It Takes Two to Tango: The Interplay between Prostate Cancer and Its Microenvironment from an Epigenetic Perspective
Abstract
Simple Summary
Abstract
1. Introduction
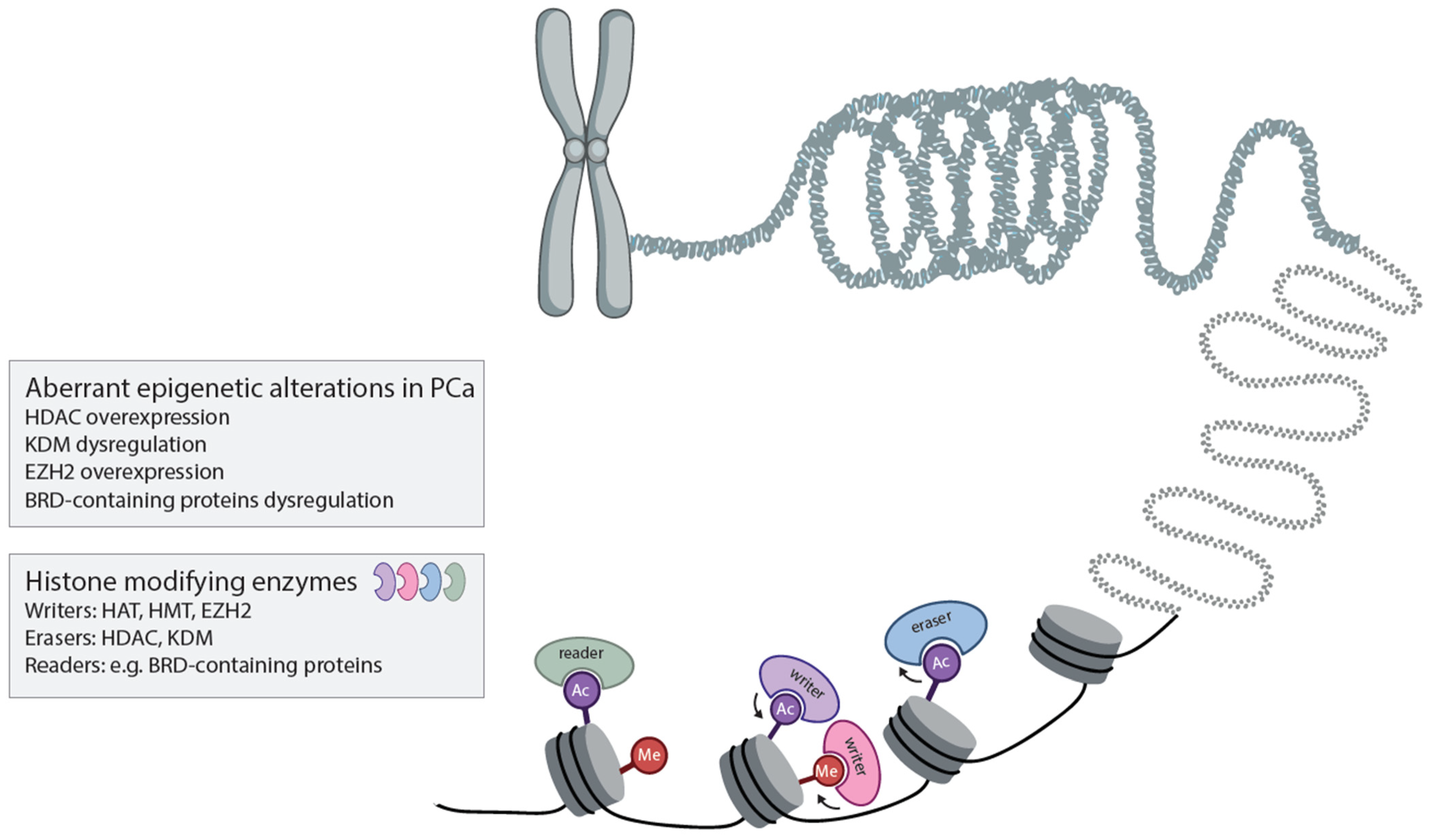
2. What Are the Roles of Various Epigenetic Modifiers in PCa Initiation, Progression and Aggressiveness?
Histone Modifiers in Prostate Cancer Development, Progression and Treatment
3. How Can Epigenetic Alterations Be Targeted in PCa?
3.1. Histone Deacetylase Inhibitors
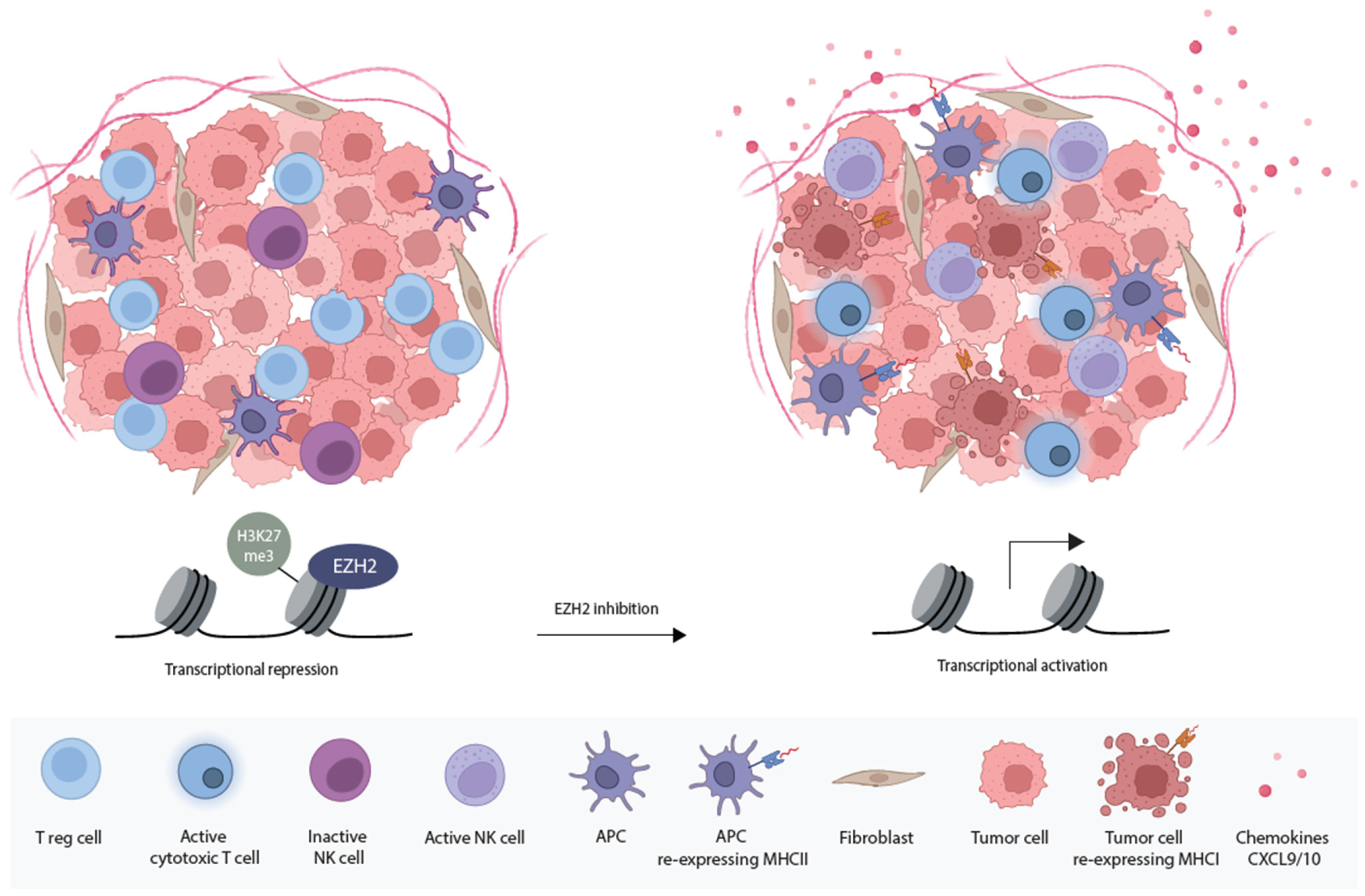
3.2. EZH2 Inhibitors
3.3. BET, p300/CBP, LSD1 and NNMT Inhibitors
4. What Is the Role of Epigenetic Aberrations in Shaping the Immunosuppressive Properties of the Prostate Tumor Microenvironment?
4.1. Epigenetics in the Prostate Tumor Microenvironment
4.2. Current Therapies Targeting the Prostate Tumor Microenvironment
4.3. Challenges of Current Therapies
5. Are Epigenome-Targeting Agents Able to Reverse Tumor-Associated Immune Evasion and Therefore Sensitize PCa to Immunotherapy?
5.1. Epigenetic Therapies Targeting the Prostate TME to Increase Tumor Immunogenicity
5.2. Enhancing Tumor Immunogenicity Using HDAC Inhibitors
5.3. Enhancing Immunogenicity with EZH2 Inhibitors
5.4. Boosting Anti-Tumor Immunity Using NNMT Inhibitors
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Bolton, E.C.; Jones, J.O. Androgens and androgen receptor signaling in prostate tumorigenesis. J. Mol. Endocrinol. 2015, 54, R15–R29. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Campbell, B.K.; Stylli, S.S.; Corcoran, N.M.; Hovens, C.M. The Prostate Cancer Immune Microenvironment, Biomarkers and Therapeutic Intervention. Uro 2022, 2, 74–92. [Google Scholar] [CrossRef]
- Harris, W.P.; Mostaghel, E.A.; Nelson, P.S.; Montgomery, B. Androgen deprivation therapy: Progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat. Clin. Pract. Urol. 2009, 6, 76–85. [Google Scholar] [CrossRef]
- Lonergan, P.; Tindall, D. Androgen receptor signaling in prostate cancer development and progression. J. Carcinog. 2011, 10, 20. [Google Scholar] [CrossRef]
- Jacob, A.; Raj, R.; Allison, D.B.; Myint, Z.W. Androgen Receptor Signaling in Prostate Cancer and Therapeutic Strategies. Cancers 2021, 13, 5417. [Google Scholar] [CrossRef]
- Zaret, K.S.; Carroll, J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes. Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef]
- Hankey, W.; Chen, Z.; Wang, Q. Shaping chromatin states in prostate cancer by pioneer transcription factors. Cancer Res. 2020, 80, 2427–2436. [Google Scholar] [CrossRef]
- Culig, Z.; Santer, F.R. Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev. 2014, 33, 413–427. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.-E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Ha, G.; Hoff, A.M.; Wala, J.A.; Carrot-Zhang, J.; Whelan, C.W.; Haradhvala, N.J.; Freeman, S.S.; Reed, S.C.; Rhoades, J.; et al. Structural Alterations Driving Castration-Resistant Prostate Cancer Revealed by Linked-Read Genome Sequencing. Cell 2018, 174, 433–447.e19. [Google Scholar] [CrossRef] [PubMed]
- Takeda, D.Y.; Spisák, S.; Seo, J.H.; Bell, C.; O’Connor, E.; Korthauer, K.; Ribli, D.; Csabai, I.; Solymosi, N.; Szállási, Z.; et al. A Somatically Acquired Enhancer of the Androgen Receptor Is a Noncoding Driver in Advanced Prostate Cancer. Cell 2018, 174, 422–432.e13. [Google Scholar] [CrossRef]
- Li, Y.; Chan, S.C.; Brand, L.J.; Hwang, T.H.; Silverstein, K.A.T.; Dehm, S.M. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013, 73, 483–489. [Google Scholar] [CrossRef]
- Guo, Z.; Yang, X.; Sun, F.; Jiang, R.; Linn, D.E.; Chen, H.; Chen, H.; Kong, X.; Melamed, J.; Tepper, C.G.; et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009, 69, 2305–2313. [Google Scholar] [CrossRef] [PubMed]
- Prekovic, S.; van den Broeck, T.; Linder, S.; van Royen, M.E.; Houtsmuller, A.B.; Handle, F.; Joniau, S.; Zwart, W.; Claessens, F. Molecular underpinnings of enzalutamide resistance. Endocr. Relat. Cancer 2018, 25, R545–R557. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, M.M.; Qiu, X.; Zhu, Y.; Takeda, D.Y.; Pan, W.; Baca, S.C.; Gusev, A.; Korthauer, K.D.; Severson, T.M.; Ha, G.; et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 2020, 52, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.B.; Mehra, R.; Chinnaiyan, A.M.; Shen, R.; Ghosh, D.; Zhou, M.; Macvicar, G.R.; Varambally, S.; Harwood, J.; Bismar, T.A.; et al. Androgen-Independent Prostate Cancer Is a Heterogeneous Group of DiseasesLessons from a Rapid Autopsy Program. Cancer Res. 2004, 64, 9209–9216. [Google Scholar] [CrossRef]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.; Varambally, S.; et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef]
- Davies, A.; Zoubeidi, A.; Selth, L.A. The epigenetic and transcriptional landscape of neuroendocrine prostate cancer. Endocr. Relat. Cancer 2020, 27, R35–R50. [Google Scholar] [CrossRef]
- Perry, A.S.; Watson, R.W.G.; Lawler, M.; Hollywood, D. The epigenome as a therapeutic target in prostate cancer. Nat. Rev. Urol. 2010, 7, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Conteduca, V.; Hess, J.; Yamada, Y.; Ku, S.Y.; Beltran, H. Epigenetics in prostate cancer: Clinical implications. Transl. Androl. Urol. 2021, 10, 3104–3116. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, A.V.; Ellis, L. Epigenetic reprogramming: A key mechanism driving therapeutic resistance. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Easwaran, H.; Tsai, H.C.; Baylin, S.B. Cancer Epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance. Mol. Cell 2014, 54, 716–727. [Google Scholar] [CrossRef]
- Mazrooei, P.; Kron, K.J.; Zhu, Y.; Zhou, S.; Grillo, G.; Mehdi, T.; Ahmed, M.; Severson, T.M.; Guilhamon, P.; Armstrong, N.S.; et al. Cistrome Partitioning Reveals Convergence of Somatic Mutations and Risk Variants on Master Transcription Regulators in Primary Prostate Tumors Cancer Cell Article Cistrome Partitioning Reveals Convergence of Somatic Mutations and Risk Variants on Master Transcription Regulators in Primary Prostate Tumors. Cancer Cell 2019, 36, 674–689. [Google Scholar] [PubMed]
- Linder, S.; Hoogstraat, M.; Stelloo, S.; Eickhoff, N.; Schuurman, K.; de Barros, H.; Alkemade, M.; Bekers, E.M.; Severson, T.M.; Sanders, J.; et al. Drug-Induced Epigenomic Plasticity Reprograms Circadian Rhythm Regulation to Drive Prostate Cancer toward Androgen Independence. Cancer Discov. 2022, 12, 2074–2097. [Google Scholar] [CrossRef]
- Albany, C.; Alva, A.S.; Aparicio, A.M.; Singal, R.; Yellapragada, S.; Sonpavde, G.; Hahn, N.M. Epigenetics in prostate cancer. Prostate Cancer 2011, 2011, 580318. [Google Scholar] [CrossRef]
- Shiao, S.L.; Chu, G.C.Y.; Chung, L.W.K. Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett. 2016, 380, 340–348. [Google Scholar] [CrossRef]
- Stultz, J.; Fong, L. How to turn up the heat on the cold immune microenvironment of metastatic prostate cancer. Prostate Cancer Prostatic Dis. 2021, 24, 697–717. [Google Scholar] [CrossRef]
- Zhou, H.; He, Q.; Li, C.; Alsharafi, B.L.M.; Deng, L.; Long, Z.; Gan, Y. Focus on the tumor microenvironment: A seedbed for neuroendocrine prostate cancer. Front. Cell Dev. Biol. 2022, 10, 955669. [Google Scholar] [CrossRef]
- Nagireddy, S.; Qureshi, R.; Best, J.; Frech, F.S.; Shah, K.; Soni, Y.; Kuchakulla, M.; Narasimman, M.; Arora, H. Current Treatment Modalities Targeting Tumor Microenvironment in Castration-Resistant Prostate Cancer. Adv. Exp. Med. Biol. 2021, 1329, 295–323. [Google Scholar]
- Yang, J.; Xu, J.; Wang, W.; Zhang, B.; Yu, X.; Shi, S. Epigenetic regulation in the tumor microenvironment: Molecular mechanisms and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.L.; Weinberg, R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013, 19, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Beard, J. Embryological Aspects and Etiology of Carcinoma. Lancet 1902, 159, 1758–1761. Available online: https://www.sciencedirect.com/science/article/pii/S0140673601899360 (accessed on 6 July 2023). [CrossRef][Green Version]
- Markert, C.L. Neoplasia: A Disease of Cell Differentiation. Cancer Res. 1968, 28, 1908–1914. Available online: https://aacrjournals.org/cancerres/article/28/9/1908/477125/Neoplasia-A-Disease-of-Cell-Differentiation1 (accessed on 6 July 2023). [PubMed]
- Goel, S.; Bhatia, V.; Biswas, T.; Ateeq, B. Epigenetic reprogramming during prostate cancer progression: A perspective from development. Semin. Cancer Biol. 2022, 83, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Hunt, C.R.; Ramnarain, D.; Horikoshi, N.; Iyengar, P.; Pandita, R.K.; Shay, J.W.; Pandita, T.K. Histone Modifications and DNA Double-Strand Break Repair after Exposure to Ionizing Radiations. Radiat. Res. 2013, 179, 383–392. [Google Scholar] [CrossRef]
- House, N.C.M.; Koch, M.R.; Freudenreich, C.H. Chromatin modifications and DNA repair: Beyond double-strand breaks. Front. Genet. 2014, 5, 68602. [Google Scholar] [CrossRef]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef]
- Abbas, A.; Gupta, S. The Role of Histone Deacetylases in Prostate Cancer. Epigenetics Off. J. DNA Methylation Soc. 2008, 3, 300–309. [Google Scholar] [CrossRef]
- Macedo-Silva, C.; Benedetti, R.; Ciardiello, F.; Cappabianca, S.; Jerónimo, C.; Altucci, L. Epigenetic mechanisms underlying prostate cancer radioresistance. Clin. Epigenetics 2021, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Röske, A.; Gekeler, V.; Beckers, T.; Stephan, C.; Jung, K.; Fritzsche, F.R.; Niesporek, S.; Denkert, C.; Dietel, M.; et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br. J. Cancer 2008, 98, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Sun, L.; Mai, A.; Chiang, Y.T.; Farrar, W.L.; Danesi, R.; Helgason, C.D. The emerging role of histone lysine demethylases in prostate cancer. Mol. Cancer 2012, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; He, H.H.; Gao, S.; Chen, S.; Yu, Z.; Gao, Y.; Chen, S.; Chen, M.W.; Zhang, J.; Ahmed, M.; et al. Lysine-Specific Demethylase 1 Has Dual Functions as a Major Regulator of Androgen Receptor Transcriptional Activity. Cell Rep. 2014, 9, 1618–1627. [Google Scholar] [CrossRef]
- Komura, K.; Jeong, S.H.; Hinohara, K.; Qu, F.; Wang, X.; Hiraki, M.; Azuma, H.; Lee, G.M.; Kantoff, P.W.; Sweeney, C.J. Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc. Natl. Acad. Sci. USA 2016, 113, 6259–6264. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.A.B.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef]
- Urbanucci, A.; Mills, I.G. Bromodomain-containing proteins in prostate cancer. Mol. Cell Endocrinol. 2018, 462, 31–40. [Google Scholar] [CrossRef]
- Chan, H.M.; La Thangue, N.B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001, 114, 2363–2373. [Google Scholar] [CrossRef]
- Raisner, R.; Kharbanda, S.; Jin, L.; Jeng, E.; Chan, E.; Merchant, M.; Haverty, P.M.; Bainer, R.; Cheung, T.; Arnott, D.; et al. Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep. 2018, 24, 1722–1729. [Google Scholar] [CrossRef]
- Weinert, B.T.; Narita, T.; Satpathy, S.; Srinivasan, B.; Hansen, B.K.; Schölz, C.; Hamilton, W.B.; Zucconi, B.E.; Wang, W.W.; Liu, W.R.; et al. Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 2018, 174, 231–244.e12. [Google Scholar] [CrossRef] [PubMed]
- Waddell, A.R.; Huang, H.; Liao, D. CBP/p300: Critical Co-Activators for Nuclear Steroid Hormone Receptors and Emerging Therapeutic Targets in Prostate and Breast Cancers. Cancers 2021, 13, 2872. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Wang, C.; Reutens, A.T.; Wang, J.; Angeletti, R.H.; Siconolfi-Baez, L.; Ogryzko, V.; Avantaggiati, M.L.; Pestell, R.G. p300 and p300/cAMP-response Element-binding Protein-associated Factor Acetylate the Androgen Receptor at Sites Governing Hormone-dependent Transactivation. J. Biol. Chem. 2000, 275, 20853–20860. [Google Scholar] [CrossRef] [PubMed]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013, 9, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.A.; Coscia, F.; Chryplewicz, A.; Chang, J.W.; Hernandez, K.M.; Pan, S.; Tienda, S.M.; Nahotko, D.A.; Li, G.; Blaženović, I.; et al. Proteomics reveals NNMT as a master metabolic regulator of cancer associated fibroblasts. Nature 2019, 569, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhu, M.; Li, X.; Er, L.; Li, S. NNMT Is an Immune-Related Prognostic Biomarker That Modulates the Tumor Microenvironment in Pan-Cancer. Dis. Markers 2023, 2023, 9226712. [Google Scholar] [CrossRef] [PubMed]
- Campagna, R.; Vignini, A. NAD+ Homeostasis and NAD+-Consuming Enzymes: Implications for Vascular Health. Antioxidants 2023, 12, 376. [Google Scholar] [CrossRef]
- You, Z.; Liu, Y.; Liu, X. Nicotinamide N-methyltransferase enhances the progression of prostate cancer by stabilizing sirtuin 1. Oncol. Lett. 2018, 15, 9195–9201. [Google Scholar] [CrossRef]
- Frønsdal, K.; Saatcioglu, F. Histone deacetylase inhibitors differentially mediate apoptosis in prostate cancer cells. Prostate 2005, 62, 299–306. [Google Scholar] [CrossRef]
- Marks, P.A.; Richon, V.M.; Rifkind, R.A. Histone deacetylase inhibitors: Inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 2000, 92, 1210–1216. [Google Scholar] [CrossRef]
- Roy, S.; Packman, K.; Jeffrey, R.; Tenniswood, M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005, 12, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Welsbie, D.S.; Xu, J.; Chen, Y.; Borsu, L.; Scher, H.I.; Rosen, N.; Sawyers, C.L. Histone Deacetylases Are Required for Androgen Receptor Function in Hormone-Sensitive and Castrate-Resistant Prostate Cancer. Cancer Res. 2009, 69, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Marzio, G.; Wagener, C.; Gutierrez, M.I.; Cartwright, P.; Helin, K.; Giacca, M. E2F family members are differentially regulated by reversible acetylation. J. Biol. Chem. 2000, 275, 10887–10892. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Kawaguchi, Y.; Lai, C.H.; Kovacs, J.J.; Higashimoto, Y.; Appella, E.; Yao, T. MDM2–HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002, 21, 6236–6245. [Google Scholar] [CrossRef]
- Gu, W.; Roeder, R.G. Activation of p53 Sequence-Specific DNA Binding by Acetylation of the p53 C-Terminal Domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef]
- Feng, S.; De Carvalho, D.D. Clinical advances in targeting epigenetics for cancer therapy. FEBS J. 2022, 289, 1214–1239. [Google Scholar] [CrossRef] [PubMed]
- Severson, T.M.; Zhu, Y.; Prekovic, S.; Schuurman, K.; Nguyen, H.M.; Brown, L.G.; Hakkola, S.; Kim, Y.; Kneppers, J.; Linder, S.; et al. Enhancer profiling identifies epigenetic markers of endocrine resistance and reveals therapeutic options for metastatic castration-resistant prostate cancer patients. medRxiv 2023. [Google Scholar] [CrossRef]
- Molife, L.R.; Attard, G.; Fong, P.C.; Karavasilis, V.; Reid, A.H.M.; Patterson, S.; Riggs, C.E., Jr.; Hiagno, C.; Stadler, W.M.; McCulloch, W.; et al. Phase II, two-stage, single-arm trial of the histone deacetylase inhibitor (HDACi) romidepsin in metastatic castration-resistant prostate cancer (CRPC). Ann. Oncol. 2010, 21, 109–113. [Google Scholar] [CrossRef]
- Bradley, D.; Rathkopf, D.; Dunn, R.; Stadler, W.M.; Liu, G.; Smith, D.C.; Pili, R.; Zwiebel, J.; Scher, H.; Hussain, M. Vorinostat in advanced prostate cancer patients progressing on prior chemotherapy (National Cancer Institute Trial 6862). Cancer 2009, 115, 5541–5549. [Google Scholar] [CrossRef]
- Ferrari, A.C.; Alumkal, J.J.; Stein, M.N.; Taplin, M.E.; Babb, J.; Barnett, E.S.; Gomez-Pinillos, A.; Liu, X.; Moore, D.; DiPaola, R.; et al. Epigenetic therapy with panobinostat combined with bicalutamide rechallenge in castration-resistant prostate cancer. Clin. Cancer Res. 2019, 25, 52–63. [Google Scholar] [CrossRef]
- Taplin, M.-E.; Hussain, A.; Shah, S.; Shore, N.D.; Agrawal, M.; Clark, W.; Edenfield, W.; Nordquist, L.T.; Sartor, O.A.; Butrynski, J.E.; et al. ProSTAR: A phase Ib/II study of CPI-1205, a small molecule inhibitor of EZH2, combined with enzalutamide (E) or abiraterone/prednisone (A/P) in patients with metastatic castration-resistant prostate cancer (mCRPC). J. Clin. Oncol. 2019, 37, TPS335. [Google Scholar] [CrossRef]
- Abida, W.; Iannotti, N.; Kocabas Argon, E.; Appleman, L.; Michaud, N.; Rajarethinam, A.; Adib, D.; Saltzstein, D.R. 586P Safety of tazemetostat (TAZ) in combination with abiraterone/prednisone (A/P) or enzalutamide (E) in patients (Pts) with metastatic castration-resistant prostate cancer (mCRPC). Ann. Oncol. 2021, 32, S636. [Google Scholar] [CrossRef]
- Aggarwal, R.R.; Schweizer, M.T.; Nanus, D.M.; Pantuck, A.J.; Heath, E.I.; Campeau, E.; Attwell, S.; Norek, K.; Snyder, M.; Bauman, L.; et al. A Phase 1b/2a Study of the Pan-BET Bromodomain Inhibitor ZEN-3694 in Combination with Enzalutamide in Patients with Metastatic Castration Resistant Prostate Cancer. Clin. Cancer Res. 2020, 26, 5338–5347. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Cojocaru, E.; Plummer, E.R.; Knurowski, T.; Clegg, K.; Ashby, F.; Pegg, N.; West, W.; Brooks, A.N. An open label phase I/IIa study to evaluate the safety and efficacy of CCS1477 as monotherapy and in combination in patients with advanced solid/metastatic tumors. J. Clin. Oncol. 2019, 37, TPS5089. [Google Scholar] [CrossRef]
- Sampson, C.M.; Dimet, A.L.; Neelakantan, H.; Ogunseye, K.O.; Stevenson, H.L.; Hommel, J.D.; Watowich, S.J. Combined nicotinamide N-methyltransferase inhibition and reduced-calorie diet normalizes body composition and enhances metabolic benefits in obese mice. Sci. Rep. 2021, 11, 5637. [Google Scholar] [CrossRef] [PubMed]
- Seifarth, W.; Frank, O.; Zeilfelder, U.; Spiess, B.; Greenwood, A.D.; Hehlmann, R.; Leib-Mösch, C. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 2005, 79, 341–352. [Google Scholar] [CrossRef]
- Grunewald, C.M.; Schulz, W.A.; Skowron, M.A.; Hoffmann, M.J.; Niegisch, G. Tumor immunotherapy—The potential of epigenetic drugs to overcome resistance. Transl. Cancer Res. 2018, 7, 1151–1160. [Google Scholar] [CrossRef]
- Liu, Z.; Ren, Y.; Weng, S.; Xu, H.; Li, L.; Han, X. A New Trend in Cancer Treatment: The Combination of Epigenetics and Immunotherapy. Front. Immunol. 2022, 13, 809761. [Google Scholar] [CrossRef] [PubMed]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef]
- Stone, M.L.; Chiappinelli, K.B.; Li, H.; Murphy, L.M.; Travers, M.E.; Topper, M.J.; Mathios, D.; Lim, M.; Shih, I.; Wang, T.; et al. Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proc. Natl. Acad. Sci. USA 2017, 114, E10981–E10990. [Google Scholar] [CrossRef]
- Sheng, W.; LaFleur, M.W.; Nguyen, T.H.; Chen, S.; Chakravarthy, A.; Conway, J.R.; Li, Y.; Chen, H.; Yang, H.; Hsu, P.; et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 2018, 174, 549–563.e19. [Google Scholar] [CrossRef] [PubMed]
- Morel, K.L.; Sheahan, A.V.; Burkhart, D.L.; Baca, S.C.; Boufaied, N.; Liu, Y.; Qiu, X.; Cañadas, I.; Roehle, K.; Heckler, M.; et al. EZH2 inhibition activates a dsRNA–STING–interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat. Cancer 2021, 2, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zhang, Y.; Mao, F.; Zhang, Z.; Li, Z.; Wang, R.; Liu, J.; Liu, X. Inhibition of EZH2 Enhances the Antitumor Efficacy of Metformin in Prostate Cancer. Mol. Cancer Ther. 2020, 19, 2490–2501. [Google Scholar] [CrossRef] [PubMed]
- Faivre, E.J.; Wilcox, D.; Lin, X.; Hessler, P.; Torrent, M.; He, W.; Uziel, T.; Albert, D.H.; McDaniel, K.; Kati, W.; et al. Exploitation of castration-resistant prostate cancer transcription factor dependencies by the novel BET inhibitor ABBV-075. Mol. Cancer Res. 2017, 15, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Welti, J.; Sharp, A.; Brooks, N.; Yuan, W.; McNair, C.; Chand, S.N.; Pal, A.; Figueiredo, I.; Riisnaes, R.; Gurel, B.; et al. Targeting the p300/cbp axis in lethal prostate cancer. Cancer Discov. 2021, 11, 1118–1137. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Martin, N.I.; van Haren, M.J. Nicotinamide N-methyl transferase (NNMT): An emerging therapeutic target. Drug Discov. Today 2021, 26, 2699–2706. [Google Scholar] [CrossRef]
- Ruf, S.; Rajagopal, S.; Kadnur, S.V.; Hallur, M.S.; Rani, S.; Kristam, R.; Swaminathan, S.; Zope, B.R.; Gondrala, P.K.; Swamy, I.; et al. Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders. Sci. Rep. 2022, 12, 15440. [Google Scholar] [CrossRef]
- Van Haren, M.J.; Zhang, Y.; Thijssen, V.; Buijs, N.; Gao, Y.; Mateuszuk, L.; Fedak, F.A.; Kij, A.; Campagna, R.; Sartini, D.; et al. Macrocyclic peptides as allosteric inhibitors of nicotinamide N-methyltransferase (NNMT). RSC Chem. Biol. 2021, 2, 1546–1555. [Google Scholar] [CrossRef]
- Reustle, A.; Menig, L.S.; Leuthold, P.; Hofmann, U.; Stühler, V.; Schmees, C.; Becker, M.; Haag, M.; Klumpp, V.; Winter, S.; et al. Nicotinamide-N-methyltransferase is a promising metabolic drug target of primary and metastatic clear cell renal cell carcinoma. Clin. Transl. Med. 2022, 12, e883. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, T.; Zeng, Y. Epigenetic Modifications in Prostate Cancer Metastasis and Microenvironment. Cancers 2023, 15, 2243. [Google Scholar] [CrossRef]
- Jones, K.; Zhang, Y.; Kong, Y.; Farah, E.; Wang, R.; Li, C.; Wang, X.; Zhang, Z.; Wang, J.; Mao, F.; et al. Epigenetics in prostate cancer treatment. J. Transl. Genet. Genom. 2021, 5, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Saleh, R.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Role of Epigenetic Modifications in Inhibitory Immune Checkpoints in Cancer Development and Progression. Front. Immunol. 2020, 11, 544793. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.N.; Xia, S.J. Stroma–epithelium crosstalk in prostate cancer. Asian J. Androl. 2009, 11, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Wang, Z.; Cheng, L. Tumor microenvironment heterogeneity an important mediator of prostate cancer progression and therapeutic resistance. npj Precis. Oncol. 2022, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Lu, Y.; Roca, H.; Keller, J.M.; Zhang, J.; McCauley, L.K.; Keller, E.T. Immune mediators in the tumor microenvironment of prostate cancer. Chin. J. Cancer 2017, 36, 29. [Google Scholar] [CrossRef]
- Fay, E.K.; Graff, J.N. Immunotherapy in Prostate Cancer. Cancers 2020, 12, 1752. [Google Scholar] [CrossRef]
- Boettcher, A.N.; Usman, A.; Morgans, A.; VanderWeele, D.J.; Sosman, J.; Wu, J.D. Past, Current, and Future of Immunotherapies for Prostate Cancer. Front. Oncol. 2019, 9, 884. [Google Scholar] [CrossRef]
- Kwon, J.T.W.; Bryant, R.J.; Parkes, E.E. The tumor microenvironment and immune responses in prostate cancer patients. Endocr. Relat. Cancer 2021, 28, T95. [Google Scholar] [CrossRef]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef]
- Sanda, M.G.; Restifo, N.P.; Walsh, J.C.; Kawakami, Y.; Nelson, W.G.; Pardoll, D.M.; Simons, J.W. Molecular Characterization of Defective Antigen Processing in Human Prostate Cancer. JNCI J. Natl. Cancer Inst. 1995, 87, 280–285. [Google Scholar] [CrossRef]
- Garcia-Lora, A.; Algarra, I.; Garrido, F. MHC class I antigens, immune surveillance, and tumor immune escape. J. Cell Physiol. 2003, 195, 346–355. [Google Scholar] [CrossRef]
- Lundholm, M.; Hägglöf, C.; Wikberg, M.L.; Stattin, P.; Egevad, L.; Bergh, A.; Wikström, P.; Palmqvist, R.; Edin, S. Secreted Factors from Colorectal and Prostate Cancer Cells Skew the Immune Response in Opposite Directions. Sci. Rep. 2015, 5, 15651. [Google Scholar] [CrossRef]
- Vitkin, N.; Nersesian, S.; Siemens, D.R.; Koti, M. The tumor immune contexture of prostate cancer. Front. Immunol. 2019, 10, 445559. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Jeon, J.W.; Sievers, C.; Allen, C.T. Antigen processing and presentation in cancer immunotherapy. J. Immunother. Cancer 2020, 8, e001111. [Google Scholar] [CrossRef] [PubMed]
- Archer, M.; Dogra, N.; Kyprianou, N. Inflammation as a Driver of Prostate Cancer Metastasis and Therapeutic Resistance. Cancers 2020, 12, 2984. [Google Scholar] [CrossRef] [PubMed]
- Koinis, F.; Xagara, A.; Chantzara, E.; Leontopoulou, V.; Aidarinis, C.; Kotsakis, A. Myeloid-Derived Suppressor Cells in Prostate Cancer: Present Knowledge and Future Perspectives. Cells 2022, 11, 20. [Google Scholar] [CrossRef]
- Yang, L.; Pang, Y.; Moses, H.L. TGF-β and immune cells: An important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010, 31, 220–227. [Google Scholar] [CrossRef]
- Kang, N.; Eccleston, M.; Clermont, P.L.; Latarani, M.; Male, D.K.; Wang, Y.; Crea, F. EZH2 inhibition: A promising strategy to prevent cancer immune editing. Epigenomics 2020, 12, 1457–1476. [Google Scholar] [CrossRef]
- Liu, V.C.; Wong, L.Y.; Jang, T.; Shah, A.H.; Park, I.; Yang, X.; Zhang, Q.; Lonning, S.; Teicher, B.A.; Lee, C. Tumor evasion of the immune system by converting CD4+CD25- T cells into CD4+CD25+ T regulatory cells: Role of tumor-derived TGF-beta. J. Immunol. 2007, 178, 2883–2892. [Google Scholar] [CrossRef]
- Sigalotti, L.; Fratta, E.; Coral, S.; Maio, M. Epigenetic drugs as immunomodulators for combination therapies in solid tumors. Pharmacol. Ther. 2014, 142, 339–350. [Google Scholar] [CrossRef]
- Kitamura, H.; Torigoe, T.; Asanuma, H.; Honma, I.; Sato, N.; Tsukamoto, T. Down-regulation of HLA class I antigens in prostate cancer tissues and up-regulation by histone deacetylase inhibition. J. Urol. 2007, 178, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Nie, D.Y.; Ba-alawi, W.; Ji, Y.S.; Zhang, Z.W.; Cruickshank, J.; Haight, J.; Ciamponi, F.E.; Chen, J.; Duan, S.; et al. PRMT inhibition induces a viral mimicry response in triple-negative breast cancer. Nat. Chem. Biol. 2022, 18, 821–830. [Google Scholar] [CrossRef]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef]
- Zheng, H.; Zhao, W.; Yan, C.; Watson, C.C.; Massengill, M.; Xie, M.; Massengill, C.; Noyes, D.R.; Martinez, G.V.; Afzal, R.; et al. HDAC Inhibitors Enhance T-Cell Chemokine Expression and Augment Response to PD-1 Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2016, 22, 4119–4132. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Errico, D.; Vento-Tormo, R.; Sieweke, M.; Ballestar, E. Epigenetic control of myeloid cell differentiation, identity and function. Nat. Rev. Immunol. 2015, 15, 7–17. [Google Scholar] [CrossRef]
- Mikyšková, R.; Indrová, M.; Vlková, V.; Bieblová, J.; Šímová, J.; Paračková, Z.; Pajtasz-Piasecka, E.; Rossowska, J.; Reiniš, M. DNA demethylating agent 5-azacytidine inhibits myeloid-derived suppressor cells induced by tumor growth and cyclophosphamide treatment. J. Leukoc. Biol. 2014, 95, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Ning, F.; Liu, Z.C.; Wu, L.; Li, Z.Q.; Qi, Y.F.; Zhang, G.; Wang, H.; Cai, S.; Du, J. Histone deacetylase inhibitors deplete myeloid-derived suppressor cells induced by 4T1 mammary tumors in vivo and in vitro. Cancer Immunol. Immunother. 2017, 66, 355–366. [Google Scholar] [CrossRef]
- Wolf, N.K.; Kissiov, D.U.; Raulet, D.H. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat. Rev. Immunol. 2022, 23, 90–105. [Google Scholar] [CrossRef]
- Zhu, S.; Denman, C.J.; Cobanoglu, Z.S.; Kiany, S.; Lau, C.C.; Gottschalk, S.M.; Hughes, D.P.M.; Kleinerman, E.S.; Lee, D.A. The narrow-spectrum HDAC inhibitor entinostat enhances NKG2D expression without NK cell toxicity, leading to enhanced recognition of cancer cells. Pharm. Res. 2015, 32, 779–792. [Google Scholar] [CrossRef]
- Kondělková, K.; Vokurková, D.; Krejsek, J.; Borská, L.; Fiala, Z.; Ctirad, A. Regulatory T cells (TREG) and their roles in immune system with respect to immunopathological disorders. Acta Medica 2010, 53, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Blank, C.U.; Haining, W.N.; Held, W.; Hogan, P.G.; Kallies, A.; Lugli, E.; Lynn, R.C.; Philip, M.; Rao, A.; Restifo, N.P.; et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019, 19, 665–674. [Google Scholar] [CrossRef]
- Karantanos, T.; Chistofides, A.; Barhdan, K.; Li, L.; Boussiotis, V.A. Regulation of T Cell Differentiation and Function by EZH2. Front. Immunol. 2016, 7, 172. [Google Scholar] [CrossRef] [PubMed]
- Bugide, S.; Green, M.R.; Wajapeyee, N. Inhibition of Enhancer of zeste homolog 2 (EZH2) induces natural killer cell-mediated eradication of hepatocellular carcinoma cells. Proc. Natl. Acad. Sci. USA 2018, 115, E3509–E3518. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Quiros, J.; Mahuron, K.; Pai, C.C.; Ranzani, V.; Young, A.; Silveria, S.; Harwin, T.; Abnousian, A.; Pagani, M.; et al. Targeting EZH2 Reprograms Intratumoral Regulatory T Cells to Enhance Cancer Immunity. Cell Rep. 2018, 23, 3262–3274. [Google Scholar] [CrossRef]
- Qiao, Y.; Kang, K.; Giannopoulou, E.; Fang, C.; Ivashkiv, L.B. IFN-γ Induces Histone 3 Lysine 27 Trimethylation in a Small Subset of Promoters to Stably Silence Gene Expression in Human Macrophages. Cell Rep. 2016, 16, 3121–3129. [Google Scholar] [CrossRef]
- Wu, J.; Lanier, L.L. Natural killer cells and cancer. Adv. Cancer Res. 2003, 90, 127–156. [Google Scholar]
- Qiu, J.; Sharma, S.; Rollins, R.A.; Paul, T.A. The complex role of EZH2 in the tumor microenvironment: Opportunities and challenges for immunotherapy combinations. Future Med. Chem. 2020, 12, 1415–1430. [Google Scholar] [CrossRef]
- Burr, M.L.; Sparbier, C.E.; Chan, K.L.; Chan, Y.C.; Kersbergen, A.; Lam, E.Y.N.; Azidis-Yates, E.; Vassiliadis, D.; Bell, C.C.; Gilan, O.; et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell 2019, 36, 385–401.e8. [Google Scholar] [CrossRef]
- Yin, J.; Leavenworth, J.W.; Li, Y.; Luo, Q.; Xie, H.; Liu, X.; Huang, S.; Yan, H.; Fu, Z.; Zhang, L.Y.; et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc. Natl. Acad. Sci. USA 2015, 112, 15988–15993. [Google Scholar] [CrossRef]
- Study Details|ORIOn-E: A Study Evaluating CPI-1205 in Patients with Advanced Solid Tumors|ClinicalTrials.gov. Available online: https://clinicaltrials.gov/study/NCT03525795 (accessed on 9 November 2023).
- Kilgour, M.K.; MacPherson, S.; Zacharias, L.G.; Ellis, A.E.; Sheldon, R.D.; Liu, E.Y.; Keyes, S.; Pauly, B.; Carleton, G.; Allard, B.; et al. 1-Methylnicotinamide is an immune regulatory metabolite in human ovarian cancer. Sci. Adv. 2021, 7, 2668–2698. [Google Scholar] [CrossRef]
- Neelakantan, H.; Vance, V.; Wetzel, M.D.; Wang, H.Y.L.; McHardy, S.F.; Finnerty, C.C.; Hommel, J.D.; Watowich, S.J. Selective and membrane-permeable small molecule inhibitors of nicotinamide N-methyltransferase reverse high fat diet-induced obesity in mice. Biochem. Pharmacol. 2018, 147, 141–152. [Google Scholar] [CrossRef]
- Kannt, A.; Rajagopal, S.; Kadnur, S.V.; Suresh, J.; Bhamidipati, R.K.; Swaminathan, S.; Hallur, M.S.; Kristam, R.; Elvert, R.; Czech, J.; et al. A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders. Sci. Rep. 2018, 8, 3660. [Google Scholar] [CrossRef] [PubMed]
- Reustle, A.; Di Marco, M.; Meyerhoff, C.; Nelde, A.; Walz, J.S.; Winter, S.; Kandabarau, S.; Büttner, F.; Haag, M.; Backert, L.; et al. Integrative -omics and HLA-ligandomics analysis to identify novel drug targets for ccRCC immunotherapy. Genome Med. 2020, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Hontecillas-Prieto, L.; Flores-Campos, R.; Silver, A.; de Álava, E.; Hajji, N.; García-Domínguez, D.J. Synergistic Enhancement of Cancer Therapy Using HDAC Inhibitors: Opportunity for Clinical Trials. Front. Genet. 2020, 11, 578011. [Google Scholar] [CrossRef] [PubMed]
- Haffner, M.C.; Zwart, W.; Roudier, M.P.; True, L.D.; Nelson, W.G.; Epstein, J.I.; De Marzo, A.M.; Nelson, P.S.; Yegnasubramanian, S. Genomic and phenotypic heterogeneity in prostate cancer. Nat. Rev. Urol. 2020, 18, 79–92. [Google Scholar] [CrossRef]
| Epigenetic Drug | Molecular Target | Cancer Type | Phase | References |
|---|---|---|---|---|
| Romidepsin | HDAC1/2i | Chemotherapy naïve mCRPC | Phase II | Molife et al. [68] |
| Vorinostat | HDACi | Progressive mCRPC after 1 prior chemotherapy treatment | Phase II | Bradeley et al. [69] |
| Epigenetic Drug | Molecular Target | Combination | Type | Cancer Type | Phase | References |
|---|---|---|---|---|---|---|
| Panibostat | Pan-HDACi | Bicalutamide | Anti-androgen | Recurrent PCa | Phase I/II | Ferrari et al. [70] |
| CPI-1205 | EZH2i | Enzalutamide/Abiraterone/Prednisone | AR/androgen biosynthesis inhibitor/glucocorticoid receptor agonist | mCRPC patients | Phase Ib/II | Taplin et al. [71] |
| Tazometostat | EZH2i | Enzalutamide/Abiraterone/Prednisone | AR/androgen biosynthesis inhibitor/glucocorticoid receptor agonist | mCRPC patients | Phase I | Abida et al. [72] |
| ZEN-3694 | BETi | Enzalutamide | ARi | CRPC patients | Phase II | Aggarwal et al. [73] |
| CCS1477 | P300/CBPi | Enzalutamide/Abiraterone/Darolutamide/Olaparib/Atezolizumab | AR/androgen biosynthesis inhibitor/PARPi/IgG1 ab | Advanced solid tumors | Phase I/II | Bono et al. [74] |
| CPI-1205 | EZH2i | Ipilimumab | Anti-CTLA4 ICB | Advanced solid tumors | Phase I/II | ClinicalTrial.gov [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaalberg, A.; Pottendorfer, E.; Zwart, W.; Bergman, A.M. It Takes Two to Tango: The Interplay between Prostate Cancer and Its Microenvironment from an Epigenetic Perspective. Cancers 2024, 16, 294. https://doi.org/10.3390/cancers16020294
Zaalberg A, Pottendorfer E, Zwart W, Bergman AM. It Takes Two to Tango: The Interplay between Prostate Cancer and Its Microenvironment from an Epigenetic Perspective. Cancers. 2024; 16(2):294. https://doi.org/10.3390/cancers16020294
Chicago/Turabian StyleZaalberg, Anniek, Elisabeth Pottendorfer, Wilbert Zwart, and Andries M. Bergman. 2024. "It Takes Two to Tango: The Interplay between Prostate Cancer and Its Microenvironment from an Epigenetic Perspective" Cancers 16, no. 2: 294. https://doi.org/10.3390/cancers16020294
APA StyleZaalberg, A., Pottendorfer, E., Zwart, W., & Bergman, A. M. (2024). It Takes Two to Tango: The Interplay between Prostate Cancer and Its Microenvironment from an Epigenetic Perspective. Cancers, 16(2), 294. https://doi.org/10.3390/cancers16020294






