Ensemble Deep Learning Model to Predict Lymphovascular Invasion in Gastric Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Patients and Tumor Samples
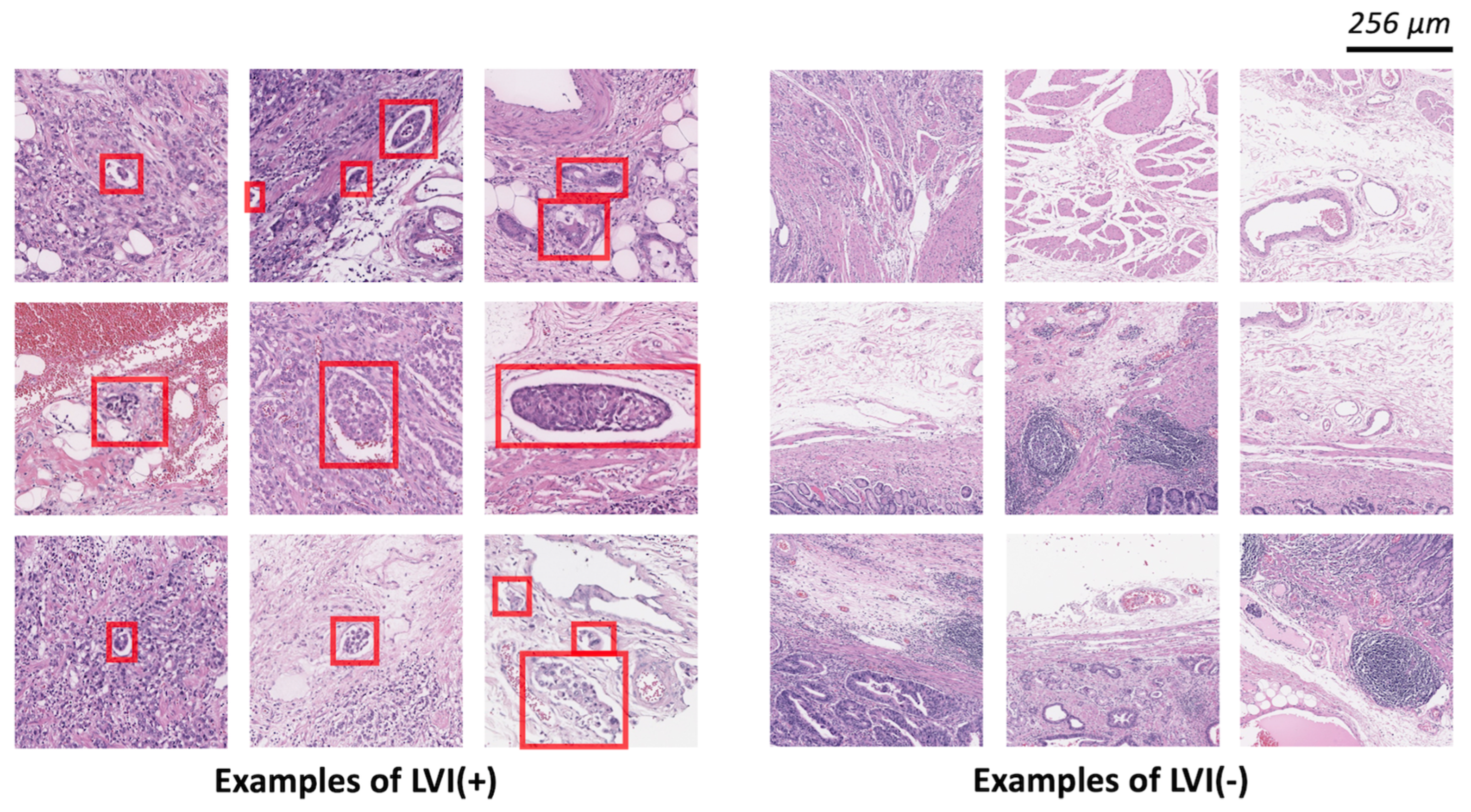
2.2. Datasets
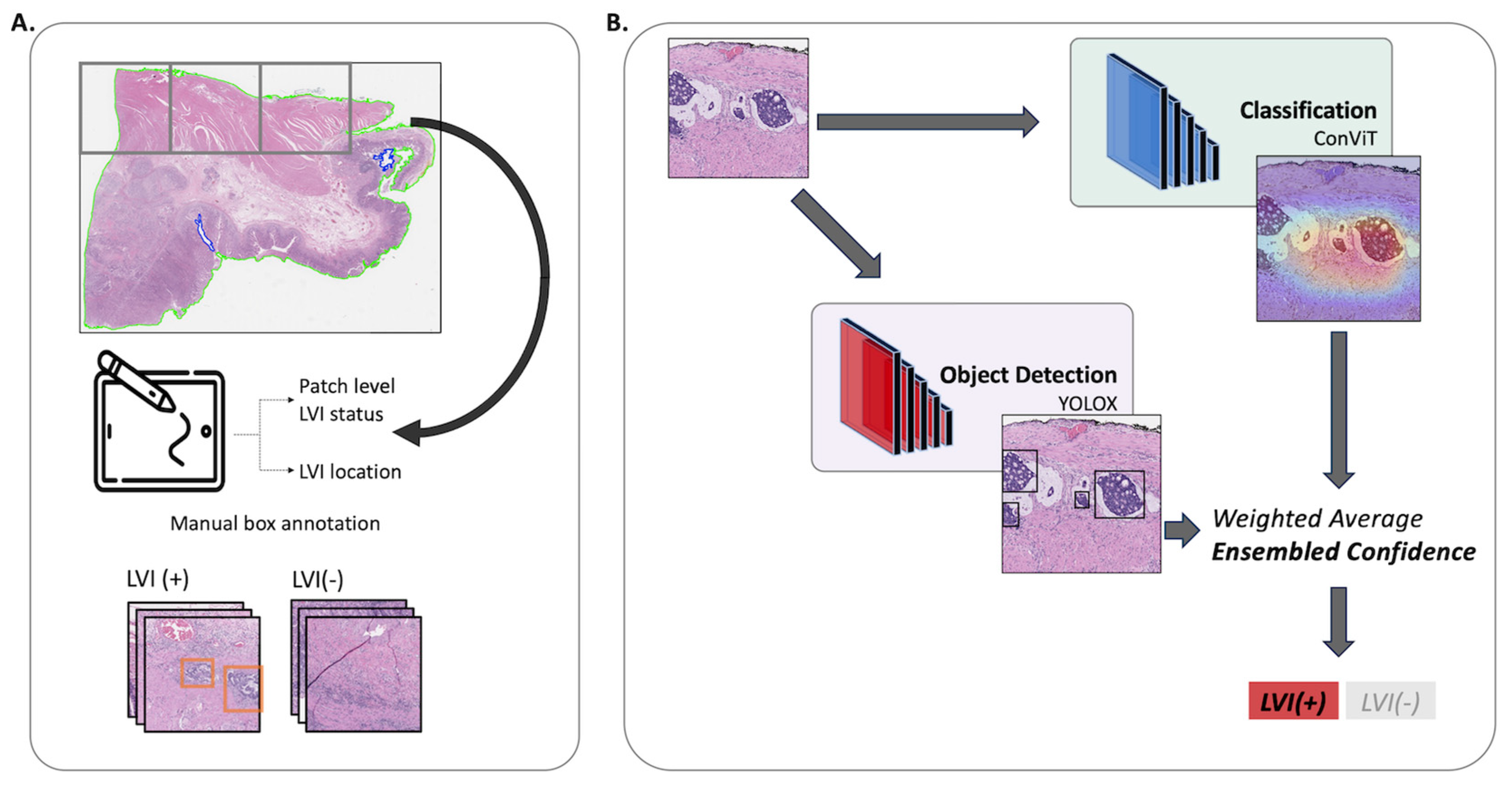
2.3. Model Development
2.4. Classification Models
2.5. Detection Models
2.6. Ensemble Model
2.7. Evaluation Metrics
3. Results
3.1. Patient Characteristics
3.2. Patch-Level Analysis
3.3. Patch-Level Analysis: Classification Models
3.4. Patch-Level Analysis: Detection Models
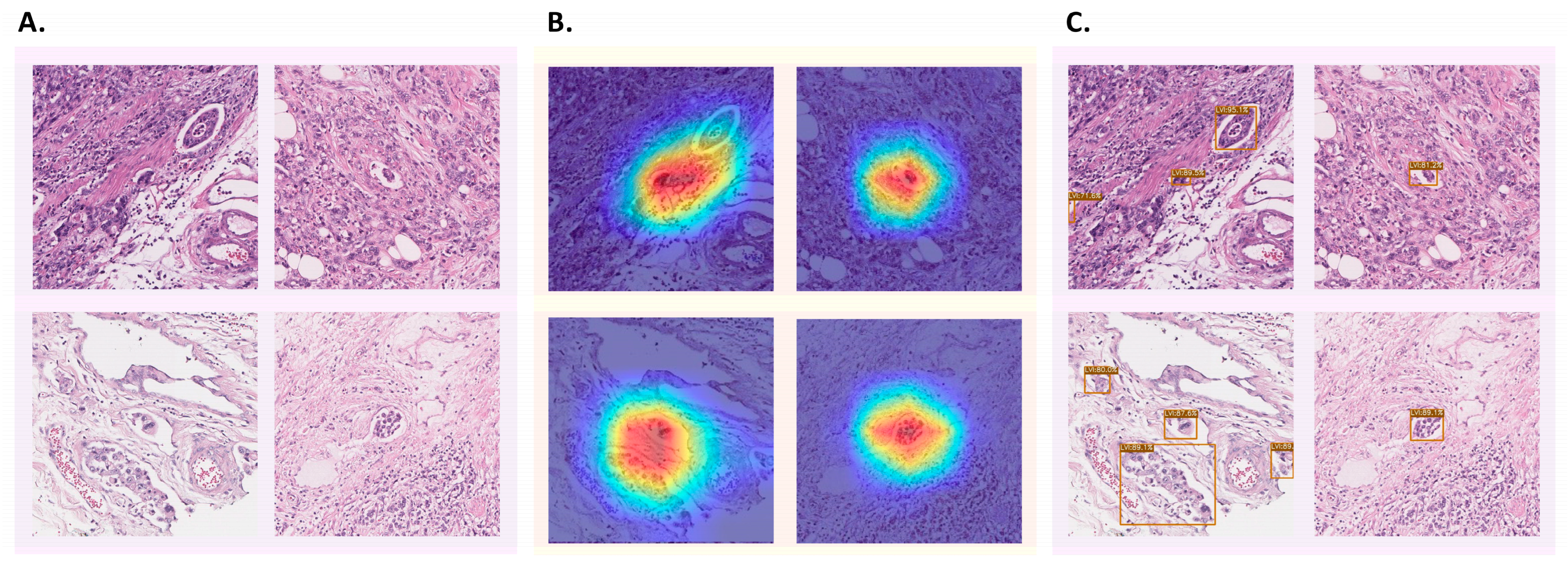
3.5. Patch-Level Analysis: Ensemble Model
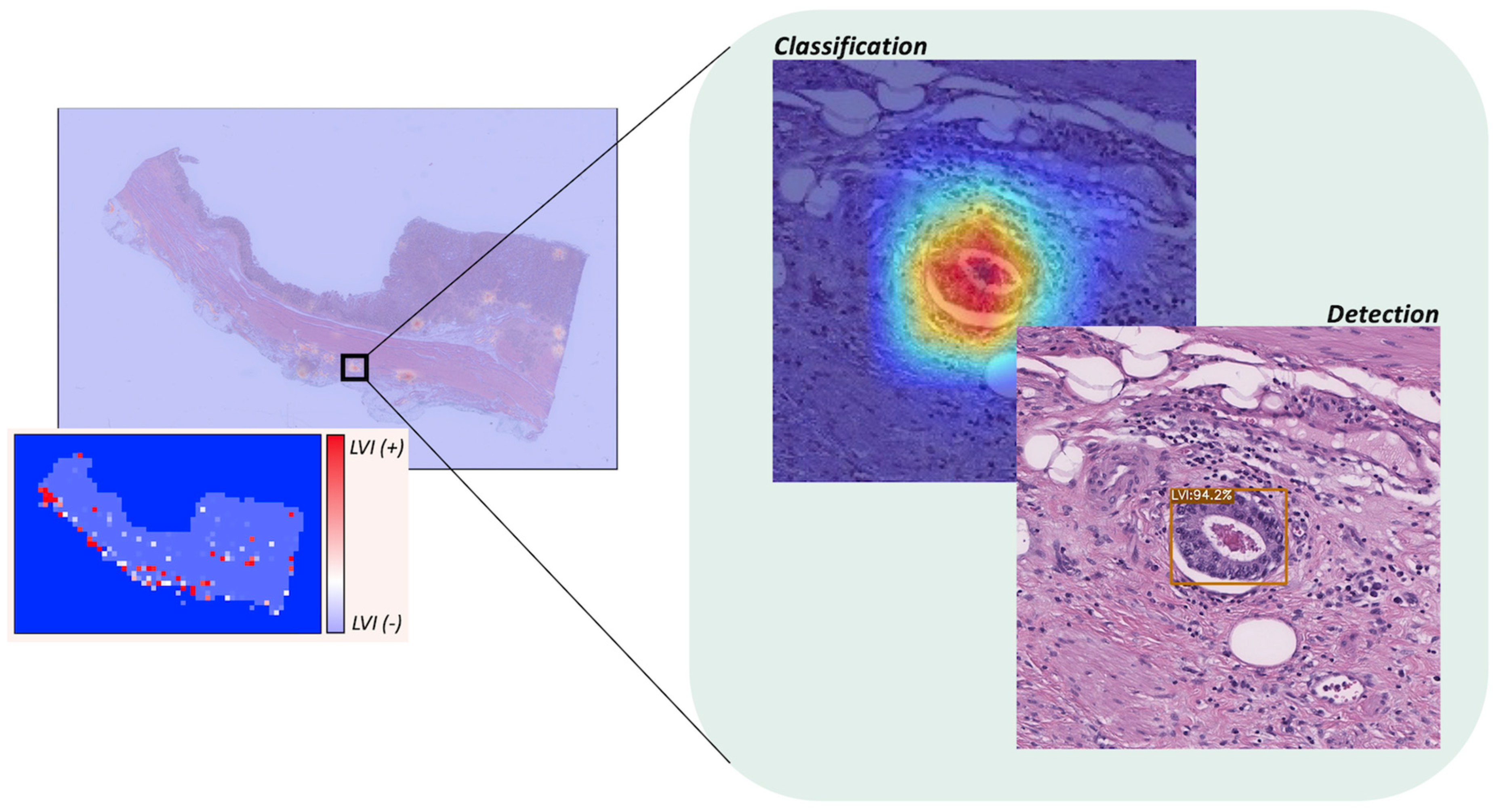
3.6. WSI-Level Analysis
3.7. External Validation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hong, S.; Won, Y.J.; Lee, J.J.; Jung, K.W.; Kong, H.J.; Im, J.S.; Seo, H.G.; The Community of Population-Based Regional Cancer Registries. Cancer statistics in Korea: Incidence, mortality, survival, and prevalence in 2018. Cancer Res. Treat. 2021, 53, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Lordick, F.; Carneiro, F.; Cascinu, S.; Fleitas, T.; Haustermans, K.; Piessen, G.; Vogel, A.; Smyth, E.C. Gastric cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 1005–1020. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M. Global Cancer Observatory: Cancer Today. International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today (accessed on 14 June 2023).
- Takada, K.; Yoshida, M.; Aizawa, D.; Sato, J.; Ono, H.; Sugino, T. Lymphovascular invasion in early gastric cancer: Impact of ancillary D2-40 and elastin staining on interobserver agreement. Histopathology 2020, 76, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Nitti, D.; Marchet, A.; Olivieri, M.; Ambros, A.; Mencarelli, R.; Belluco, C.; Lise, M. Ratio between metastatic and examined lymph nodes is an independent prognostic factor after D2 resection for gastric cancer: Analysis of a large European monoinstitutional experience. Ann. Surg. Oncol. 2003, 10, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, M.; Oda, I.; Taniguchi, H.; Suzuki, H.; Morita, S.; Fukagawa, T.; Sekine, S.; Kushima, R.; Katai, H. Risk stratification and predictive risk-scoring model for lymph node metastasis in early gastric cancer. J. Gastroenterol. 2016, 51, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Gotoda, T.; Yanagisawa, A.; Sasako, M.; Ono, H.; Nakanishi, Y.; Shimoda, T.; Kato, Y. Incidence of lymph node metastasis from early gastric cancer: Estimation with a large number of cases at two large centers. Gastric Cancer 2000, 3, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, H.; Koumori, K.; Watanabe, H.; Kano, K.; Shimoda, Y.; Aoyama, T.; Yamada, T.; Hiroshi, T.; Yamamoto, N.; Cho, H.; et al. The clinical significance of lymphovascular invasion in gastric cancer. In Vivo 2020, 34, 1533–1539. [Google Scholar] [CrossRef]
- Song, Y.J.; Shin, S.H.; Cho, J.S.; Park, M.H.; Yoon, J.H.; Jegal, Y.J. The role of lymphovascular invasion as a prognostic factor in patients with lymph node-positive operable invasive breast cancer. J. Breast Cancer 2011, 14, 198–203. [Google Scholar] [CrossRef]
- Talamonti, M.S.; Kim, S.P.; Yao, K.A.; Wayne, J.D.; Feinglass, J.; Bennett, C.L.; Rao, S. Surgical outcomes of patients with gastric carcinoma: The importance of primary tumor location and microvessel invasion. Surgery 2003, 134, 720–727, Discussion 727–729. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Kim, Y.I.; Kook, M.C.; Choi, J.E.; Lee, J.Y.; Kim, C.G.; Eom, B.W.; Yoon, H.M.; Ryu, K.W.; Kim, Y.W.; Choi, I.J. Evaluation of submucosal or lymphovascular invasion detection rates in early gastric cancer based on pathology section interval. J. Gastric Cancer 2020, 20, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Kwee, R.M.; Kwee, T.C. Predicting lymph node status in early gastric cancer. Gastric Cancer 2008, 11, 134–148. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.H.; Park, J.C.; Lee, Y.C.; Noh, S.H.; Kim, H. Lymphovascular invasion is an important predictor of lymph node metastasis in endoscopically resected early gastric cancers. Oncol. Rep. 2011, 25, 1589–1595. [Google Scholar] [CrossRef]
- Lee, S.Y.; Yoshida, N.; Dohi, O.; Lee, S.P.; Ichikawa, D.; Kim, J.H.; Sung, I.K.; Park, H.S.; Otsuji, E.; Itoh, Y.; et al. Differences in prevalence of lymphovascular invasion among early gastric cancers between Korea and Japan. Gut Liver 2017, 11, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Zaorsky, N.G.; Patil, N.; Freedman, G.M.; Tuluc, M. Differentiating lymphovascular invasion from retraction artifact on histological specimen of breast carcinoma and their implications on prognosis. J. Breast Cancer 2012, 15, 478–480. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, K.W.; Gould, V.E.; Hirschl, S.; Imbriglia, J.E.; Patchefsky, A.S.; Penner, D.W.; Pickren, J.; Schwartz, I.S.; Wheeler, J.E.; Barnes, J.M.; et al. Interobserver variation in the identification of breast carcinoma in intramammary lymphatics. Hum. Pathol. 1982, 13, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Gresta, L.T.; Rodrigues-Junior, I.A.; de Castro, L.P.; Cassali, G.D.; Cabral, M.M. Assessment of vascular invasion in gastric cancer: A comparative study. World J. Gastroenterol. 2013, 19, 3761–3769. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Sirinukunwattana, K.; Khalid Alham, N.; Browning, L.; Colling, R.; Protheroe, A.; Protheroe, E.; Jones, S.; Aberdeen, A.; Rittscher, J.; et al. The potential of artificial intelligence to detect lymphovascular invasion in testicular cancer. Cancers 2021, 13, 1325. [Google Scholar] [CrossRef]
- Yonemura, Y.; Endou, Y.; Tabachi, K.; Kawamura, T.; Yun, H.Y.; Kameya, T.; Hayashi, I.; Bandou, E.; Sasaki, T.; Miura, M. Evaluation of lymphatic invasion in primary gastric cancer by a new monoclonal antibody, D2-40. Hum. Pathol. 2006, 37, 1193–1199. [Google Scholar] [CrossRef]
- Arigami, T.; Natsugoe, S.; Uenosono, Y.; Arima, H.; Mataki, Y.; Ehi, K.; Yanagida, S.; Ishigami, S.; Hokita, S.; Aikou, T. Lymphatic invasion using D2-40 monoclonal antibody and its relationship to lymph node micrometastasis in pN0 gastric cancer. Br. J. Cancer 2005, 93, 688–693. [Google Scholar] [CrossRef]
- Sako, A.; Kitayama, J.; Ishikawa, M.; Yamashita, H.; Nagawa, H. Impact of immunohistochemically identified lymphatic invasion on nodal metastasis in early gastric cancer. Gastric Cancer 2006, 9, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Araki, I.; Hosoda, K.; Yamashita, K.; Katada, N.; Sakuramoto, S.; Moriya, H.; Mieno, H.; Ema, A.; Kikuchi, S.; Mikami, T.; et al. Prognostic impact of venous invasion in stage IB node-negative gastric cancer. Gastric Cancer 2015, 18, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.I.; Lewin, D.N.; Wang, H.L.; Lauwers, G.Y.; Srivastava, A.; Shyr, Y.; Shakhtour, B.; Revetta, F.; Washington, M.K. Lymphovascular invasion in colorectal cancer: An interobserver variability study. Am. J. Surg. Pathol. 2008, 32, 1816–1821. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, R.; Messenger, D.E.; Riddell, R.H.; Pollett, A.; Cook, M.; Al-Haddad, S.; Streutker, C.J.; Divaris, D.X.; Pandit, R.; Newell, K.J.; et al. Venous invasion in colorectal cancer impact of an elastin stain on detection and interobserver agreement among gastrointestinal and nongastrointestinal pathologists. Am. J. Surg. Pathol. 2013, 37, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.; Chong, Y.; Jung, C.K.; Kwak, T.Y.; Lee, J.Y.; Park, J.; Rho, M.J.; Go, H. Introduction to digital pathology and computer-aided pathology. J. Pathol. Transl. Med. 2020, 54, 125–134. [Google Scholar] [CrossRef]
- Ahmad, Z.; Rahim, S.; Zubair, M.; Abdul-Ghafar, J. Artificial intelligence (AI) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: Present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. A comprehensive review. Diagn. Pathol. 2021, 16, 24. [Google Scholar] [CrossRef]
- Joshi, G.; Jain, A.; Araveeti, S.R.; Adhikari, S.; Garg, H.; Bhandari, M. FDA approved Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices: An updated landscape. medRxiv 2022. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Quiroga-Garza, G.M.; Bien, L.; Heled, R.; Laifenfeld, D.; Linhart, C.; Sandbank, J.; Albrecht Shach, A.; Shalev, V.; Vecsler, M.; et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: A blinded clinical validation and deployment study. Lancet Digit. Health 2020, 2, e407–e416. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Turkki, R.; Byckhov, D.; Lundin, M.; Isola, J.; Nordling, S.; Kovanen, P.E.; Verrill, C.; von Smitten, K.; Joensuu, H.; Lundin, J.; et al. Breast cancer outcome prediction with tumour tissue images and machine learning. Breast Cancer Res. Treat. 2019, 177, 41–52. [Google Scholar] [CrossRef]
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018, 8, 3395. [Google Scholar] [CrossRef]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y.; et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019, 53, 1800986. [Google Scholar] [CrossRef] [PubMed]
- Hinata, M.; Ushiku, T. Detecting immunotherapy-sensitive subtype in gastric cancer using histologic image-based deep learning. Sci. Rep. 2021, 11, 22636. [Google Scholar] [CrossRef] [PubMed]
- Bejnordi, B.E.; Veta, M.; van Diest, P.J.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; Consortium, C. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Steiner, D.F.; MacDonald, R.; Liu, Y.; Truszkowski, P.; Hipp, J.D.; Gammage, C.; Thng, F.; Peng, L.; Stumpe, M.C. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am. J. Surg. Pathol. 2018, 42, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ahn, S.; Kim, H.S.; An, J.; Sim, J. A robust model training strategy using hard negative mining in a weakly labeled dataset for lymphatic invasion in gastric cancer. J. Pathol. Clin. Res. 2024, 10, e355. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- d’Ascoli, S.; Touvron, H.; Leavitt, M.L.; Morcos, A.S.; Biroli, G.; Sagun, L. Convit: Improving vision transformers with soft convolutional inductive biases. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 2286–2296. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLOv3: An incremental improvement. arXiv 2018. [Google Scholar] [CrossRef]
- Ge, Z.; Liu, S.; Wang, F.; Li, Z.; Sun, J. YOLOX: Exceeding YOLO series in 2021. arXiv 2021. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common objects in context. In Computer Vision—ECCV 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer: Cham, Switzerland, 2014; Volume 8693, pp. 740–755. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017. [Google Scholar] [CrossRef]
- Piansaddhayanaon, C.; Santisukwongchote, S.; Shuangshoti, S.; Tao, Q.; Sriswasdi, S.; Chuangsuwanich, E. ReCasNet: Improving consistency within the two-stage mitosis detection framework. Artif. Intell. Med. 2023, 135, 102462. [Google Scholar] [CrossRef]
| Variable | Total (N = 63) |
|---|---|
| Age 1 | 69.6 (10.2) |
| Sex 2 | |
| Male | 46 (73.0%) |
| Female | 17 (27.0%) |
| Lauren Classification 2 | |
| Intestinal | 36 (57.1%) |
| Diffuse | 12 (19.0%) |
| Mixed | 15 (23.8%) |
| Grade 2 | |
| Well differentiated | 3 (4.8%) |
| Moderately differentiated | 31 (49.2%) |
| Poorly differentiated | 29 (46.0%) |
| T Staging 2 | |
| pT1a | 2 (3.2%) |
| pT1b | 17 (27.0%) |
| pT2 | 5 (7.9%) |
| pT3 | 13 (20.6%) |
| pT4a | 23 (36.5%) |
| pT4b | 3 (4.8%) |
| N Staging 2 | |
| pN0 | 10 (18.2%) |
| pN1 | 6 (10.9%) |
| pN2 | 13 (23.6%) |
| pN3a | 8 (14.5%) |
| pN3b | 18 (32.7%) |
| LN Involvement 1 | 12.0 (13.7) |
| Perineural Invasion 2 | |
| Present | 39 (61.9%) |
| Not identified | 24 (38.1%) |
| IHC Expression of C-erb B2 2 | |
| 0 | 36 (57.1%) |
| 1+ | 11 (17.5%) |
| 2+ | 6 (9.5%) |
| 3+ | 7 (11.1%) |
| Not available | 3 (4.8%) |
| # of WSI | # of Positive Per WSI 1 | # of Negative Per WSI 1 | |
|---|---|---|---|
| Train set | 64 | 68.77 (90.04) | 159.23 (87.54) |
| Valid set | 16 | 28.50 (25.61) | 161.12 (60.69) |
| Test set | 20 | 105.3 (91.73) | 201.4 (102.48) |
| Method | Model | AUROC | AUPRC | Accuracy | F1 Score |
|---|---|---|---|---|---|
| Classification | ResNet50 | 0.9762 (0.9726–0.9798) | 0.9593 (0.9447–0.9739) | 0.9319 (0.9254–0.9384) | 0.8992 (0.8895–0.9089) |
| EfficientNetB3 | 0.9731 (0.9693–0.9769) | 0.9551 (0.935–0.9752) | 0.9281 (0.9217–0.9345) | 0.8929 (0.8827–0.9031) | |
| ConViT | 0.9796 (0.9765–0.9827) | 0.9648 (0.9592–0.9704) | 0.9348 (0.9288–0.9408) | 0.9025 (0.8935–0.9115) | |
| Detection | YOLOv3 | 0.9666 (0.9623–0.9709) | 0.9302 (0.9203–0.9401) | 0.927 (0.9196–0.9344) | 0.8977 (0.8868–0.9086) |
| YOLOX | 0.9702 (0.9648–0.9756) | 0.9423 (0.9323–0.9523) | 0.9353 (0.9278–0.9428) | 0.9064 (0.8962–0.9166) | |
| Ensemble | 0.988 (0.9852–0.9908) | 0.9769 (0.9717–0.9821) | 0.9514 (0.9459–0.9569) | 0.928 (0.9198–0.9362) | |
| Method | True Negative Rate 1 | False Positive Rate 1 | False Negative Rate 1 | True Positive Rate 1 |
|---|---|---|---|---|
| ConViT | 96.63 (0.03) | 3.37 (0.03) | 11.88 (0.09) | 88.12 (0.09) |
| YOLOX | 94.99 (0.03) | 5.01 (0.03) | 10.88 (0.12) | 89.12 (0.12) |
| Ensemble | 97.56 (0.02) | 2.44 (0.02) | 10.21 (0.07) | 89.79 (0.07) |
| Method | Model | AUROC | AUPRC | Accuracy | F1 Score |
|---|---|---|---|---|---|
| Classification | ConViT | 0.9184 (0.8975–0.9393) | 0.869 (0.8338–0.9041) | 0.8674 (0.8465–0.8883) | 0.7896 (0.7543–0.8248) |
| Detection | YOLOX | 0.8915 (0.8638–0.9192) | 0.8319 (0.7876–0.8763) | 0.8592 (0.8364–0.882) | 0.7934 (0.7577–0.8291) |
| Ensemble | 0.9438 (0.9258–0.9619) | 0.9132 (0.8875–0.939) | 0.8983 (0.879–0.9175) | 0.8358 (0.8035–0.8681) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Cha, S.; Kim, J.; Kim, J.J.; Kim, N.; Jae Gal, S.G.; Kim, J.H.; Lee, J.H.; Choi, Y.-D.; Kang, S.-R.; et al. Ensemble Deep Learning Model to Predict Lymphovascular Invasion in Gastric Cancer. Cancers 2024, 16, 430. https://doi.org/10.3390/cancers16020430
Lee J, Cha S, Kim J, Kim JJ, Kim N, Jae Gal SG, Kim JH, Lee JH, Choi Y-D, Kang S-R, et al. Ensemble Deep Learning Model to Predict Lymphovascular Invasion in Gastric Cancer. Cancers. 2024; 16(2):430. https://doi.org/10.3390/cancers16020430
Chicago/Turabian StyleLee, Jonghyun, Seunghyun Cha, Jiwon Kim, Jung Joo Kim, Namkug Kim, Seong Gyu Jae Gal, Ju Han Kim, Jeong Hoon Lee, Yoo-Duk Choi, Sae-Ryung Kang, and et al. 2024. "Ensemble Deep Learning Model to Predict Lymphovascular Invasion in Gastric Cancer" Cancers 16, no. 2: 430. https://doi.org/10.3390/cancers16020430
APA StyleLee, J., Cha, S., Kim, J., Kim, J. J., Kim, N., Jae Gal, S. G., Kim, J. H., Lee, J. H., Choi, Y.-D., Kang, S.-R., Song, G.-Y., Yang, D.-H., Lee, J.-H., Lee, K.-H., Ahn, S., Moon, K. M., & Noh, M.-G. (2024). Ensemble Deep Learning Model to Predict Lymphovascular Invasion in Gastric Cancer. Cancers, 16(2), 430. https://doi.org/10.3390/cancers16020430







