Intramedullary Spinal Cord Tumors: Whole-Genome Sequencing to Assist Management and Prognosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Statistical Analysis
4. Results
4.1. Characteristics of Patients and Tumors
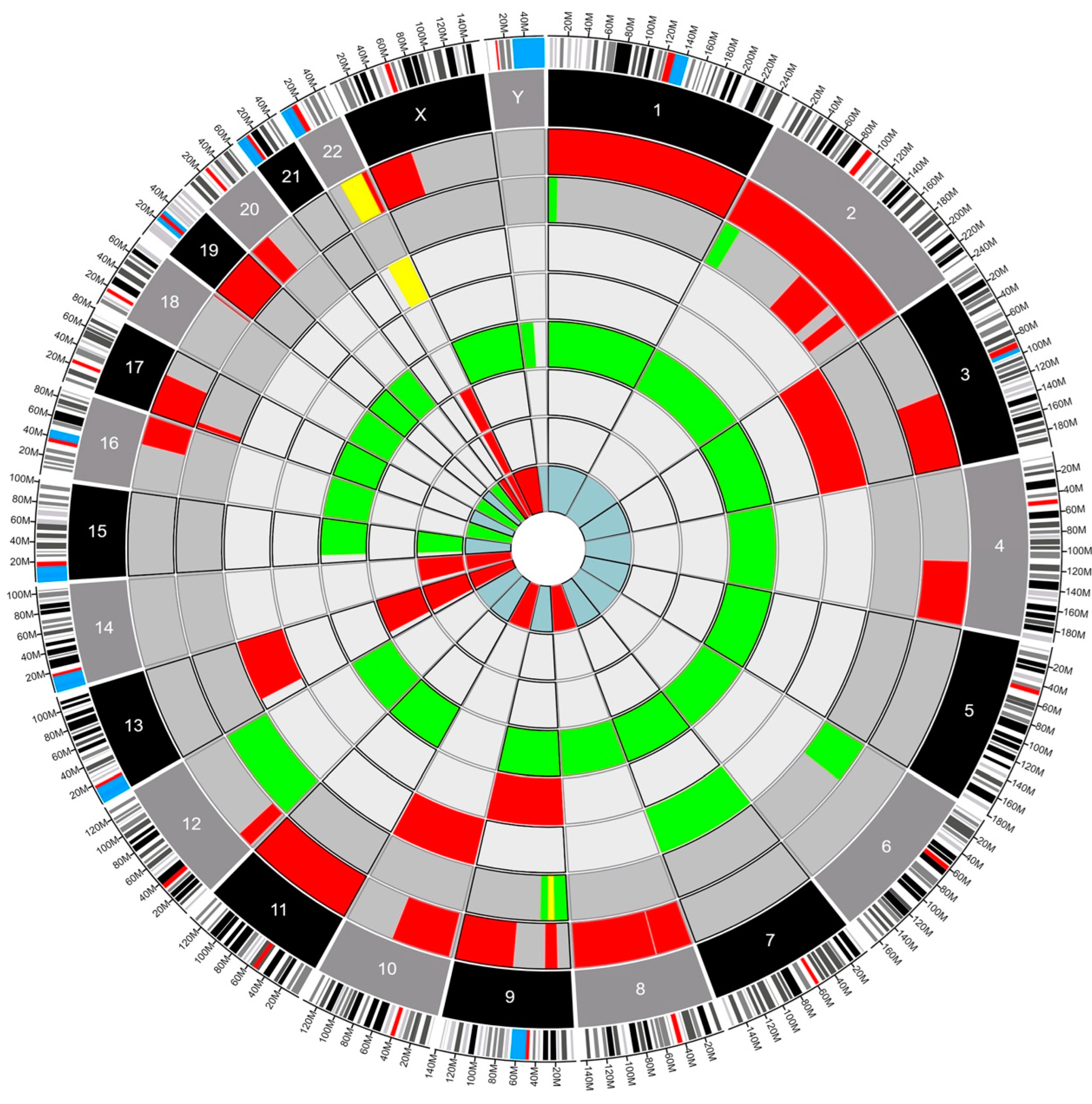
4.2. Genetic Analysis
- Deletion
- Gain
- Loss of Heterozygosity
- Glioblastoma
- Ependymoma
- Myxopapillary Ependymoma
5. Discussion
Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Das, J.M.; Hoang, S.; Mesfin, F.B. Intramedullary Spinal Cord Tumors. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK442031/ (accessed on 24 April 2023).
- Samartzis, D.; Gillis, C.C.; Shih, P.; O’Toole, J.E.; Fessler, R.G. Intramedullary Spinal Cord Tumors: Part I-Epidemiology, Pathophysiology, and Diagnosis. Glob. Spine J. 2015, 5, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, M.C.; Tredway, T.L. Adult primary intradural spinal cord tumors: A review. Curr. Neurol. Neurosci. Rep. 2011, 11, 320–328. [Google Scholar] [CrossRef]
- Knafo, S.; Aghakhani, N.; David, P.; Parker, F. Management of intramedullary spinal cord tumors: A single-center experience of 247 patients. Rev. Neurol. 2021, 177, 508–514. [Google Scholar] [CrossRef]
- Baig Mirza, A.; Gebreyohanes, A.; Knight, J.; Bartram, J.; Vastani, A.; Kalaitzoglou, D.; Lavrador, J.P.; Kailaya-Vasan, A.; Maratos, E.; Bell, D.; et al. Prognostic factors for surgically managed intramedullary spinal cord tumours: A single-centre case series. Acta Neurochir. 2022, 164, 2605–2622. [Google Scholar] [CrossRef]
- WHO. Classification of Tumours Editorial Board. In Central Nervous System Tumours: WHO Classification of Tumours; WHO Classification of Tumours; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Zadnik, P.L.; Gokaslan, Z.L.; Burger, P.C.; Bettegowda, C. Spinal cord tumours: Advances in genetics and their implications for treatment. Nat. Rev. Neurol. 2013, 9, 257–266. [Google Scholar] [CrossRef]
- Crespo, I.; Tao, H.; Nieto, A.B.; Rebelo, O.; Domingues, P.; Vital, A.L.; Patino, M.D.C.; Barbosa, M.; Lopes, M.C.; Oliveira, C.R.; et al. Amplified and homozygously deleted genes in glioblastoma: Impact on gene expression levels. PLoS ONE 2012, 7, e46088. [Google Scholar] [CrossRef]
- Rao, S.K.; Edwards, J.; Joshi, A.D.; Siu, I.M.; Riggins, G.J. A survey of glioblastoma genomic amplifications and deletions. J. Neurooncol. 2010, 96, 169–179. [Google Scholar] [CrossRef]
- Cowell, J.K.; Lo, K.C. Application of oligonucleotides arrays for coincident comparative genomic hybridization, ploidy status and loss of heterozygosity studies in human cancers. Methods Mol. Biol. 2009, 556, 47–65. [Google Scholar] [PubMed]
- Yin, D.; Ogawa, S.; Kawamata, N.; Tunici, P.; Finocchiaro, G.; Eoli, M.; Ruckert, C.; Huynh, T.; Liu, G.; Kato, M.; et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol. Cancer Res. 2009, 7, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Pajtler, K.W.; Witt, H.; Sill, M.; Jones, D.T.; Hovestadt, V.; Kratochwil, F.; Wani, K.; Tatevossian, R.; Punchihewa, C.; Johann, P.; et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell. 2015, 27, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Nishizaki, T.; Ozaki, S.; Harada, K.; Ito, H.; Arai, H.; Beppu, T.; Sasaki, K. Investigation of genetic alterations associated with the grade of astrocytic tumor by comparative genomic hybridization. Genes Chromosomes Cancer 1998, 21, 340–346. [Google Scholar] [CrossRef]
- Ciriello, G.; Miller, M.L.; Aksoy, B.A.; Senbabaoglu, Y.; Schultz, N.; Sander, C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013, 45, 1127–1133. [Google Scholar] [CrossRef]
- Van Loo, P.; Nordgard, S.H.; Lingjærde, O.C.; Russnes, H.G.; Rye, I.H.; Sun, W.; Weigman, V.J.; Marynen, P.; Zetterberg, A.; Naume, B.; et al. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. USA 2010, 107, 16910–16915. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Ciani, Y.; Fedrizzi, T.; Prandi, D.; Lorenzin, F.; Locallo, A.; Gasperini, P.; Franceschini, G.M.; Benelli, M.; Elemento, O.; Fava, L.L.; et al. Allele-specific genomic data elucidate the role of somatic gain and copy-number neutral loss of heterozygosity in cancer. Cell Syst. 2022, 13, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sjöblom, T. Targeting Loss of Heterozygosity: A Novel Paradigm for Cancer Therapy. Pharmaceuticals 2021, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Raffeld, M.; Abdullaev, Z.; Pack, S.D.; Xi, L.; Nagaraj, S.; Briceno, N.; Vera, E.; Pittaluga, S.; Lopes Abath Neto, O.; Quezado, M.; et al. High level MYCN amplification and distinct methylation signature define an aggressive subtype of spinal cord ependymoma. Acta Neuropathol. Commun. 2020, 8, 101. [Google Scholar] [CrossRef]
- Buchwald, Z.S.; Tian, S.; Rossi, M.; Smith, G.H.; Switchenko, J.; Hauenstein, J.E.; Moreno, C.S.; Press, R.H.; Prabhu, R.S.; Zhong, J.; et al. Genomic copy number variation correlates with survival outcomes in WHO grade IV glioma. Sci. Rep. 2020, 10, 7355. [Google Scholar] [CrossRef]
- Claus, E.B.; Walsh, K.M.; Wiencke, J.K.; Molinaro, A.M.; Wiemels, J.L.; Schildkraut, J.M.; Bondy, M.L.; Berger, M.; Jenkins, R.; Wrensch, M. Survival and low-grade glioma: The emergence of genetic information. Neurosurg. Focus FOC 2015, 38, E6. Available online: https://thejns.org/focus/view/journals/neurosurg-focus/38/1/article-pE6.xml (accessed on 28 June 2022). [CrossRef]
- Hawkins, C.; Walker, E.; Mohamed, N.; Zhang, C.; Jacob, K.; Shirinian, M.; Alon, N.; Kahn, D.; Fried, I.; Scheinemann, K.; et al. BRAF-KIAA1549 fusion predicts better clinical outcome in pediatric low-grade astrocytoma. Clin. Cancer Res. 2011, 17, 4790–4798. [Google Scholar] [CrossRef]
- Horbinski, C. To BRAF or not to BRAF: Is that even a question anymore? J. Neuropathol. Exp. Neurol. 2013, 72, 2–7. [Google Scholar] [CrossRef]
- Lebrun, L.; Meléndez, B.; Blanchard, O.; De Nève, N.; Van Campenhout, C.; Lelotte, J.; Balériaux, D.; Riva, M.; Brotchi, J.; Bruneau, M.; et al. Clinical, radiological, and molecular characterization of intramedullary astrocytomas. Acta Neuropathol. Commun. 2020, 8, 128. [Google Scholar] [CrossRef]
- Iwata, K.; Nakagawa, H.; Hashizume, Y. Significance of MIB-1, PCNA indices, and p53 protein over-expression in intramedullary tumors of the spinal cord. Noshuyo Byori. 1996, 13, 73–78. [Google Scholar] [PubMed]
- Strik, H.M.; Effenberger, O.; Schäfer, O.; Risch, U.; Wickboldt, J.; Meyermann, R. A case of spinal glioblastoma multiforme: Immunohistochemical study and review of the literature. J. Neurooncol. 2000, 50, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Alvi, M.A.; Ida, C.M.; Paolini, M.A.; Kerezoudis, P.; Meyer, J.; Fritcher, E.G.B.; Goncalves, S.; Meyer, F.B.; Bydon, M.; Raghunathan, A. Spinal cord high-grade infiltrating gliomas in adults: Clinico-pathological and molecular evaluation. Mod. Pathol. 2019, 32, 1236–1243. [Google Scholar] [CrossRef]
- Fujisawa, H.; Reis, R.M.; Nakamura, M.; Colella, S.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Loss of heterozygosity on chromosome 10 is more extensive in primary (de novo) than in secondary glioblastomas. Lab. Investig. 2000, 80, 65–72. [Google Scholar] [CrossRef]
- Nakamura, M.; Yang, F.; Fujisawa, H.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Loss of heterozygosity on chromosome 19 in secondary glioblastomas. J. Neuropathol. Exp. Neurol. 2000, 59, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.G.; Sun, Q.F.; Tirakotai, W.; Zhao, W.G.; Shen, J.K.; Luo, Q.Z.; Bertalanffy, H. Loss of heterozygosity on chromosome 22 in sporadic schwannoma and its relation to the proliferation of tumor cells. Chin. Med. J. 2005, 118, 1517–1524. [Google Scholar]
- Arslantas, A.L.İ.; Artan, S.; Öner, Ü.; Durmaz, R.; Müslümanoglu, H.; Atasoy, M.A.; Basaran, N.; Tel, E. Detection of chromosomal imbalances in spinal meningiomas by comparative genomic hybridization. Neurol. Med. Chir. 2003, 43, 12–18; discussion 19. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, L.; Bizet, M.; Melendez, B.; Alexiou, B.; Absil, L.; Van Campenhout, C.; D’Haene, N.; Rorive, S.; Fuks, F.; Decaestecker, C.; et al. Analyses of DNA Methylation Profiling in the Diagnosis of Intramedullary Astrocytomas. J. Neuropathol. Exp. Neurol. 2021, 80, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Sequist, L.V.; Heist, R.S.; Shaw, A.T.; Fidias, P.; Rosovsky, R.; Temel, J.S.; Lennes, I.T.; Digumarthy, S.; Waltman, B.A.; Bast, E.; et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann. Oncol. 2011, 22, 2616–2624. [Google Scholar] [CrossRef] [PubMed]
- Hussain, I.; Parker, W.E.; Barzilai, O.; Bilsky, M.H. Surgical Management of Intramedullary Spinal Cord Tumors. Neurosurg. Clin. N. Am. 2020, 31, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Iyer, R.R.; Azad, T.D.; Wang, Q.; Garzon-Muvdi, T.; Wang, J.; Liu, A.; Burger, P.; Eberhart, C.; Rodriguez, F.J.; et al. Genomic Landscape of Intramedullary Spinal Cord Gliomas. Sci. Rep. 2019, 9, 18722. [Google Scholar] [CrossRef] [PubMed]
| Variable | Level | n (%) = 61 |
|---|---|---|
| Sex | Female | 35 (57.4) |
| Male | 26 (42.6) | |
| Age at surgery | Mean | 47 |
| Median | 46 | |
| Minimum | 21 | |
| Maximum | 76 | |
| Std Dev | 15 | |
| Hospital days | Mean | 8 |
| Std Dev | 4.47 | |
| Tumor location | Cervical | 37 (60.7) |
| Cervicothoracic | 1 (1.6) | |
| Lumbar | 5 (8.2) | |
| Thoracic | 17 (27.9) | |
| Thoracolumbar | 1 (1.6) | |
| Resection % | Biopsy | 6 (9.8) |
| 100 | 25 (41.0) | |
| 50–78 | 4 (6.6) | |
| 78–99 | 20 (32.8) | |
| <50 | 6 (9.8) | |
| Outcomes | Mortality | 1 (1.6) |
| Morbidity | 19 (31.2) | |
| KPS | Mean | 77 |
| Std Dev | 14 | |
| Post Op KPS | Mean | 71 |
| Std Dev | 16 | |
| FU Total Days | Mean | 1287 |
| Median | 721 | |
| Minimum | 4 | |
| Maximum | 6819 | |
| Std Dev | 1600 |
| Variable | Level | n (%) = 61 |
|---|---|---|
| Pathology | Ependymoma | 24 (39.3) |
| Hemangioblastoma | 14 (23.0) | |
| Myxopapillary Ependymoma | 5 (8.2) | |
| Low grade astrocytoma | 4 (6.6) | |
| Anaplastic Astrocytoma | 2 (3.3) | |
| Glioblastoma | 2 (3.3) | |
| Lipoma | 2 (3.3) | |
| Mature Teratoma | 2 (3.3) | |
| Anaplastic Ependymoma | 1 (1.6) | |
| B cell lymphoma | 1 (1.6) | |
| Benign Neuroepithelial Cyst | 1 (1.6) | |
| Cavernous Hemangioma | 1 (1.6) | |
| Metastatic Carcinoma | 1 (1.6) | |
| Schwannoma | 1 (1.6) |
| PFS (Mths) | ||||
|---|---|---|---|---|
| Covariate | Level | N | Hazard Ratio (95% CI) | p Value |
| Gain | No | 3 | 0.44 (0.01–5.49) | 0.343 |
| Yes | 5 | - | - | |
| Loss | No | 1 | 1.09 (0.01–13.45) | 0.545 |
| Yes | 7 | - | - | |
| Copy Neutral Loss of Heterozygosity | No | 5 | 0.05 (0.01–0.69) | 0.008 |
| Yes | 3 | - | - | |
| p53 status. | Not Over-expressed | 5 | 0.29 (0.02–3.58) | 0.351 |
| Over-expressed | 2 | - | - | |
| BRAFV600E | Negative | 9 | 0.92 (0.07–126.63) | 0.545 |
| Positive | 1 | - | - | |
| KRAS results | Negative | 6 | 0.11 (0.00–2.08) | 0.083 |
| Positive | 1 | - | - | |
| Group | ||||
|---|---|---|---|---|
| Covariate | Level | Not Progressed | Progressed | p Value * |
| Loss Count | ≤3 | 4 | 0 | 0.143 |
| >3 | 1 | 3 | ||
| Copy Neutral | No | 5 | 0 | 0.0179 |
| Yes | 0 | 3 | ||
| Total | ≤8 | 3 | 0 | 0.196 |
| >8 | 2 | 3 | ||
| p53 | Not Over-expressed | 4 | 1 | 0.143 |
| Over-expressed | 0 | 2 | ||
| ATRX | Not Deleted | 3 | 0 | 0.1 |
| Deleted | 0 | 2 | ||
| Gain Mutation | No | 2 | 1 | 1 |
| Yes | 3 | 2 | ||
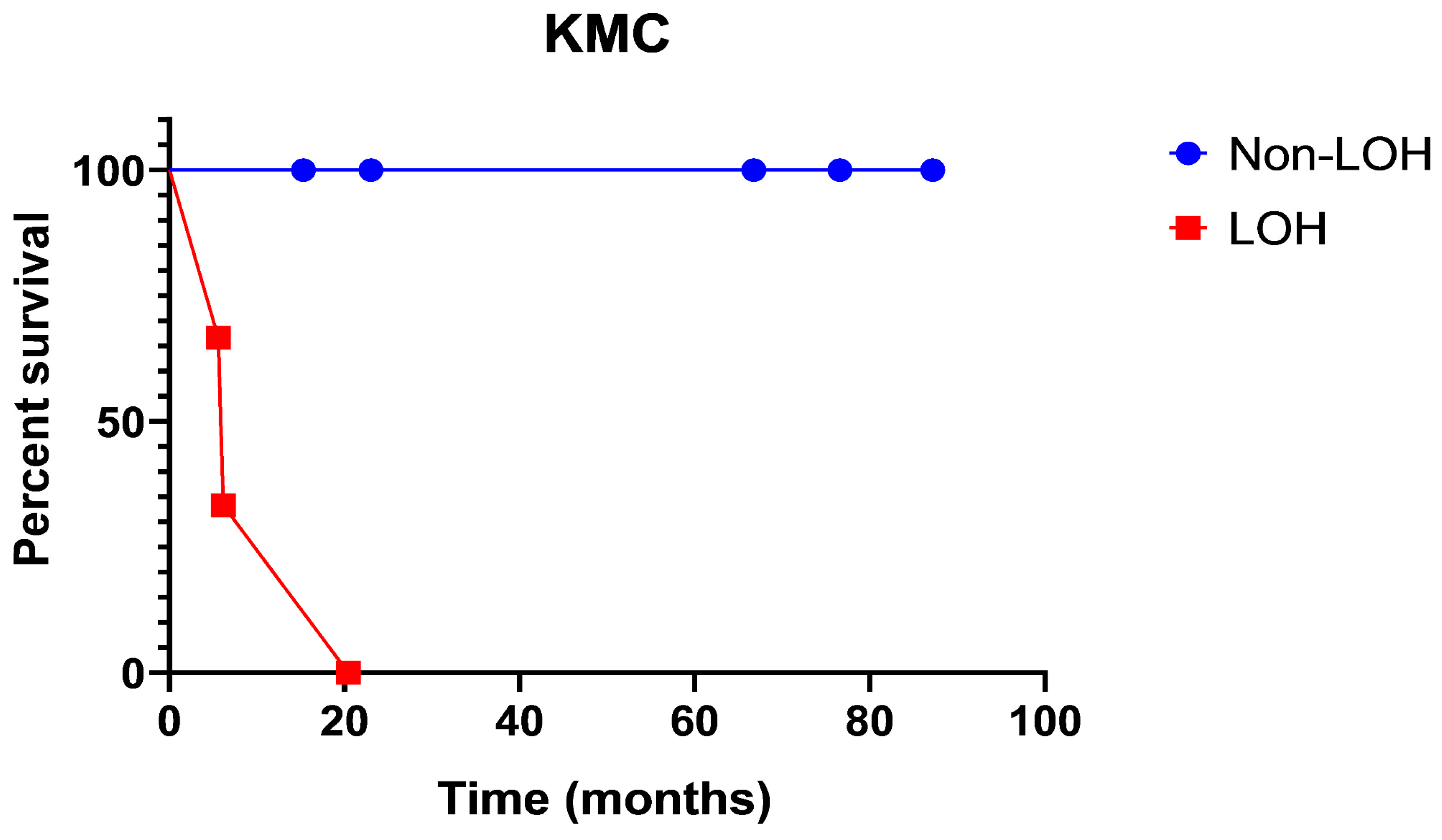
| Comparison of Survival Curves | ||
| Log-rank (Mantel–Cox) test | ||
| Chi square | 7.647 | |
| df | 1 | |
| p value | 0.0057 | |
| p value summary | ** | |
| Are the survival curves sig different? | Yes | |
| Gehan–Breslow–Wilcoxon test | ||
| Chi square | 6.759 | |
| df | 1 | |
| p value | 0.0093 | |
| p value summary | ** | |
| Are the survival curves sig different? | Yes | |
| Median survival | ||
| Dataset-A | Undefined | |
| Dataset-B | 6.143 | |
| Hazard Ratio (Mantel-Haenszel) | A/B | B/A |
| Ratio (and its reciprocal) | 0.02802 | 35.68 |
| 95% Cl of ratio | 0.002224 to 0.3531 | 2.832 to 449.6 |
| Hazard Ratio (logrank) | A/B | B/A |
| Ratio (and its reciprocal) | 0.000 | |
| 95% Cl of ratio | −1.000 to −1.000 | −1.000 to −1.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayol del Valle, M.; Morales, B.; Philbrick, B.; Adeagbo, S.; Goyal, S.; Newman, S.; Frontera, N.L.; Nduom, E.; Olson, J.; Neill, S.; et al. Intramedullary Spinal Cord Tumors: Whole-Genome Sequencing to Assist Management and Prognosis. Cancers 2024, 16, 404. https://doi.org/10.3390/cancers16020404
Mayol del Valle M, Morales B, Philbrick B, Adeagbo S, Goyal S, Newman S, Frontera NL, Nduom E, Olson J, Neill S, et al. Intramedullary Spinal Cord Tumors: Whole-Genome Sequencing to Assist Management and Prognosis. Cancers. 2024; 16(2):404. https://doi.org/10.3390/cancers16020404
Chicago/Turabian StyleMayol del Valle, Miguel, Bryan Morales, Brandon Philbrick, Segun Adeagbo, Subir Goyal, Sarah Newman, Natasha L. Frontera, Edjah Nduom, Jeffrey Olson, Stewart Neill, and et al. 2024. "Intramedullary Spinal Cord Tumors: Whole-Genome Sequencing to Assist Management and Prognosis" Cancers 16, no. 2: 404. https://doi.org/10.3390/cancers16020404
APA StyleMayol del Valle, M., Morales, B., Philbrick, B., Adeagbo, S., Goyal, S., Newman, S., Frontera, N. L., Nduom, E., Olson, J., Neill, S., & Hoang, K. (2024). Intramedullary Spinal Cord Tumors: Whole-Genome Sequencing to Assist Management and Prognosis. Cancers, 16(2), 404. https://doi.org/10.3390/cancers16020404






