Liver X Receptors Enhance Epithelial to Mesenchymal Transition in Metastatic Prostate Cancer Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Culture and Treatment
2.2. Western Blot Analysis
2.3. RT-qPCR
2.4. Migration and Invasion Assay
2.5. Histological and Immunofluorescence Analysis
2.6. Animals and Tumor Xenografts
2.7. RNA Sequencing Analysis
2.8. Statistical Analysis
3. Results
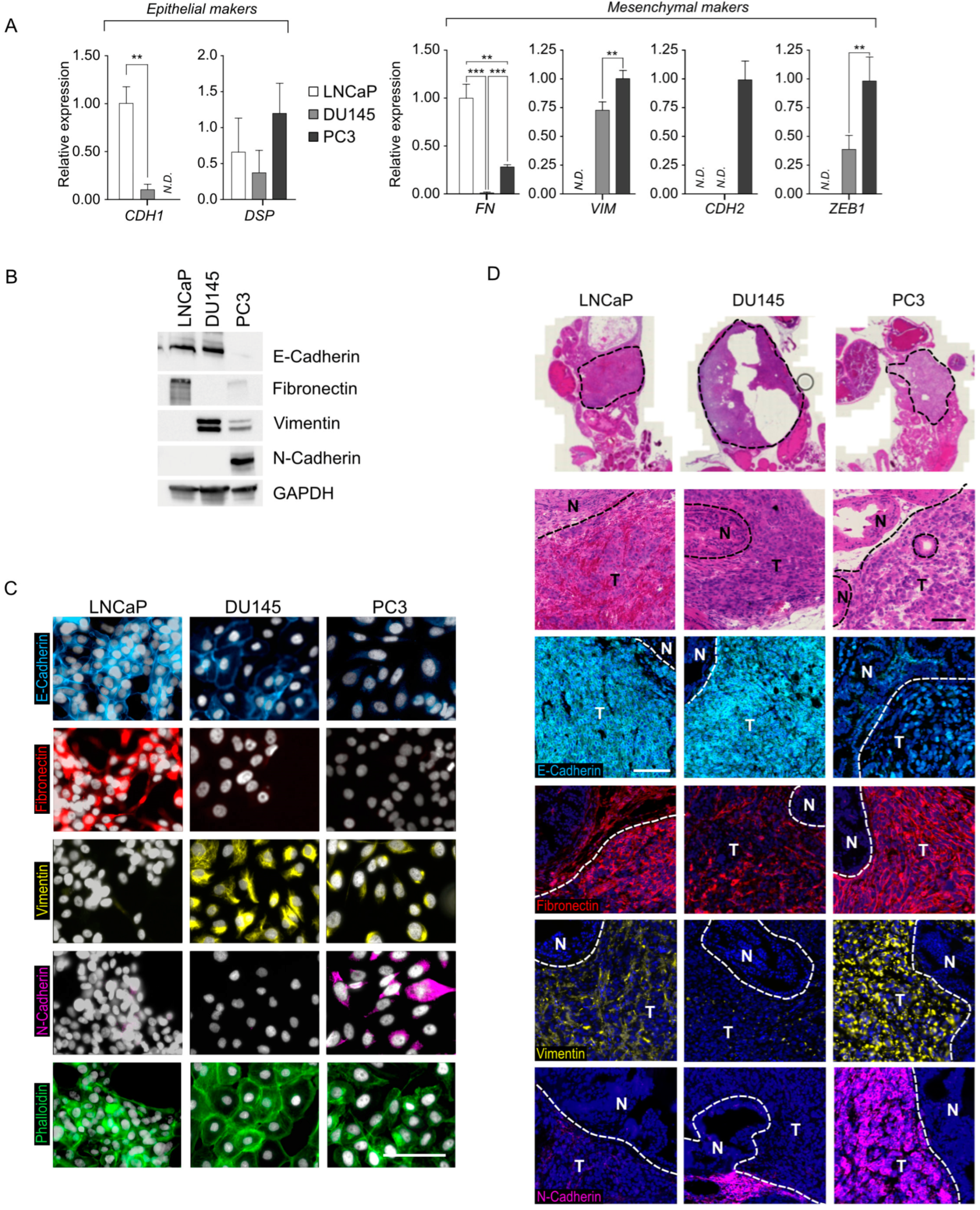
3.1. Characterization of the Prostatic Tumor Cell Lines LNCaP, DU145, and PC3 Reveals Varying Degrees of EMT Status
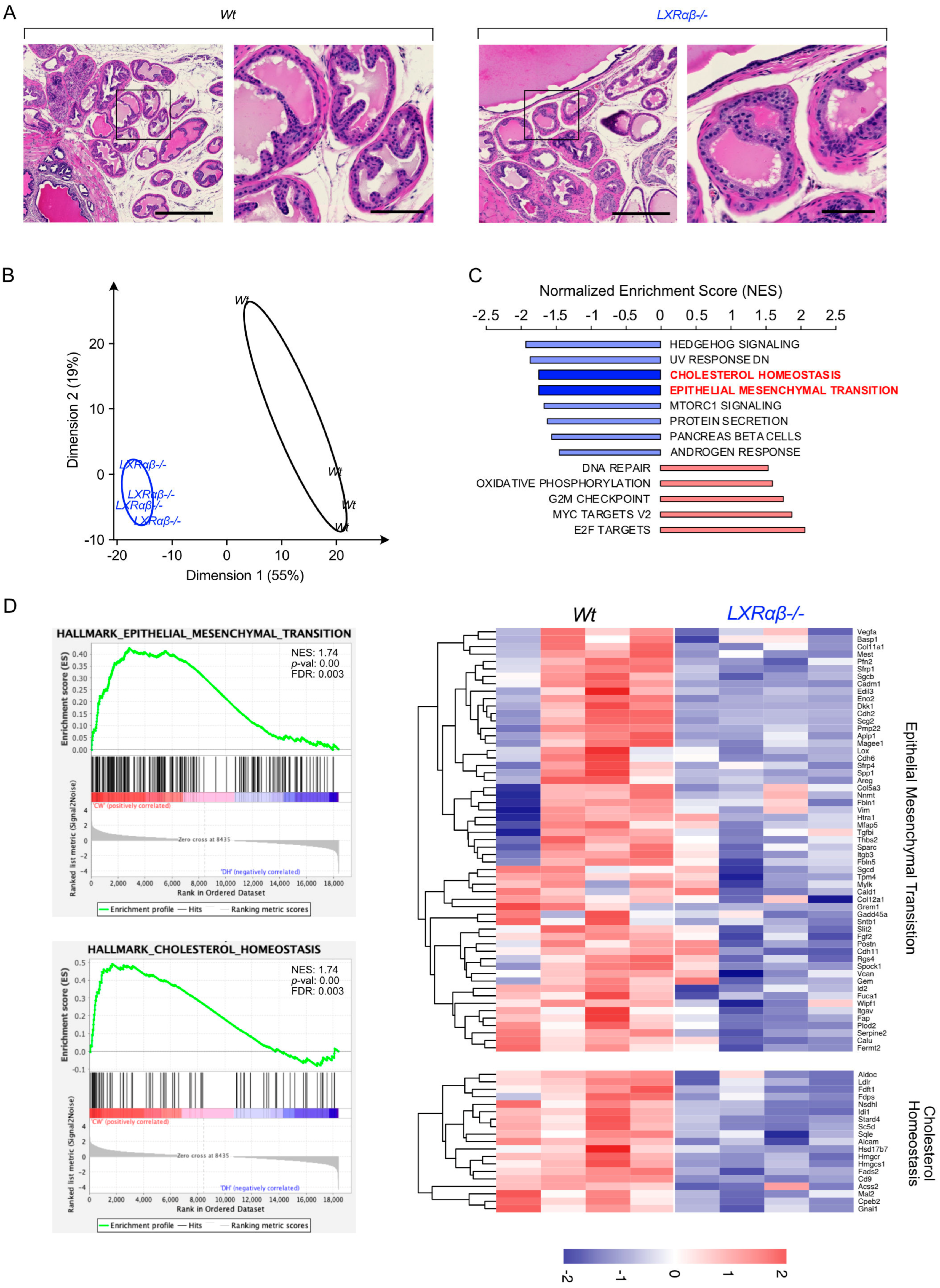
3.2. LXR Signaling Is Functional in Prostate Tumor Cell Lines
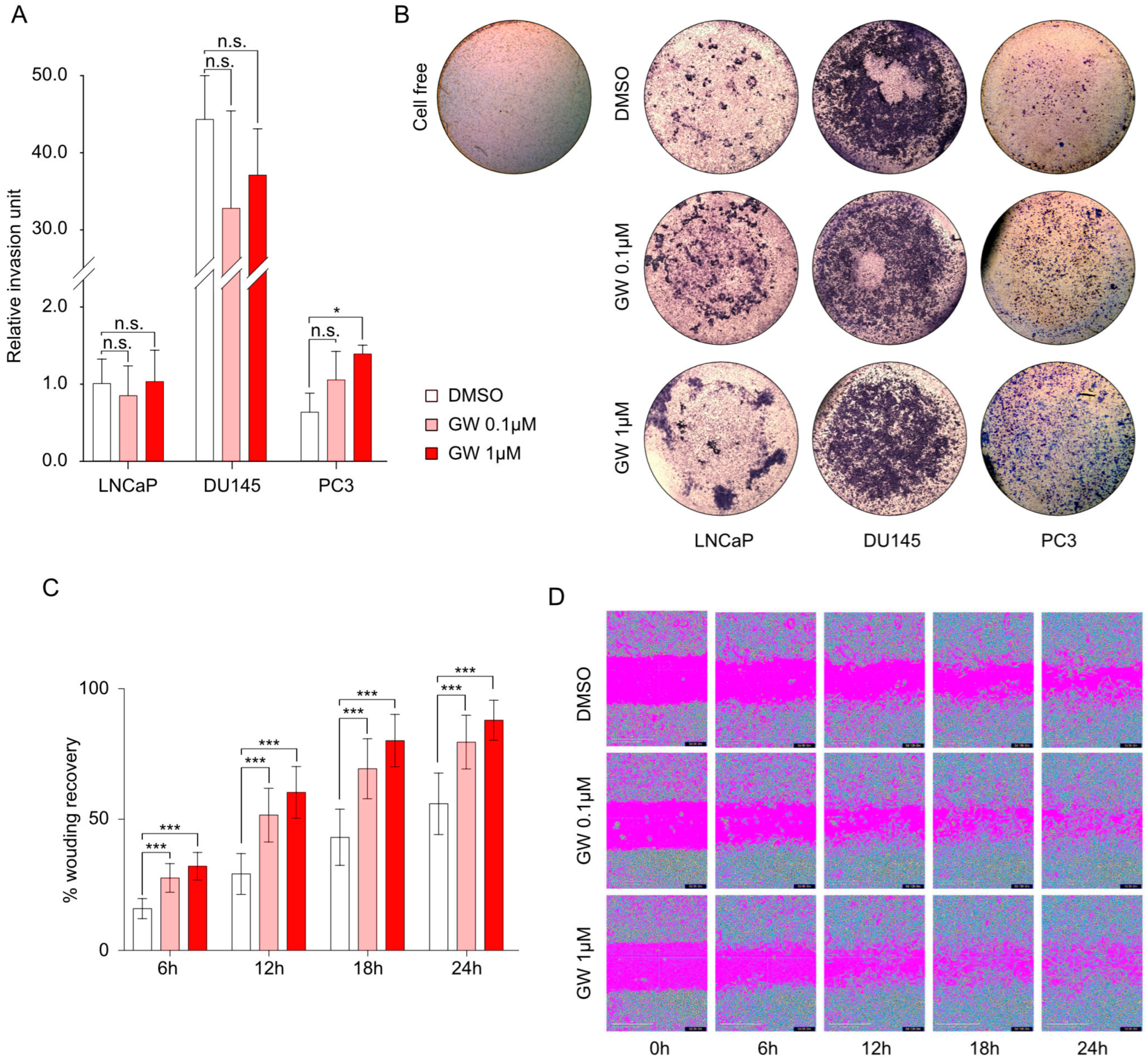
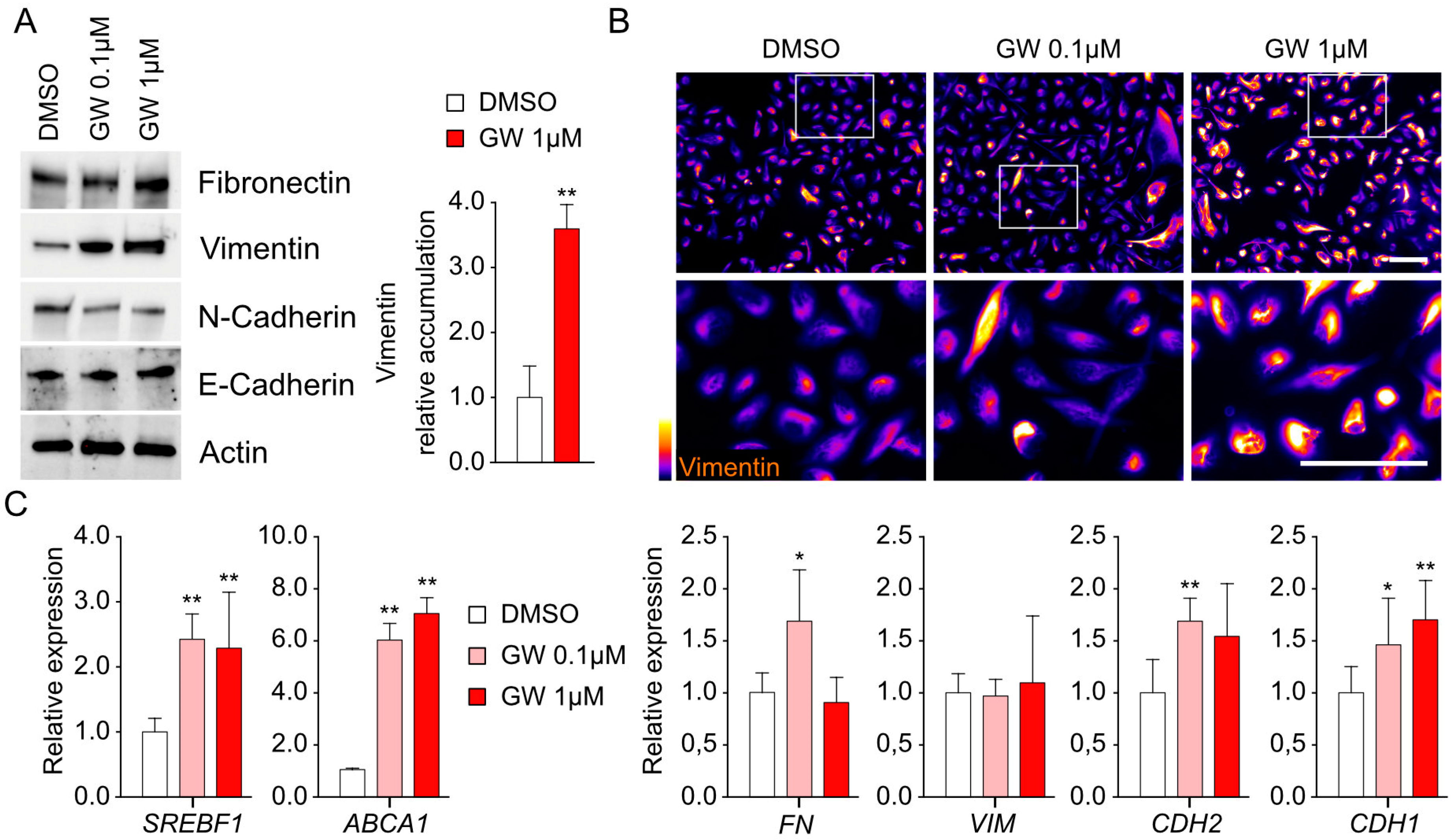
3.3. Migration and Invasiveness Capacities Are Enhanced by LXR Activation and Correlated with Vimentin Accumulation
3.4. Transcriptomic Signature Associated with LXR Stimulation Reveals Amphiregulin-Encoded Gene as a Potential Target
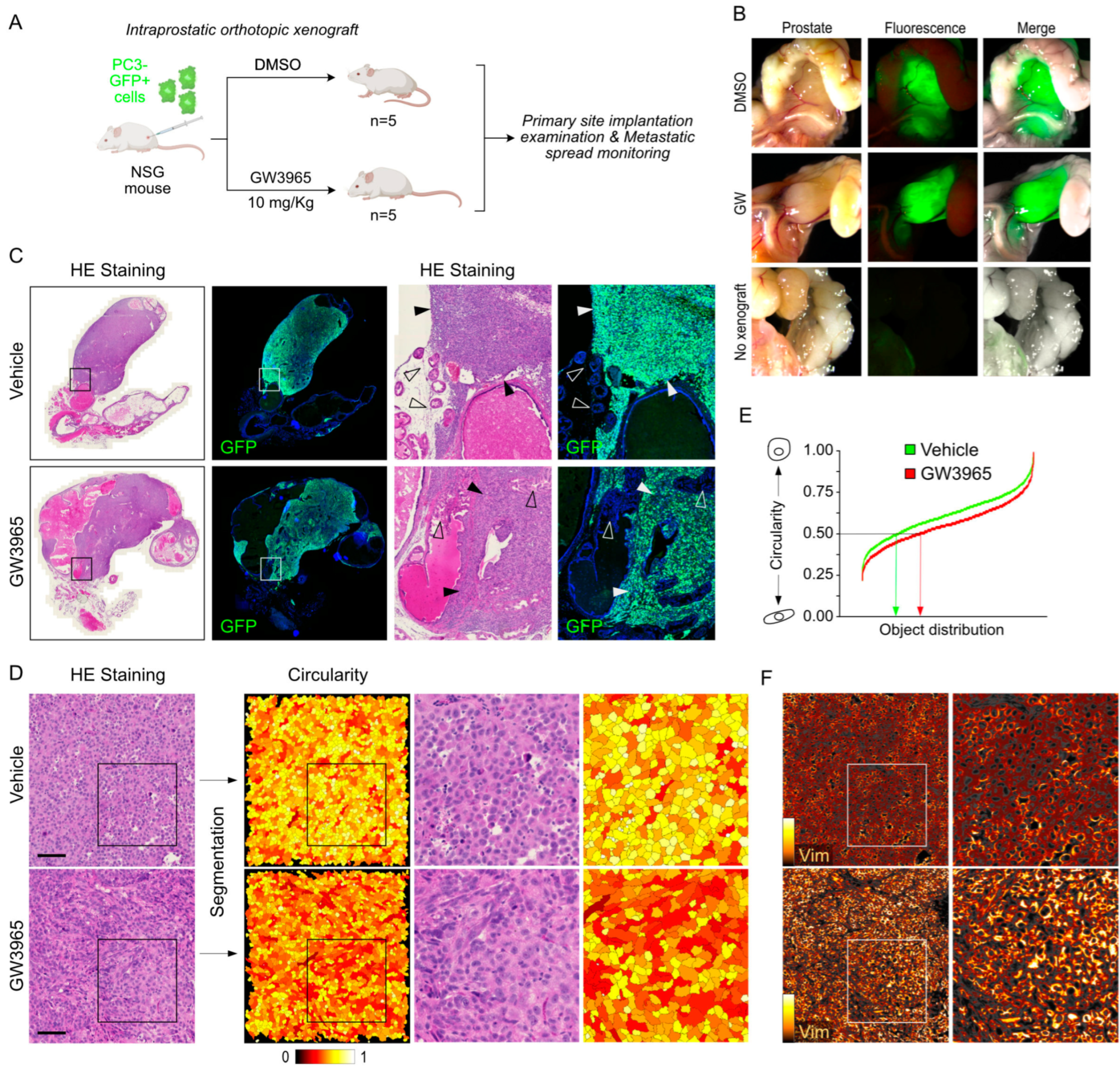
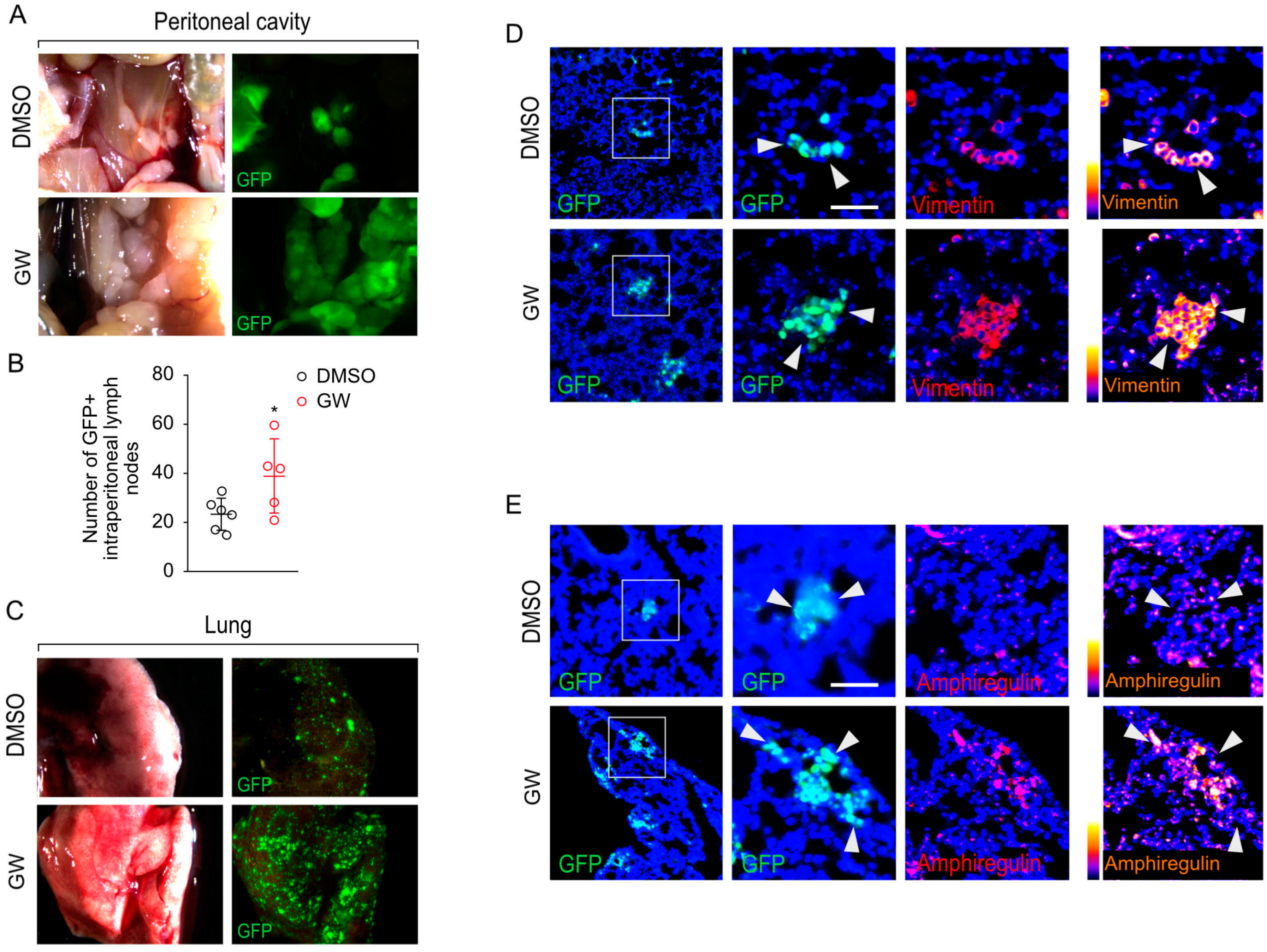
3.5. Pharmacological Targeting of LXR Results in Increased Metastatic Properties of PC3 Cells In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in Cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Mock, K.; Preca, B.-T.; Brummer, T.; Brabletz, S.; Stemmler, M.P.; Brabletz, T. The EMT-Activator ZEB1 Induces Bone Metastasis Associated Genes Including BMP-Inhibitors. Oncotarget 2015, 6, 14399–14412. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Chen, Y.; Song, H.; Chen, L.; Wang, R. Inhibition of ZEB1 Reverses EMT and Chemoresistance in Docetaxel-Resistant Human Lung Adenocarcinoma Cell Line. J. Cell Biochem. 2013, 114, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and Drug Resistance: The Mechanistic Link and Clinical Implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, N.; Denecker, G.; Bruneel, K.; Blancke, G.; Akay, Ö.; Taminau, J.; De Coninck, J.; De Smedt, E.; Skrypek, N.; Van Loocke, W.; et al. The EMT Transcription Factor ZEB2 Promotes Proliferation of Primary and Metastatic Melanoma While Suppressing an Invasive, Mesenchymal-Like Phenotype. Cancer Res. 2020, 80, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Zelcer, N.; Hong, C.; Boyadjian, R.; Tontonoz, P. LXR Regulates Cholesterol Uptake through Idol-Dependent Ubiquitination of the LDL Receptor. Science 2009, 325, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.R.; Tu, H.; Luk, A.; Repa, J.J.; Medina, J.C.; Li, L.; Schwendner, S.; Wang, S.; Thoolen, M.; Mangelsdorf, D.J.; et al. Role of LXRs in Control of Lipogenesis. Genes. Dev. 2000, 14, 2831–2838. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.B.; Bradley, M.N.; Castrillo, A.; Bruhn, K.W.; Mak, P.A.; Pei, L.; Hogenesch, J.; O’connell, R.M.; Cheng, G.; Saez, E.; et al. LXR-Dependent Gene Expression Is Important for Macrophage Survival and the Innate Immune Response. Cell 2004, 119, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Poupeau, A.; Postic, C. Cross-Regulation of Hepatic Glucose Metabolism via ChREBP and Nuclear Receptors. Biochim. Biophys. Acta 2011, 1812, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Viennois, E.; Pommier, A.J.C.; Mouzat, K.; Oumeddour, A.; El Hajjaji, F.-Z.; Dufour, J.; Caira, F.; Volle, D.H.; Baron, S.; Lobaccaro, J.-M.A. Targeting Liver X Receptors in Human Health: Deadlock or Promising Trail? Expert Opin. Ther. Targets 2011, 15, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Shichino, S.; Ueha, S.; Hashimoto, S.; Otsuji, M.; Abe, J.; Tsukui, T.; Deshimaru, S.; Nakajima, T.; Kosugi-Kanaya, M.; Shand, F.H.; et al. Transcriptome Network Analysis Identifies Protective Role of the LXR/SREBP-1c Axis in Murine Pulmonary Fibrosis. JCI Insight 2019, 4, e122163. [Google Scholar] [CrossRef] [PubMed]
- Carthy, J.M.; Stöter, M.; Bellomo, C.; Vanlandewijck, M.; Heldin, A.; Morén, A.; Kardassis, D.; Gahman, T.C.; Shiau, A.K.; Bickle, M.; et al. Chemical Regulators of Epithelial Plasticity Reveal a Nuclear Receptor Pathway Controlling Myofibroblast Differentiation. Sci. Rep. 2016, 6, 29868. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zang, J.; Cao, H.; Wu, Y.; Yan, D.; Qin, X.; Zhou, L.; Fan, F.; Ni, J.; Xu, X.; et al. Liver X Receptors Agonist GW3965 Re-Sensitizes Gefitinib-Resistant Human Non-Small Cell Lung Cancer Cell to Gefitinib Treatment by Inhibiting NF-κB in Vitro. Oncotarget 2017, 8, 15802–15814. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Andersson, L.C.; Bouton, D.; Warner, M.; Gustafsson, J.-A. Stromal Growth and Epithelial Cell Proliferation in Ventral Prostates of Liver X Receptor Knockout Mice. Proc. Natl. Acad. Sci. USA 2009, 106, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, N.; Daigo, Y.; Takano, A.; Taniwaki, M.; Kato, T.; Hayama, S.; Murakami, H.; Takeshima, Y.; Inai, K.; Nishimura, H.; et al. Increases of Amphiregulin and Transforming Growth Factor-Alpha in Serum as Predictors of Poor Response to Gefitinib among Patients with Advanced Non-Small Cell Lung Cancers. Cancer Res. 2005, 65, 9176–9184. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Pommier, A.J.C.; Dufour, J.; Alves, G.; Viennois, E.; De Boussac, H.; Trousson, A.; Volle, D.H.; Caira, F.; Val, P.; Arnaud, P.; et al. Liver x Receptors Protect from Development of Prostatic Intra-Epithelial Neoplasia in Mice. PLoS Genet. 2013, 9, e1003483. [Google Scholar] [CrossRef] [PubMed]
- Pisignano, G.; Napoli, S.; Magistri, M.; Mapelli, S.N.; Pastori, C.; Di Marco, S.; Civenni, G.; Albino, D.; Enriquez, C.; Allegrini, S.; et al. A Promoter-Proximal Transcript Targeted by Genetic Polymorphism Controls E-Cadherin Silencing in Human Cancers. Nat. Commun. 2017, 8, 15622. [Google Scholar] [CrossRef]
- Armstrong, H.K.; Gillis, J.L.; Johnson, I.R.D.; Nassar, Z.D.; Moldovan, M.; Levrier, C.; Sadowski, M.C.; Chin, M.Y.; Tomlinson Guns, E.S.; Tarulli, G.; et al. Dysregulated Fibronectin Trafficking by Hsp90 Inhibition Restricts Prostate Cancer Cell Invasion. Sci. Rep. 2018, 8, 2090. [Google Scholar] [CrossRef]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Sommariva, M.; Limonta, P.; Gagliano, N. Epithelial-To-Mesenchymal Transition Markers and CD44 Isoforms Are Differently Expressed in 2D and 3D Cell Cultures of Prostate Cancer Cells. Cells 2019, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yamada, S. N-Cadherin Dependent Collective Cell Invasion of Prostate Cancer Cells Is Regulated by the N-Terminus of α-Catenin. PLoS ONE 2013, 8, e55069. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.E.; Sakai, S.; Lambert, G.; Nicol, C.J.; Matsusue, K.; Pimprale, S.; Lee, Y.-H.; Ricote, M.; Glass, C.K.; Brewer, H.B.; et al. Conditional Disruption of the Peroxisome Proliferator-Activated Receptor Gamma Gene in Mice Results in Lowered Expression of ABCA1, ABCG1, and apoE in Macrophages and Reduced Cholesterol Efflux. Mol. Cell Biol. 2002, 22, 2607–2619. [Google Scholar] [CrossRef] [PubMed]
- Repa, J.J.; Liang, G.; Ou, J.; Bashmakov, Y.; Lobaccaro, J.M.; Shimomura, I.; Shan, B.; Brown, M.S.; Goldstein, J.L.; Mangelsdorf, D.J. Regulation of Mouse Sterol Regulatory Element-Binding Protein-1c Gene (SREBP-1c) by Oxysterol Receptors, LXRalpha and LXRbeta. Genes. Dev. 2000, 14, 2819–2830. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.; Deb, T.B.; Thompson, S.A.; Wells, A.; Johnson, G.R. A Differential Requirement for the COOH-Terminal Region of the Epidermal Growth Factor (EGF) Receptor in Amphiregulin and EGF Mitogenic Signaling. J. Biol. Chem. 1999, 274, 8900–8909. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.S.; Chauhan, S.B.; Kumar, A.; Kumar, S.; Engwerda, C.R.; Sundar, S.; Kumar, R. Amphiregulin in Cellular Physiology, Health, and Disease: Potential Use as a Biomarker and Therapeutic Target. J. Cell Physiol. 2022, 237, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Berasain, C.; Avila, M.A. The EGFR Signalling System in the Liver: From Hepatoprotection to Hepatocarcinogenesis. J. Gastroenterol. 2014, 49, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Zhang, H.; Lu, J.; Zhang, Z.; Wu, H.; Liang, Z. AREG Mediates the Epithelial-mesenchymal Transition in Pancreatic Cancer Cells via the EGFR/ERK/NF-κB Signalling Pathway. Oncol. Rep. 2020, 43, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Soukupova, J.; Malfettone, A.; Bertran, E.; Hernández-Alvarez, M.I.; Peñuelas-Haro, I.; Dituri, F.; Giannelli, G.; Zorzano, A.; Fabregat, I. Epithelial-Mesenchymal Transition (EMT) Induced by TGF-β in Hepatocellular Carcinoma Cells Reprograms Lipid Metabolism. Int. J. Mol. Sci. 2021, 22, 5543. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wang, X.; Song, D.; Liu, X.; Gu, Y.; Xu, Z.; Wang, X.; Zhang, X.; Ye, Q.; Tong, Z.; et al. Cholesterol Induces Epithelial-to-Mesenchymal Transition of Prostate Cancer Cells by Suppressing Degradation of EGFR through APMAP. Cancer Res. 2019, 79, 3063–3075. [Google Scholar] [CrossRef] [PubMed]
- Viennois, E.; Esposito, T.; Dufour, J.; Pommier, A.; Fabre, S.; Kemeny, J.-L.; Guy, L.; Morel, L.; Lobaccaro, J.-M.; Baron, S. Lxrα Regulates the Androgen Response in Prostate Epithelium. Endocrinology 2012, 153, 3211–3223. [Google Scholar] [CrossRef] [PubMed]
- Bousset, L.; Septier, A.; Bunay, J.; Voisin, A.; Guiton, R.; Damon-Soubeyrant, C.; Renaud, Y.; De Haze, A.; Sapin, V.; Fogli, A.; et al. Absence of Nuclear Receptors LXRs Impairs Immune Response to Androgen Deprivation and Leads to Prostate Neoplasia. PLoS Biol. 2020, 18, e3000948. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Zhang, B.; Zhao, G. Liver X Receptor α (LXRα) Promoted Invasion and EMT of Gastric Cancer Cells by Regulation of NF-κB Activity. Hum. Cell 2017, 30, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Kosako, H.; Tanabe, K.; Yanagida, M.; Sakurai, M.; Amano, M.; Kaibuchi, K.; Inagaki, M. Phosphorylation of Vimentin by Rho-Associated Kinase at a Unique Amino-Terminal Site That Is Specifically Phosphorylated during Cytokinesis. J. Biol. Chem. 1998, 273, 11728–11736. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Yasui, Y.; Kawajiri, A.; Nigg, E.A.; Terada, Y.; Tatsuka, M.; Nagata, K.; Inagaki, M. Aurora-B Regulates the Cleavage Furrow-Specific Vimentin Phosphorylation in the Cytokinetic Process. J. Biol. Chem. 2003, 278, 8526–8530. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.E.; He, T.; Trejo-Skalli, A.V.; Härmälä-Braskén, A.-S.; Hellman, J.; Chou, Y.-H.; Goldman, R.D. Specific in Vivo Phosphorylation Sites Determine the Assembly Dynamics of Vimentin Intermediate Filaments. J. Cell Sci. 2004, 117, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; McManus, F.P.; Plutoni, C.; Pascariu, C.M.; Nelson, T.; Alberici Delsin, L.E.; Emery, G.; Thibault, P. Quantitative SUMO Proteomics Identifies PIAS1 Substrates Involved in Cell Migration and Motility. Nat. Commun. 2020, 11, 834. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.; Park, J.; Ahn, S.G.; Lee, J.; Park, Y.; Ooshima, A.; Mizuno, S.; Yamashita, S.; Park, K.-S.; Lee, S.-Y.; et al. RNF208, an Estrogen-Inducible E3 Ligase, Targets Soluble Vimentin to Suppress Metastasis in Triple-Negative Breast Cancers. Nat. Commun. 2019, 10, 5805. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Shimano, H.; Inoue, N.; Nakagawa, Y.; Matsuzaka, T.; Takahashi, A.; Yahagi, N.; Sone, H.; Suzuki, H.; Toyoshima, H.; et al. Protein Kinase A Suppresses Sterol Regulatory Element-Binding Protein-1C Expression via Phosphorylation of Liver X Receptor in the Liver. J. Biol. Chem. 2007, 282, 11687–11695. [Google Scholar] [CrossRef] [PubMed]
- Cook, P.W.; Mattox, P.A.; Keeble, W.W.; Pittelkow, M.R.; Plowman, G.D.; Shoyab, M.; Adelman, J.P.; Shipley, G.D. A Heparin Sulfate-Regulated Human Keratinocyte Autocrine Factor Is Similar or Identical to Amphiregulin. Mol. Cell. Biol. 1991, 11, 2547–2557. [Google Scholar] [CrossRef] [PubMed]
- Skiriutė, D.; Vaitkienė, P.; Ašmonienė, V.; Steponaitis, G.; Deltuva, V.P.; Tamašauskas, A. Promoter Methylation of AREG, HOXA11, hMLH1, NDRG2, NPTX2 and Tes Genes in Glioblastoma. J. Neurooncol. 2013, 113, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Busser, B.; Sancey, L.; Brambilla, E.; Coll, J.-L.; Hurbin, A. The Multiple Roles of Amphiregulin in Human Cancer. Biochim. Biophys. Acta 2011, 1816, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Panupinthu, N.; Yu, S.; Zhang, D.; Zhang, F.; Gagea, M.; Lu, Y.; Grandis, J.R.; Dunn, S.E.; Lee, H.Y.; Mills, G.B. Self-Reinforcing Loop of Amphiregulin and Y-Box Binding Protein-1 Contributes to Poor Outcomes in Ovarian Cancer. Oncogene 2014, 33, 2846–2856. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Ying, M.; Chen, G.; Lin, A.; Xie, Y.; Ohara, N.; Zhou, D. ADAM17 Is Associated with EMMPRIN and Predicts Poor Prognosis in Patients with Uterine Cervical Carcinoma. Tumour Biol. 2014, 35, 7575–7586. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-W.; Kim, D.K.; Min, A.; Lee, K.-H.; Nam, H.-J.; Kim, J.H.; Kim, J.-S.; Kim, T.-Y.; Im, S.-A.; Park, I.A. Amphiregulin Confers Trastuzumab Resistance via AKT and ERK Activation in HER2-Positive Breast Cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, H.; Wang, L.; Lu, J.; Duan, H.; Liu, X.; Liang, Z. Expression of Amphiregulin Predicts Poor Outcome in Patients with Pancreatic Ductal Adenocarcinoma. Diagn. Pathol. 2016, 11, 60. [Google Scholar] [CrossRef]
- Tørring, N.; Hansen, F.D.; Sørensen, B.S.; Ørntoft, T.F.; Nexø, E. Increase in Amphiregulin and Epiregulin in Prostate Cancer Xenograft after Androgen Deprivation-Impact of Specific HER1 Inhibition. Prostate 2005, 64, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Long, Q.; Zhu, D.; Fu, D.; Zhang, B.; Han, L.; Qian, M.; Guo, J.; Xu, J.; Cao, L.; et al. Targeting Amphiregulin (AREG) Derived from Senescent Stromal Cells Diminishes Cancer Resistance and Averts Programmed Cell Death 1 Ligand (PD-L1)-Mediated Immunosuppression. Aging Cell 2019, 18, e13027. [Google Scholar] [CrossRef] [PubMed]


| Antigen | Source | WB Working Dilution | IF Working Dilution | Reference | Manufacturer |
|---|---|---|---|---|---|
| Fibronectin | Rabbit | 1/1000 | 1/200 | Ab2413 | Abcam Cambridge UK |
| Vimentin | Rabbit | 1/1000 | 1/200 | D21H3 | Cell Signaling Technology, Ozyme, Saint-Cyr- l’Ecole, France |
| N-Cadherin | Rabbit | 1/1000 | 1/200 | D4R1H | Cell Signaling Technology |
| E-Cadherin | Rabbit | 1/1000 | 1/200 | 610182 | BD Transduction Laboratories, Le Pont de Claix, France |
| Amphiregulin | Rabbit | 1/1000 | 1/200 | 16036-1-AP | Proteintech, Stockport, UK |
| GAPDH | Mouse | 1/10,000 | - | D4C6R | Cell Signaling Technology |
| GFP | Rabbit | - | 1/1000 | A11122 | Invitrogen, Villebon Sur Yvette, France |
| Actin | Rabbit | 1/10,000 | - | A2066 | Sigma-Aldrich, Saint-Quentin-Fallavier, France |
| Gene | Primer Sequence Forward | Primer Sequence Reverse |
|---|---|---|
| CDH2 | TGCCCTCAATGGGATCTTGA | AGGCCATATGTGGGATTGCC |
| VIM | AGAGGAAGCCGAAAACACCC | TTGCAAAGATTCCACTTTGCGT |
| FN | TCCCTCGGAACATCAGAAAC | CAGTGGGAGACCTCGAGAAG |
| SNAI1 | CTCTAGGCCCTGGCTGCTAC | GCTTGTGGAGCAGGGACATT |
| SNAI2 | TTGTGTTTGCAAGATCTGCGG | GCAAATGCTCTGTTGCAGTGA |
| ZEB1 | ATCCTGGGGCCTGAAGCT | TGGTGTGCCCTGCCTCTGGT |
| CDH1 | ATGAGTGTCCCCCGGTATCT | ACGAGCAGAGAATCATAAGGGGCG |
| DSP | GATCACCGACCAGAACTCGG | AAGTTCTGCACCTGACGCTT |
| ABCG1 | CAGGAAGATTAGACACTGTG | GAAAGGGGAATGGAGAGAAG |
| SREBF1 | GGAGGGGTAGGGCCAACGGCCT | CATGTCTTCGAAAGTGCAATCC |
| NR1H3 | AGGAGTGTCGGCTTCGCAAA | CTCTTCTTGCCGCTTCAGTTT |
| NR1H2 | AACAAACGCTCCTTCTCCGA | GGTGATACACTCTGTCTCGT |
| AREG | TCCCCTGTGAGTGAAATGCC | GTTACTGCTTCCAGGTGCTCT |
| ENS | Gene Name | Base Mean | log2FoldChange | FC | lfcSE | Stat | p-Value | Padj |
|---|---|---|---|---|---|---|---|---|
| ENSG00000120708 | TGFBI | 0.55747659 | 1.78535814 | 3.44704023 | 1.90412944 | 0.93762436 | 0.34843749 | NA |
| ENSG00000109321 | AREG | 490.424353 | 1.16715752 | 2.24568803 | 0.10414778 | 11.2067441 | 3.78 × 10−29 | 9.96 × 10−26 |
| ENSG00000140092 | FBLN5 | 13.0413403 | 0.39755778 | 1.31727611 | 0.38800452 | 1.02462151 | 0.30554181 | NA |
| ENSG00000115935 | WIPF1 | 14.8540542 | 0.3936751 | 1.31373574 | 0.36984887 | 1.06442154 | 0.28713778 | NA |
| ENSG00000115738 | ID2 | 633.907513 | 0.29972602 | 1.23091063 | 0.14101883 | 2.12543265 | 0.03355053 | 0.61122854 |
| ENSG00000166923 | GREM1 | 13.0693234 | 0.29423341 | 1.22623323 | 0.40445516 | 0.72748092 | 0.4669314 | NA |
| ENSG00000135919 | SERPINE2 | 2599.06039 | 0.28987312 | 1.22253275 | 0.08467547 | 3.42334244 | 0.00061856 | 0.05965824 |
| ENSG00000167460 | TPM4 | 4732.16738 | 0.27491589 | 1.20992355 | 0.07462908 | 3.6837634 | 0.00022982 | 0.02818358 |
| ENSG00000117152 | RGS4 | 90.6207624 | 0.259963 | 1.19744799 | 0.18138207 | 1.43323428 | 0.1517909 | 0.90553331 |
| ENSG00000164949 | GEM | 331.016875 | 0.25879656 | 1.19648023 | 0.1070989 | 2.4164259 | 0.01567372 | 0.45082373 |
| ENSG00000085063 | CD59 | 18866.2581 | 0.25583504 | 1.19402665 | 0.06778507 | 3.77420929 | 0.00016052 | 0.02069425 |
| ENSG00000140937 | CDH11 | 7637.94552 | 0.25487472 | 1.19323212 | 0.10960447 | 2.32540447 | 0.02005034 | 0.48799449 |
| ENSG00000122786 | CALD1 | 1470.33888 | 0.21993651 | 1.16468233 | 0.12820702 | 1.71547945 | 0.08625736 | 0.80643802 |
| ENSG00000104332 | SFRP1 | 25.8588417 | 0.21976976 | 1.16454772 | 0.26265947 | 0.83670983 | 0.40275569 | NA |
| ENSG00000176788 | BASP1 | 2800.41858 | 0.21759661 | 1.16279487 | 0.11227704 | 1.93803304 | 0.05261919 | 0.70623589 |
| ENSG00000170558 | CDH2 | 4236.84196 | 0.19929013 | 1.14813328 | 0.1124252 | 1.77264635 | 0.07628733 | 0.7912538 |
| ENSG00000065534 | MYLK | 1140.93967 | 0.19162924 | 1.14205271 | 0.09693455 | 1.9768931 | 0.04805372 | 0.68797265 |
| ENSG00000026025 | VIM | 12641.7501 | 0.16067385 | 1.11780912 | 0.08217763 | 1.95520179 | 0.05055926 | 0.70112503 |
| ENSG00000166741 | NNMT | 1964.88143 | 0.14971687 | 1.10935174 | 0.10141317 | 1.47630602 | 0.13986176 | 0.8928307 |
| ENSG00000073712 | FERMT2 | 4345.12245 | 0.14738122 | 1.10755721 | 0.05953327 | 2.47561126 | 0.01330083 | 0.41427885 |
| ENSG00000145147 | SLIT2 | 2457.46308 | 0.12358147 | 1.08943602 | 0.11657471 | 1.06010534 | 0.28909668 | 0.96733795 |
| ENSG00000128595 | CALU | 7375.79195 | 0.11542364 | 1.0832931 | 0.07137564 | 1.61712924 | 0.10585039 | 0.85045867 |
| ENSG00000112715 | VEGFA | 1458.65656 | 0.10757865 | 1.07741843 | 0.0978457 | 1.0994725 | 0.27156202 | 0.96186384 |
| ENSG00000163069 | SGCB | 2110.832 | 0.09116566 | 1.06523051 | 0.07435268 | 1.22612481 | 0.22015171 | 0.94409558 |
| ENSG00000113140 | SPARC | 7116.58414 | 0.06749718 | 1.04789719 | 0.3734534 | 0.1807379 | 0.85657331 | 0.99872891 |
| ENSG00000164176 | EDIL3 | 3250.12928 | 0.04869492 | 1.03432883 | 0.10049241 | 0.48456319 | 0.62798623 | 0.99872891 |
| ENSG00000111799 | COL12A1 | 5108.27642 | 0.04519455 | 1.03182231 | 0.08228866 | 0.54921966 | 0.58285471 | 0.99872891 |
| ENSG00000106483 | SFRP4 | 3.74887888 | −0.1991432 | 0.8710677 | 0.79958454 | −0.2490584 | 0.80331561 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouchareb, E.; Dallel, S.; De Haze, A.; Damon-Soubeyrand, C.; Renaud, Y.; Baabdaty, E.; Vialat, M.; Fabre, J.; Pouchin, P.; De Joussineau, C.; et al. Liver X Receptors Enhance Epithelial to Mesenchymal Transition in Metastatic Prostate Cancer Cells. Cancers 2024, 16, 2776. https://doi.org/10.3390/cancers16162776
Bouchareb E, Dallel S, De Haze A, Damon-Soubeyrand C, Renaud Y, Baabdaty E, Vialat M, Fabre J, Pouchin P, De Joussineau C, et al. Liver X Receptors Enhance Epithelial to Mesenchymal Transition in Metastatic Prostate Cancer Cells. Cancers. 2024; 16(16):2776. https://doi.org/10.3390/cancers16162776
Chicago/Turabian StyleBouchareb, Erwan, Sarah Dallel, Angélique De Haze, Christelle Damon-Soubeyrand, Yoan Renaud, Elissa Baabdaty, Marine Vialat, Julien Fabre, Pierre Pouchin, Cyrille De Joussineau, and et al. 2024. "Liver X Receptors Enhance Epithelial to Mesenchymal Transition in Metastatic Prostate Cancer Cells" Cancers 16, no. 16: 2776. https://doi.org/10.3390/cancers16162776
APA StyleBouchareb, E., Dallel, S., De Haze, A., Damon-Soubeyrand, C., Renaud, Y., Baabdaty, E., Vialat, M., Fabre, J., Pouchin, P., De Joussineau, C., Degoul, F., Sanmukh, S., Gendronneau, J., Sanchez, P., Gonthier-Gueret, C., Trousson, A., Morel, L., Lobaccaro, J. M., Kocer, A., & Baron, S. (2024). Liver X Receptors Enhance Epithelial to Mesenchymal Transition in Metastatic Prostate Cancer Cells. Cancers, 16(16), 2776. https://doi.org/10.3390/cancers16162776







