
| Additive model | 1.578 (0.688–3.618) | 0.282 | |||||
| miR-196a2 rs11614913 | CC | 55 | 51.89 | 56 | 54.90 | 1 | Reference |
| CT | 41 | 38.68 | 29 | 28.43 | 0.760 (0.411–1.405) | 0.381 | |
| TT | 10 | 9.43 | 17 | 16.67 | 1.663 (0.687–4.025) | 0.260 | |
| Additive model | 1.115 (0.756–1.645) | 0.582 | |||||
| Recessive model-mt vs. wt + ht (Ref.) | 1.404 (0.923–2.137) | 0.113 | |||||
| Dominant model-wt vs. ht + mt (Ref.) | 1.845 (0.785–4.334) | 0.160 | |||||
| Over-dominant model-ht vs. wt + mt (Ref.) | 0.646 (0.359–1.160) | 0.143 | |||||
| miR-146a rs2910164 | GG | 72 | 67.92 | 72 | 70.59 | 1 | Reference |
| GC | 27 | 25.47 | 20 | 19.61 | 0.817 (0.415–1.609) | 0.558 | |
| CC | 7 | 6.60 | 10 | 9.80 | 1.329 (0.471–3.747) | 0.591 | |
| Additive model | 1.001 (0.650–1.540) | 0.997 | |||||
| Recessive model-mt vs. wt + ht (Ref.) | 1.178 (0.649–2.137) | 0.591 | |||||
| Dominant model-wt vs. ht + mt (Ref.) | 1.391 (0.498–3.881) | 0.529 | |||||
| Over-dominant model-ht vs. wt + mt (Ref.) | 0.664 (0.340–1.297) | 0.231 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karajovic, J.; Kovacevic, B.; Uzelac, B.; Stefik, D.; Jovanovic, B.; Ristic, P.; Cerovic, S.; Supic, G. Association of HOTAIR, MIR155HG, TERC, miR-155, -196a2, and -146a Genes Polymorphisms with Papillary Thyroid Cancer Susceptibility and Prognosis. Cancers 2024, 16, 485. https://doi.org/10.3390/cancers16030485
Karajovic J, Kovacevic B, Uzelac B, Stefik D, Jovanovic B, Ristic P, Cerovic S, Supic G. Association of HOTAIR, MIR155HG, TERC, miR-155, -196a2, and -146a Genes Polymorphisms with Papillary Thyroid Cancer Susceptibility and Prognosis. Cancers. 2024; 16(3):485. https://doi.org/10.3390/cancers16030485
Chicago/Turabian StyleKarajovic, Jelena, Bozidar Kovacevic, Bojana Uzelac, Debora Stefik, Bojana Jovanovic, Petar Ristic, Snezana Cerovic, and Gordana Supic. 2024. "Association of HOTAIR, MIR155HG, TERC, miR-155, -196a2, and -146a Genes Polymorphisms with Papillary Thyroid Cancer Susceptibility and Prognosis" Cancers 16, no. 3: 485. https://doi.org/10.3390/cancers16030485
APA StyleKarajovic, J., Kovacevic, B., Uzelac, B., Stefik, D., Jovanovic, B., Ristic, P., Cerovic, S., & Supic, G. (2024). Association of HOTAIR, MIR155HG, TERC, miR-155, -196a2, and -146a Genes Polymorphisms with Papillary Thyroid Cancer Susceptibility and Prognosis. Cancers, 16(3), 485. https://doi.org/10.3390/cancers16030485






