Histopathological Spectrum and Molecular Characterization of Liver Tumors in the Setting of Fontan-Associated Liver Disease
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Case Selection
2.2. Clinical Features
2.3. Histology
2.4. DNA Analysis
2.5. RNA Analysis
3. Results
3.1. Cases
3.2. Histological Features
3.3. Molecular Features
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McRae, M.E. Long-term issues after the Fontan procedure. AACN Adv. Crit. Care 2013, 24, 264–282, quiz 283–284. [Google Scholar] [CrossRef]
- Gordon-Walker, T.T.; Bove, K.; Veldtman, G. Fontan-associated liver disease: A review. J. Cardiol. 2019, 74, 223–232. [Google Scholar] [CrossRef]
- Lautt, W.W.; Sheldon, R.D.; Laughlin, M.H.; Rector, R.S.; Jakob, S.M.; Bracht, H.; Porta, F.; Balsiger, B.M.; Brander, L.; Knuesel, R.; et al. Mechanism and role of intrinsic regulation of hepatic arterial blood flow: Hepatic arterial buffer response. Am. J. Physiol. Liver Physiol. 1985, 249 Pt 1, G549–G556. [Google Scholar] [CrossRef] [PubMed]
- Daniels, C.J.; Bradley, E.A.; Landzberg, M.J.; Aboulhosn, J.; Beekman, R.H.; Book, W.; Gurvitz, M.; John, A.; John, B.; Marelli, A.; et al. Fontan-Associated Liver Disease: Proceedings from the American College of Cardiology Stakeholders Meeting, October 1 to 2, 2015, Washington DC. J. Am. Coll. Cardiol. 2017, 70, 3173–3194. [Google Scholar] [CrossRef]
- Munsterman, I.D.; Duijnhouwer, A.L.; Kendall, T.J.; Bronkhorst, C.M.; Ronot, M.; van Wettere, M.; van Dijk, A.P.J.; Drenth, J.P.H.; Tjwa, E.T.T.L.; Nijmegen Fontan Initiative; et al. The clinical spectrum of Fontan-associated liver disease: Results from a prospective multimodality screening cohort. Eur. Heart J. 2019, 40, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, E.M.; Trout, A.T.; Sheridan, R.M.; Veldtman, G.R.; Dillman, J.R. Focal liver lesions following Fontan palliation of single ventricle physiology: A radiology-pathology case series. Congenit. Heart Dis. 2019, 14, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Asrani, S.K.; Asrani, N.S.; Freese, D.K.; Phillips, S.D.; Warnes, C.A.; Heimbach, J.; Kamath, P.S. Congenital heart disease and the liver. Hepatology 2012, 56, 1160–1169. [Google Scholar] [CrossRef]
- Kiesewetter, C.H.; Sheron, N.; Vettukattill, J.J.; Hacking, N.; Stedman, B.; Millward-Sadler, H.; Haw, M.; Cope, R.; Salmon, A.P.; Sivaprakasam, M.C.; et al. Hepatic changes in the failing Fontan circulation. Heart 2007, 93, 579–584. [Google Scholar] [CrossRef]
- Asrani, S.K.; Warnes, C.A.; Kamath, P.S. Hepatocellular Carcinoma after Fontan procedure. N. Engl. J. Med. 2013, 368, 1756–1757. [Google Scholar] [CrossRef]
- Hilscher, M.B.; Wells, M.L.; Venkatesh, S.K.; Cetta, F.; Kamath, P.S. Fontan-associated liver disease. Hepatology 2022, 75, 1300–1321. [Google Scholar] [CrossRef]
- Zhao, C.; Jiang, T.; Hyun Ju, J.; Zhang, S.; Tao, J.; Fu, Y.; Lococo, J.; Dockter, J.; Pawlowski, T.; Bilke, S. TruSight Oncology 500: Enabling Comprehensive Genomic Profiling and Biomarker Reporting with Targeted Sequencing. 2020. Available online: https://biorxiv.org/content/early/2020/10/22/2020.10.21.349100 (accessed on 8 December 2023).
- Sahraeian, S.M.E.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Bani Asadi, N.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Nicorici, D.; Şatalan, M.; Edgren, H.; Kangaspeska, S.; Murumägi, A.; Kallioniemi, O.; Virtanen, S.; Kilkku, O. FusionCatcher—A tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 2014. [Google Scholar] [CrossRef]
- Xu, C.; Xu, Z.; Zhang, Y.; Evert, M.; Calvisi, D.F.; Chen, X. β-Catenin signaling in hepatocellular carcinoma. J. Clin. Investig. 2022, 132, e154515. [Google Scholar] [CrossRef]
- Seitz, T.; Hellerbrand, C. Role of fibroblast growth factor signalling in hepatic fibrosis. Liver Int. 2021, 41, 1201–1215. [Google Scholar] [CrossRef]
- Fearon, A.E.; Slabber, C.F.; Kuklin, A.; Bachofner, M.; Tortola, L.; Pohlmeier, L.; Pantasis, S.; Hornemann, T.; Chen, L.; Kopf, M.; et al. Fibroblast growth factor receptor 3 in hepatocytes protects from toxin-induced liver injury and fibrosis. iScience 2021, 24, 103143. [Google Scholar] [CrossRef] [PubMed]
- Sumazin, P.; Peters, T.L.; Sarabia, S.F.; Kim, H.R.; Urbicain, M.; Hollingsworth, E.F.; Alvarez, K.R.; Perez, C.R.; Pozza, A.; Panah, M.J.N.; et al. Hepatoblastomas with carcinoma features represent a biological spectrum of aggressive neoplasms in children and young adults. J. Hepatol. 2022, 77, 1026–1037. [Google Scholar] [CrossRef]
- Ding, H.; Zhang, X.; Su, Y.; Jia, C.; Dai, C. GNAS promotes inflammation-related hepatocellular carcinoma progression by promoting STAT3 activation. Cell. Mol. Biol. Lett. 2020, 25, 8. [Google Scholar] [CrossRef]
- Nault, J.C.; Fabre, M.; Couchy, G.; Pilati, C.; Jeannot, E.; Van Nhieu, J.T.; Saint-Paul, M.-C.; De Muret, A.; Redon, M.-J.; Buffet, C.; et al. GNAS-activating mutations define a rare subgroup of inflammatory liver tumors characterized by STAT3 activation. J. Hepatol. 2012, 56, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Wallihan, D.B.; Podberesky, D.J. Hepatic pathology after Fontan palliation: Spectrum of imaging findings. Pediatr. Radiol. 2013, 43, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.; Kim, J.T.; Cox, B.; Larson, B.K.; Kim, S.; Waters, K.M.; Vail, E.; Guindi, M. Evaluation of tumor mutational burden in small early hepatocellular carcinoma and progressed hepatocellular carcinoma. Hepatic Oncol. 2021, 8, HEP39. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Sempoux, C.; Balabaud, C.; Paradis, V.; Bioulac-Sage, P. Hepatocellular nodules in vascular liver diseases. Virchows Arch. 2018, 473, 33–44. [Google Scholar] [CrossRef]
- Téllez, L.; Payancé, A.; Tjwa, E.; del Cerro, M.J.; Idorn, L.; Ovroutski, S.; De Bruyne, R.; Verkade, H.J.; De Rita, F.; de Lange, C.; et al. EASL-ERN position paper on liver involvement in patients with Fontan-type circulation. J. Hepatol. 2023, 79, 1270–1301. [Google Scholar] [CrossRef]
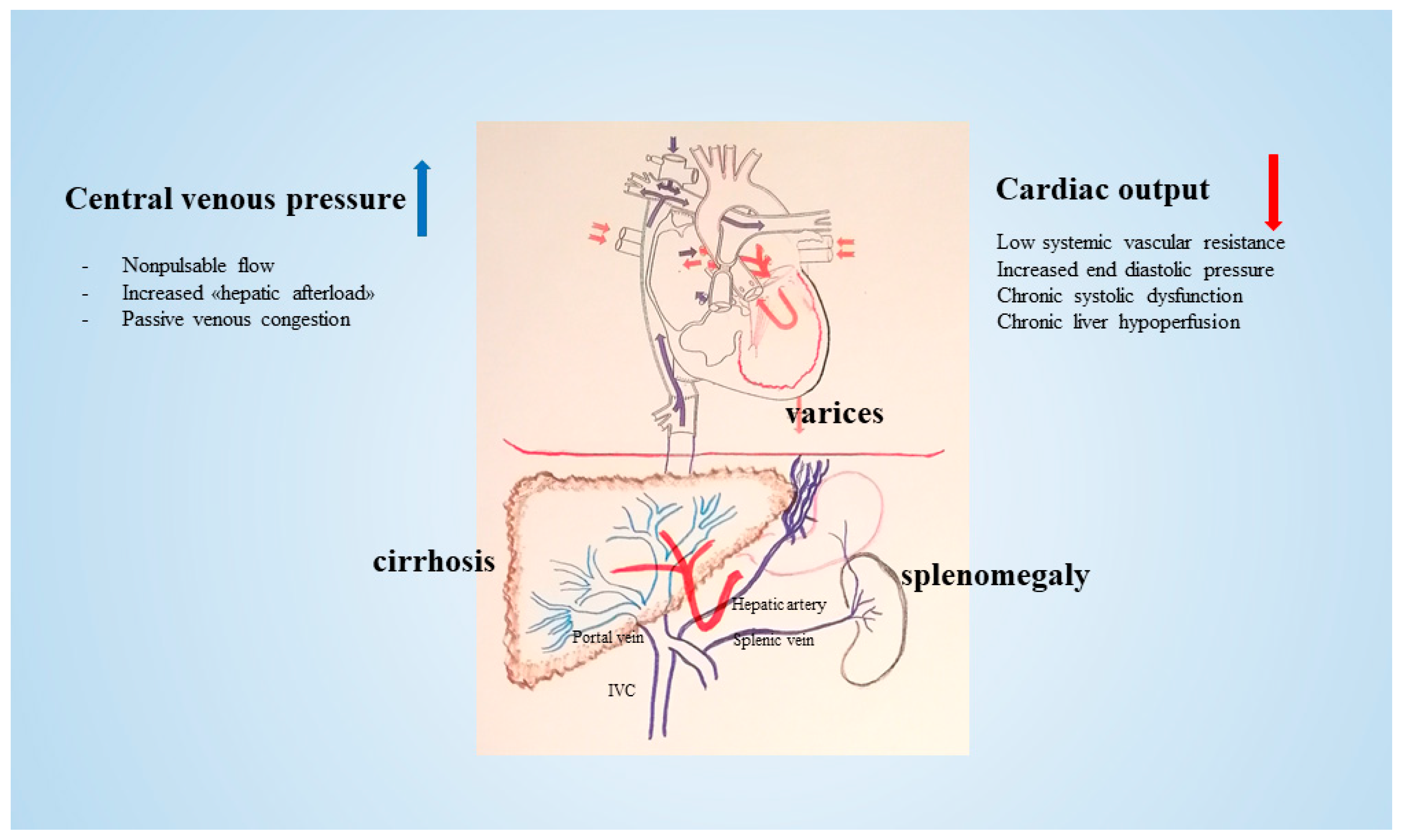
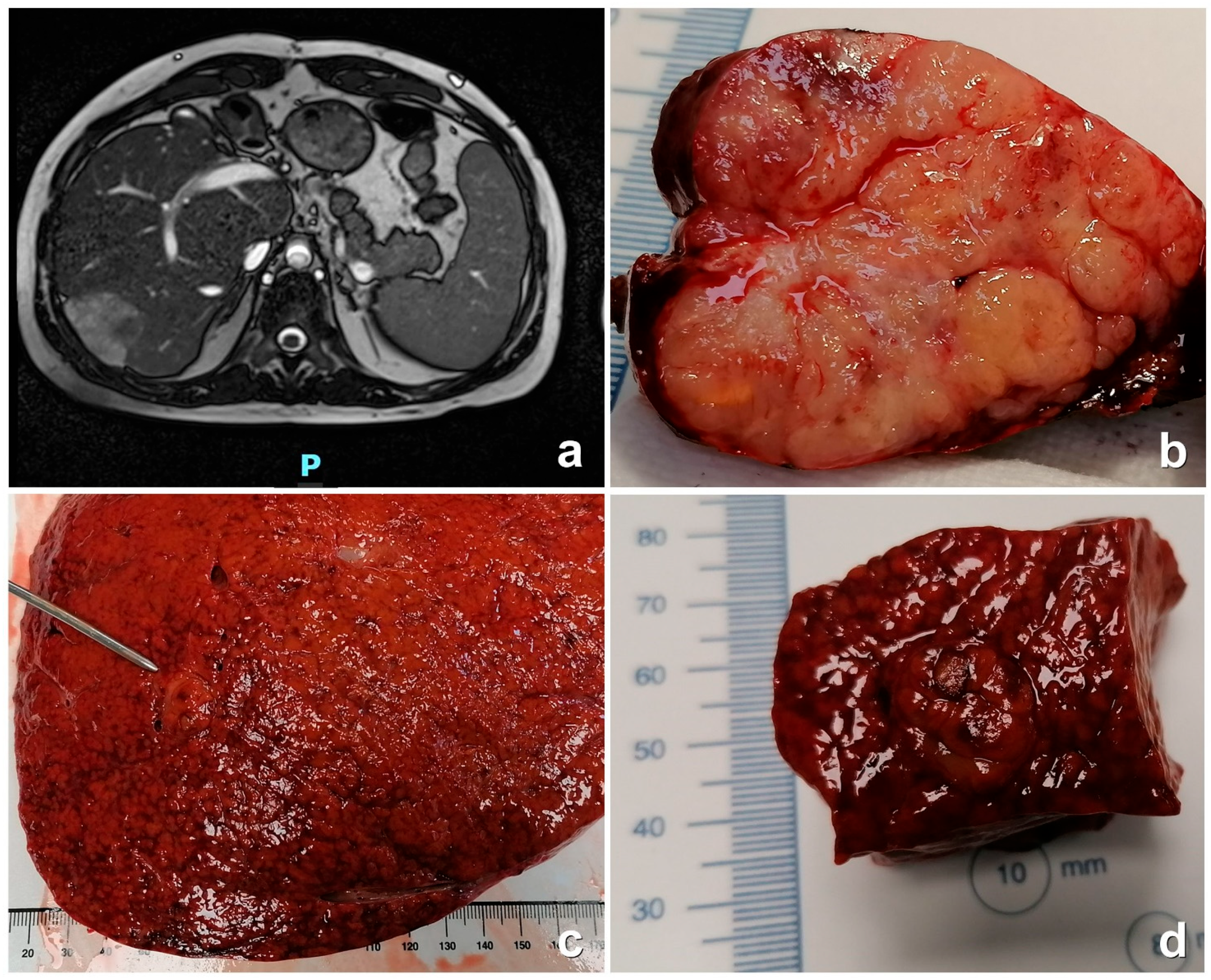

| Parameter | Number |
|---|---|
| N° patients | 31 |
| Gender F/M | 17F 14M |
| Age at Fontan (years) | 2–22 years |
| CHD | |
| TA | 10 |
| Univentricular | 6 |
| PA with intact IS | 4 |
| ccTGA | 4 |
| AVSD | 3 |
| HLHS | 2 |
| DORV | 1 |
| DILV | 1 |
| Fontan type | |
| Extracardiac | 28 |
| Intracardiac | 3 |
| Laboratory test | |
| ALT average (range) | 37.5 (14–61) |
| AST average (range) | 31.5 (14–47) |
| GGT average (range) | 127 (22–232) |
| Bil tot average (range) | 1.41 (0.35–2.48) |
| AFP (range) | 0.9–11,478 |
| Abdominal ultrasound | |
| Hepatomegaly Y/N | 16/13 * |
| Splenomegaly Y/N | 12/17 * |
| Patient | M/F | Age at FP (y) | CHD | Age at Diagnosis (y) | Interval Fontan-FALD (y) | GGT n.v. 11–65 | Platelet n.v. 150–450 | Serum AFP n.v. < 8.10 | Tumor Size | Prognosis/Death |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | F | 4 | Complete AVSD—RV dominant | 20 | 16 | 100 | 208 × 103/uL | 2.43 ng/mL | 4.5 cm | Alive HTx 2019 |
| P2 | F | 3 | TA-PA | 22 | 22 | 99 | 175 × 103/uL | 11,458.00 ng/mL | 4.5 cm | † (6 m post-FALD) |
| P3 | M | 2 | DILV and TGA | 18 22 | 16 * 20 ** | 37 | 173 × 103/uL | ** 447.8 ng/mL | 8 cm | Alive HTx 2014 |
| P4 | F | 6 | TA-PSt-TGA | 34 | 28 | 154 | 182 × 103/uL | 3005.00 ng/mL | 3 cm | † (1 y post-FALD) |
| P5 | M | 8 | PA with intact IS | 32 | 16 | 94 | 111 × 103/uL | 9.43 ng/mL | 2.3 cm | Alive |
| US | MDCT | MRI | |||||
|---|---|---|---|---|---|---|---|
| Arterial Phase | Portal Phase | Late Phase | Arterial Phase | Portal Phase | Late Phase | ||
| P1 FLCA | Iso-echoic | Slightly hyper-dense Wash-in | Hypo-dense Wash-out | Slightly hypo-dense with hyperdense ring | Hyper-intense Wash-in | Hypo-intense Wash-out | Hypo-intense |
| P2 HCC | Hypo-echoic | Hyper-dense Wash-in | Hypo-dense Wash-out | Hypo-dense | Hyper-intense Wash-in | Hypo-intense Wash-out | Hypo-intense |
| P3 ICC | Hypo-echoic | Hypo-dense No wash-in | Hyper-dense | Hypo-dense | ND | ND | ND |
| P4 HCC | Iso-echoic | Slightly hyper-dense Wash-in | Hypo-dense Wash-out | Slightly hypo-dense with hyperdense ring | ND | ND | ND |
| P5 HA | Hyper-echoic | Slightly hyper-dense No wash-in | Hyper-dense | Hypo-dense Wash-out | Iso-intense | Hyper-intense | Hypo-intense |
| P1 FLC | P1 Non-Lesional Tissue | P2 HCC | P2 Non-Lesional Tissue | P3 CC | P3 Non-Lesional Tissue | P4 HCC | P5 Adenoma | |
|---|---|---|---|---|---|---|---|---|
| DNA Analysis | ||||||||
| TMB | 3.9 | 0.8 | 7.8 | 5.5 | 3.9 | 3.1 | 0.8 | 3.1 |
| MSI | 3.30% | 2.50% | 1.90% | 0.00% | 1.10% | 1.90% | 0.90% | 5% |
| CNA | FGFR3 (NM_000142.4) 1 allele loss | None | FGFR3 (NM_000142.4) 1 allele loss | FGFR3 (NM_000142.4) 1 allele loss | FGFR3 (NM_000142.4) 1 allele loss | FGFR3 (NM_000142.4) 1 allele loss | None | None |
| Variants | GNAS (NM_080425.2) c.1299A > G; p.(Gln371Arg) [VAF 3%] | wt | FGFR2 (NM_022970.3) c.1127A > G; p.(Tyr376Cys) [VAF 6%] | FGFR2 (NM_022970.3) c.1127A > G; p.(Tyr376Cys) [VAF 13%] | LRP1B (NM_018557.2) c.9482dup; p.(Asp3161GlufsTer15) [VAF 7%] | LRP1B (NM_018557.2) c.9482dup; p.(Asp3161GlufsTer15) [VAF 6%] | CTNNB1 (NM_001904.3) c.110C > T; p.(Ser37Phe) [VAF: 12%] | HNF1A (NM_000545.5) c.489C > A; p.(Tyr163Ter) [VAF 39%) |
| RASA1 (NM_002890.2) c.2708G > C; p.(Arg903Pro) [VAF 11%] | wt | GNAS (NM_080425.2) c.1531C > T; p.(Arg511Cys) [VAF 57%] | GNAS (NM_080425.2) c.1531C > T; p.(Arg511Cys) [VAF 57%] | CTNNB1 (NM_001904.3) c.569G > A; p.(Arg190His) [VAF 21%] | CTNNB1 (NM_001904.3) c.569G > A; p.(Arg190His) [VAF 21%] | NRAS (NM_002524.4) c.182A > G; p.(Gln61Arg) [VAF: 12%] | MSH3 (NM_002439.4) c.2623G > A; p.(Asp875Asn) [VAF 44%) | |
| MAP2K2 (NM_030662.3) c.142G > C; p.(Glu48Lys) [VAF 19%] | wt | MSH3 (NM_002439.4) c.302del; p.(Val101GlufsTer25) [VAF 6%] | wt | KDM6A (NM_021140.2) c.2689C > G; p.(Leu897Val) [VAF 99%] | KDM6A (NM_021140.2) c.2689C > G; p.(Leu897Val) [VAF 99%] | NOTCH1 (NM_017617.3) c.4843A > T; p.(Met1615Leu) [VAF:48.1%] + c.3739C > T; p.(Gln1247Ter) [VAF:47%] | ||
| ATRX (NM_000489.3) c.3634_3636del; p.(Asp1212del) [VAF 37%] | ATRX (NM_000489.3) c.3634_3636del; p. (Asp1212del) [VAF 40%] | GNAS (NM_080425.2) c.1299A > G; p.(Gln371Arg) [VAF 24.9%] | GNAS (NM_080425.2) c.1299A > G; p.(Gln371Arg) [VAF 25%] | |||||
| PALB2 (NM_024675.3) c.3103A > G; p.(Ile1035Val) [VAF 43%] | PALB2 (NM_024675.3) c.3103A > G; p.(Ile1035Val) [VAF 52%] | RANBP2 (NM_006267.4) c.7216G > A; p.(Ala2406Thr) [VAF 10%] | RANBP2 (NM_006267.4) c.7216G > A; p.(Ala2406Thr) [VAF 10%] | |||||
| RNA Analysis | ||||||||
| Fusion genes | DNAJB1-PRKACA | Negative | Negative | Not analyzed | Negative | Not analyzed | Not available | Negative |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Francalanci, P.; Giovannoni, I.; Tancredi, C.; Gagliardi, M.G.; Palmieri, R.; Brancaccio, G.; Spada, M.; Maggiore, G.; Pietrobattista, A.; Monti, L.; et al. Histopathological Spectrum and Molecular Characterization of Liver Tumors in the Setting of Fontan-Associated Liver Disease. Cancers 2024, 16, 307. https://doi.org/10.3390/cancers16020307
Francalanci P, Giovannoni I, Tancredi C, Gagliardi MG, Palmieri R, Brancaccio G, Spada M, Maggiore G, Pietrobattista A, Monti L, et al. Histopathological Spectrum and Molecular Characterization of Liver Tumors in the Setting of Fontan-Associated Liver Disease. Cancers. 2024; 16(2):307. https://doi.org/10.3390/cancers16020307
Chicago/Turabian StyleFrancalanci, Paola, Isabella Giovannoni, Chantal Tancredi, Maria Giulia Gagliardi, Rosalinda Palmieri, Gianluca Brancaccio, Marco Spada, Giuseppe Maggiore, Andrea Pietrobattista, Lidia Monti, and et al. 2024. "Histopathological Spectrum and Molecular Characterization of Liver Tumors in the Setting of Fontan-Associated Liver Disease" Cancers 16, no. 2: 307. https://doi.org/10.3390/cancers16020307
APA StyleFrancalanci, P., Giovannoni, I., Tancredi, C., Gagliardi, M. G., Palmieri, R., Brancaccio, G., Spada, M., Maggiore, G., Pietrobattista, A., Monti, L., Castellano, A., Giustiniani, M. C., Onetti Muda, A., & Alaggio, R. (2024). Histopathological Spectrum and Molecular Characterization of Liver Tumors in the Setting of Fontan-Associated Liver Disease. Cancers, 16(2), 307. https://doi.org/10.3390/cancers16020307








