Exploratory Study Identifies Matrix Metalloproteinase-14 and -9 as Potential Biomarkers of Regorafenib Efficacy in Metastatic Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Patients
2.1.1. STEP I: Preclinical Investigation
2.1.2. STEP II: Clinical Validation
2.2. Analysis of Serum Factor Levels
2.3. Statistical Analysis
3. Results
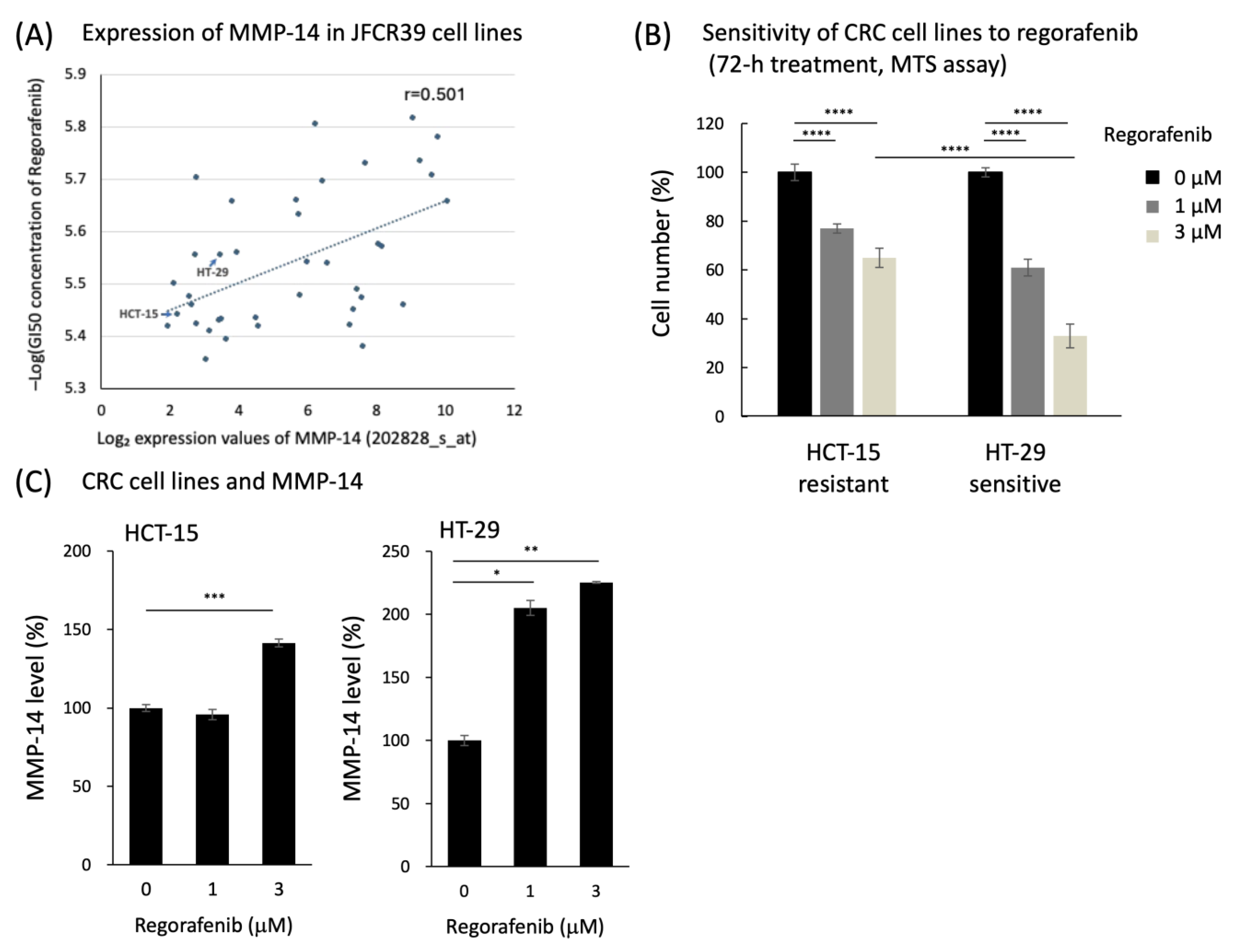
3.1. Preclinical Data Analysis
3.2. Patient and Tumor BL Characteristics
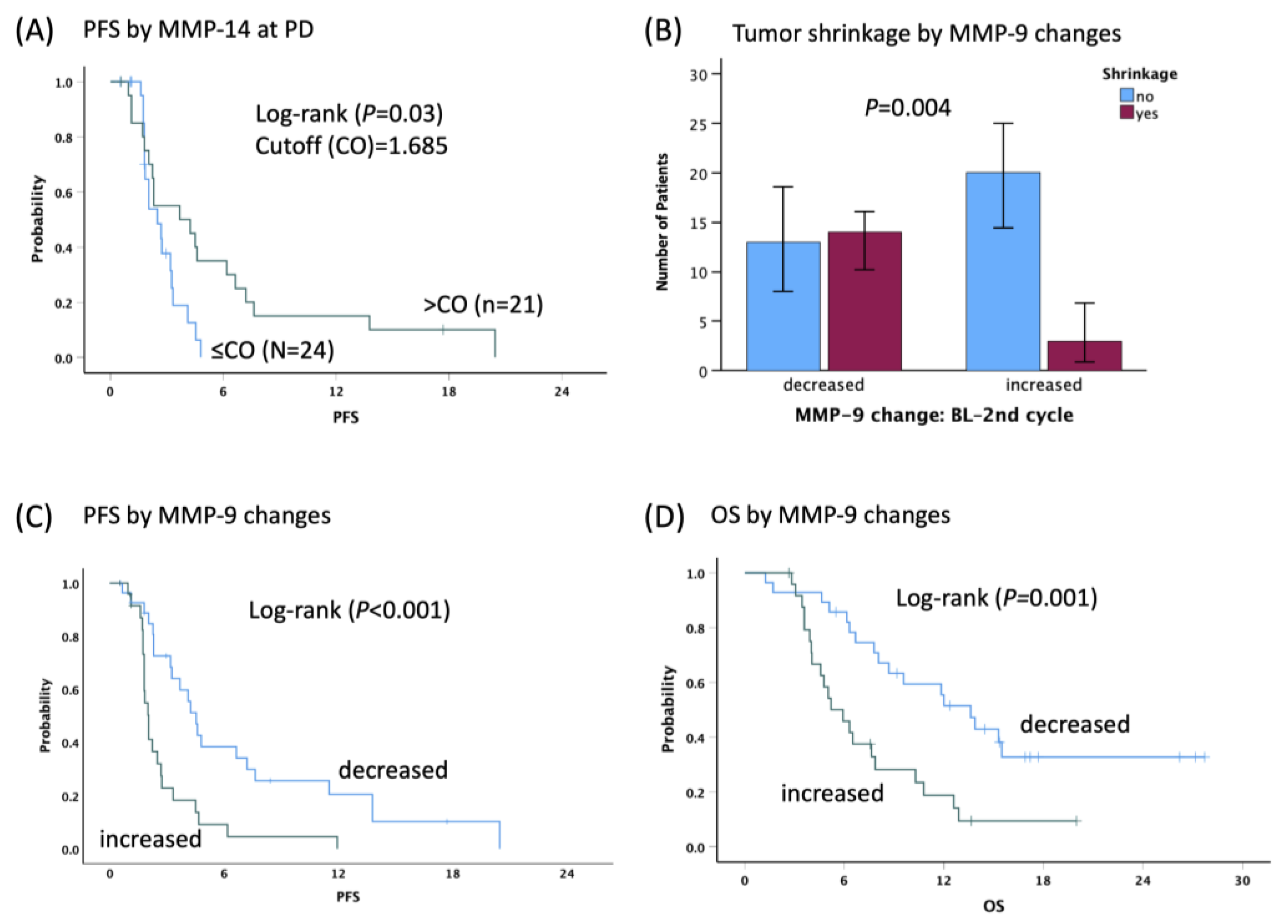
3.3. Association between Serum Factor and Clinical Outcomes
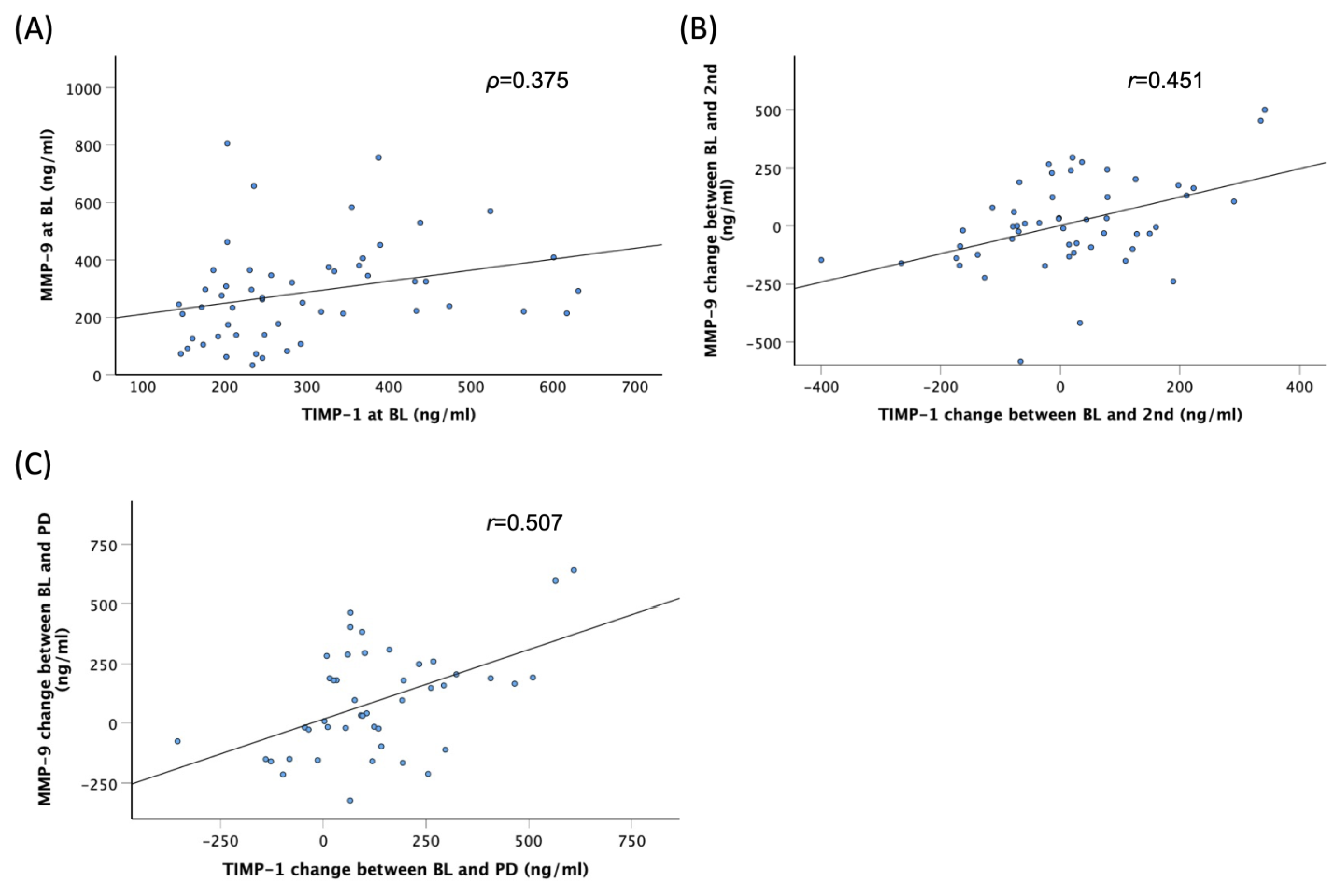
3.4. Correlations among Serum Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grothey, A.; Van Cutsem, E.; Sobrero, A.; Siena, S.; Falcone, A.; Ychou, M.; Humblet, Y.; Bouché, O.; Mineur, L.; Barone, C.; et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): An international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2013, 381, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qin, S.; Xu, R.; Yau, T.C.; Ma, B.; Pan, H.; Xu, J.; Bai, Y.; Chi, Y.; Wang, L.; et al. Regorafenib plus best supportive care versus placebo plus best supportive care in Asian patients with previously treated metastatic colorectal cancer (CONCUR): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2015, 16, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, J.; Lenz, H.J.; Siena, S.; Sobrero, A.; Falcone, A.; Ychou, M.; Humblet, Y.; Bouché, O.; Mineur, L.; Barone, C.; et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: A retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015, 16, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Seiki, M. The cell surface: The stage for matrix metalloproteinase regulation of migration. Curr. Opin. Cell Biol. 2002, 14, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Takino, T.; Okada, Y.; Cao, J.; Shinagawa, A.; Yamamoto, E.; Seiki, M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature 1994, 370, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Apte, S.S.; Soininen, R.; Cao, R.; Baaklini, G.Y.; Rauser, R.W.; Wang, J.; Cao, Y.; Tryggvason, K. Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc. Natl. Acad. Sci. USA 2000, 97, 4052–4057. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Takamura, A.; Ito, N.; Maru, Y.; Sato, H.; Suenaga, N.; Aoki, T.; Seiki, M. Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J. 2001, 20, 4782–4793. [Google Scholar] [CrossRef]
- Kajita, M.; Itoh, Y.; Chiba, T.; Mori, H.; Okada, A.; Kinoh, H.; Seiki, M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 2001, 153, 893–904. [Google Scholar] [CrossRef]
- Bergers, G.; Brekken, R.; McMahon, G.; Vu, T.H.; Itoh, T.; Tamaki, K.; Tanzawa, K.; Thorpe, P.; Itohara, S.; Werb, Z.; et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2000, 2, 737–744. [Google Scholar] [CrossRef]
- Tabouret, E.; Bertucci, F.; Pierga, J.Y.; Petit, T.; Levy, C.; Ferrero, J.M.; Campone, M.; Gligorov, J.; Lerebours, F.; Roché, H.; et al. MMP2 and MMP9 serum levels are associated with favorable outcome in patients with inflammatory breast cancer treated with bevacizumab-based neoadjuvant chemotherapy in the BEVERLY-2 study. Oncotarget 2016, 7, 18531–18540. [Google Scholar] [CrossRef] [PubMed]
- Gialeli, C.; Theocharis, A.D.; Karamanos, N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011, 278, 16–27. [Google Scholar] [CrossRef]
- Dan, S.; Tsunoda, T.; Kitahara, O.; Yanagawa, R.; Zembutsu, H.; Katagiri, T.; Yamazaki, K.; Nakamura, Y.; Yamori, T. An integrated database of chemosensitivity to 55 anticancer drugs and gene expression profiles of 39 human cancer cell lines. Cancer Res. 2002, 62, 1139–1147. [Google Scholar]
- Suenaga, M.; Mashima, T.; Kawata, N.; Wakatsuki, T.; Horiike, Y.; Matsusaka, S.; Dan, S.; Shinozaki, E.; Seimiya, H.; Mizunuma, N.; et al. Serum VEGF-A and CCL5 levels as candidate biomarkers for efficacy and toxicity of regorafenib in patients with metastatic colorectal cancer. Oncotarget 2016, 7, 34811–34823. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Nikolcheva, T.; Krensky, A.M. Transcriptional regulation of RANTES expression in T lymphocytes. Immunol. Rev. 2000, 177, 236–245. [Google Scholar] [CrossRef]
- Song, A.; Patel, A.; Thamatrakoln, K.; Liu, C.; Feng, D.; Clayberger, C.; Krensky, A.M. Functional domains and DNA-binding sequences of RFLAT-1/KLF13, a Krüppel-like transcription factor of activated T lymphocytes. J. Biol. Chem. 2002, 277, 30055–30065. [Google Scholar] [CrossRef]
- Balkwill, F. Cancer and the chemokine network. Nat. Rev. Cancer 2004, 4, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Schlecker, E.; Stojanovic, A.; Eisen, C.; Quack, C.; Falk, C.S.; Umansky, V.; Cerwenka, A. Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth. J. Immunol. 2012, 189, 5602–5611. [Google Scholar] [CrossRef]
- De la Fuente López, M.; Landskron, G.; Parada, D.; Dubois-Camacho, K.; Simian, D.; Martinez, M.; Romero, D.; Roa, J.C.; Chahuán, I.; Gutiérrez, R.; et al. The relationship between chemokines CCL2, CCL3, and CCL4 with the tumor microenvironment and tumor-associated macrophage markers in colorectal cancer. Tumour Biol. 2018, 40, 1010428318810059. [Google Scholar] [CrossRef]
- Suenaga, M.; Schirripa, M.; Cao, S.; Zhang, W.; Yang, D.; Ning, Y.; Cremolini, C.; Antoniotti, C.; Borelli, B.; Mashima, T.; et al. Gene polymorphisms in the CCL5/CCR5 pathway as genetic biomarker for outcome and hand-foot skin reaction in metastatic colorectal cancer patients treated with regorafenib. Clin. Colorectal Cancer 2018, 17, e395–e414. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, M.; Zhang, W.; Mashima, T.; Schirripa, M.; Cao, S.; Okazaki, S.; Berger, M.D.; Miyamoto, Y.; Barzi, A.; Yamaguchi, T.; et al. Potential Molecular Cross Talk Among CCR5 Pathway Predicts Regorafenib Responsiveness in Metastatic Colorectal Cancer Patients. Cancer Genom. Proteom. 2021, 18, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Coussens, L.M.; Fingleton, B.; Matrisian, L.M. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science 2002, 295, 2387–2392. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.M.; Dumas, J.; Adnane, L.; Lynch, M.; Carter, C.A.; Schütz, G.; Thierauch, K.H.; Zopf, D. Regorafenib (BAY 73-4506): A new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer 2011, 129, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Iniesta, P.; Morán, A.; De Juan, C.; Gómez, A.; Hernando, F.; García-Aranda, C.; Frías, C.; Díaz-López, A.; Rodríguez-Jiménez, F.J.; Balibrea, J.L.; et al. Biological and clinical significance of MMP-2, MMP-9, TIMP-1 and TIMP-2 in non-small cell lung cancer. Oncol. Rep. 2007, 17, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Lambert, E.; Dassé, E.; Haye, B.; Petitfrère, E. TIMPs as multifacial proteins. Crit. Rev. Oncol. Hematol. 2004, 49, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.A.; Starodub, A.; Sharma, S.; Berlin, J.; Patel, M.; Wainberg, Z.A.; Chaves, J.; Gordon, M.; Windsor, K.; Brachmann, C.B.; et al. Andecaliximab/GS-5745 Alone and Combined with mFOLFOX6 in Advanced Gastric and Gastroesophageal Junction Adenocarcinoma: Results from a Phase I Study. Clin. Cancer Res. 2018, 24, 3829–3837. [Google Scholar] [CrossRef]
- Yoshikawa, A.K.; Yamaguchi, K.; Muro, K.; Takashima, A.; Ichimura, T.; Sakai, D.; Kadowaki, S.; Chin, K.; Kudo, T.; Mitani, S.; et al. Safety and tolerability of andecaliximab as monotherapy and in combination with an anti-PD-1 antibody in Japanese patients with gastric or gastroesophageal junction adenocarcinoma: A phase 1b study. J. Immunother. Cancer 2022, 10, e003518. [Google Scholar] [CrossRef]

| Cohort | Discovery Cohort (n = 53) | Control Cohort (n = 16) | |||
|---|---|---|---|---|---|
| N | % | N | % | p-Value | |
| Sex | 0.016 | ||||
| Male | 28 | 52.8 | 3 | 18.8 | |
| Female | 25 | 47.2 | 13 | 81.3 | |
| Age (year) | 0.41 | ||||
| Median (range) | 65 (34–78) | 61 (45–77) | |||
| ≤65 | 26 | 49.1 | 10 | 62.5 | |
| >65 | 27 | 50.9 | 6 | 37.5 | |
| ECOG Performance status | 0.99 | ||||
| ECOG 0 | 33 | 62.3 | 10 | 62.5 | |
| ECOG 1 | 20 | 37.7 | 6 | 37.5 | |
| Primary tumor site | 0.019 | ||||
| Right | 37 | 69.8 | 6 | 37.5 | |
| Left | 16 | 30.2 | 10 | 62.5 | |
| Liver metastasis | 0.42 | ||||
| Yes | 34 | 64.2 | 12 | 75 | |
| No | 19 | 35.8 | 4 | 25 | |
| Lung metastasis | 0.042 | ||||
| Yes | 28 | 52.8 | 13 | 81.3 | |
| No | 25 | 47.2 | 3 | 18.8 | |
| Lymph node metastasis | 0.582 | ||||
| Yes | 29 | 54.7 | 10 | 62.5 | |
| No | 24 | 45.3 | 6 | 37.5 | |
| Peritoneal metastasis | 1.0 | ||||
| Yes | 12 | 22.6 | 4 | 25 | |
| No | 41 | 77.4 | 12 | 75 | |
| Number of metastases | 0.052 | ||||
| <2 | 18 | 34.0 | 1 | 6.3 | |
| ≥2 | 35 | 66.0 | 15 | 93.8 | |
| Primary tumor resected | 1.0 | ||||
| Yes | 44 | 83.0 | 14 | 87.5 | |
| No | 9 | 17.0 | 2 | 12.5 | |
| Adjuvant history | 0.145 | ||||
| Yes | 19 | 35.8 | 9 | 56.3 | |
| No | 34 | 64.2 | 7 | 43.8 | |
| MMPs | Point | Non-TS (n = 33) | TS (n = 17) | p-Value * | Non-DC (n = 24) | DC (n = 27) | p-Value * |
|---|---|---|---|---|---|---|---|
| MMP-9, | BL | 272.58 ± 172.63 | 329.29 ± 177.06 | 0.28 | 250.86 ± 131.02 | 330.33 ± 197.98 | 0.10 |
| (mean ± SD, ng/mL) | 2nd | 345.05 ± 217.60 | 231.21 ± 113.50 | 0.019 | 352.49 ± 192.27 | 267.32 ± 188.83 | 0.12 |
| PD | 405.66 ± 228.78 | 354.82 ± 224.69 | 0.51 | 400.84 ± 172.13 | 377.93 ± 267.04 | 0.74 | |
| MMP-9, | BL–2nd | 72.47 ± 177.013 | −98.07 ± 193.79 | 0.003 | 101.63 ± 178.81 | −63.01 ± 180.82 | 0.002 |
| (mean ± SD, ng/mL) | BL–PD | 130.24 ± 209.37 | 29.32 ± 250.31 | 0.18 | 152.01 ± 96.70 | 44.32 ± 237.07 | 0.11 |
| MMP-14, | BL | 1.91 ± 0.81 | 2.66 ± 1.01 | 0.006 | 2.54 ± 2.37 | 2.23 ± 1.08 | 0.55 |
| (mean ± SD, ng/mL) | 2nd | 2.03 ± 0.99 | 2.42 ± 0.73 | 0.16 | 2.02 ± 1.05 | 2.30 ± 0.076 | 0.29 |
| PD | 1.71 ± 0.86 | 2.34 ± 0.57 | 0.02 | 1.71 ± 0.92 | 2.06 ± 0.72 | 0.17 | |
| MMP-14, | BL–2nd | 0.15 ± 0.78 | −0.23 ± 1.24 | 0.19 | −0.52 ± 2.31 | 0.09 ± 1.19 | 0.25 |
| (mean ± SD, ng/mL) | BL–PD | −0.04 ± 0.64 | −0.24 ± 1.0 | 0.44 | −0.75 ± 2.62 | −0.02 ± 0.84 | 0.22 |
| MMP-2, | BL | 345.10 ± 142.75 | 354.43 ± 114.68 | 0.82 | 341.49 ± 143.82 | 359.11 ± 124.09 | 0.64 |
| (mean ± SD, ng/mL) | 2nd | 277.79 ± 138.51 | 289.11 ± 100.37 | 0.77 | 273.89 ± 117.14 | 296.44 ± 137.49 | 0.54 |
| PD | 334.86 ± 120.84 | 295.05 ± 127.09 | 0.34 | 320.36 ± 96.14 | 327.38 ± 143.95 | 0.85 | |
| MMP-2, | BL–2nd | −72.08 ± 144.68 | −65.32 ± 124.85 | 0.87 | −67.60 ± 133.50 | −69.08 ± 140.50 | 0.97 |
| (mean ± SD, ng/mL) | BL–PD | −23.00 ± 134.42 | −45.80 ± 105.37 | 0.59 | −31.22 ± 121.08 | −31.87 ± 129.95 | 0.99 |
| TIMP-1, | BL | 307.79 ± 122.57 | 287.63 ± 151.96 | 0.61 | 303.80 ± 125.73 | 296.75 ± 137.62 | 0.85 |
| (mean ± SD, ng/mL) | 2nd | 358.80157.13300 | 239.22 ± 81.43 | 0.001 | 380.85 ± 164.64 | 254.98 ± 93.39 | 0.002 |
| PD | 464.90194.60728 | 362.91 ± 292.72 | 0.19 | 483.44 ± 218.41 | 377.60 ± 232.69 | 0.13 | |
| TIMP-1, | BL–2nd | 48.84 ± 146.24 | −48.40 ± 127.21 | 0.025 | 77.05 ± 139.07 | −44.01 ± 126.74 | 0.002 |
| (mean ± SD, ng/mL) | BL–PD | 171.08 ± 167.43 | 54.43 ± 232.49 | 0.07 | 197.76 ± 178.05 | 69.04 ± 190.33 | 0.027 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suenaga, M.; Mashima, T.; Kawata, N.; Dan, S.; Seimiya, H.; Yamaguchi, K. Exploratory Study Identifies Matrix Metalloproteinase-14 and -9 as Potential Biomarkers of Regorafenib Efficacy in Metastatic Colorectal Cancer. Cancers 2024, 16, 2855. https://doi.org/10.3390/cancers16162855
Suenaga M, Mashima T, Kawata N, Dan S, Seimiya H, Yamaguchi K. Exploratory Study Identifies Matrix Metalloproteinase-14 and -9 as Potential Biomarkers of Regorafenib Efficacy in Metastatic Colorectal Cancer. Cancers. 2024; 16(16):2855. https://doi.org/10.3390/cancers16162855
Chicago/Turabian StyleSuenaga, Mitsukuni, Tetsuo Mashima, Naomi Kawata, Shingo Dan, Hiroyuki Seimiya, and Kensei Yamaguchi. 2024. "Exploratory Study Identifies Matrix Metalloproteinase-14 and -9 as Potential Biomarkers of Regorafenib Efficacy in Metastatic Colorectal Cancer" Cancers 16, no. 16: 2855. https://doi.org/10.3390/cancers16162855
APA StyleSuenaga, M., Mashima, T., Kawata, N., Dan, S., Seimiya, H., & Yamaguchi, K. (2024). Exploratory Study Identifies Matrix Metalloproteinase-14 and -9 as Potential Biomarkers of Regorafenib Efficacy in Metastatic Colorectal Cancer. Cancers, 16(16), 2855. https://doi.org/10.3390/cancers16162855






