Sex- and Age-Associated Differences in Genomic Alterations among Patients with Advanced Non-Small Cell Lung Cancer (NSCLC)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Genomic Profile Differences in Advanced NSCLC by Sex
3.2. Genomic Profile Differences in Advanced NSCLC by Age
3.3. Sex and Age Differences in Advanced NSCLC
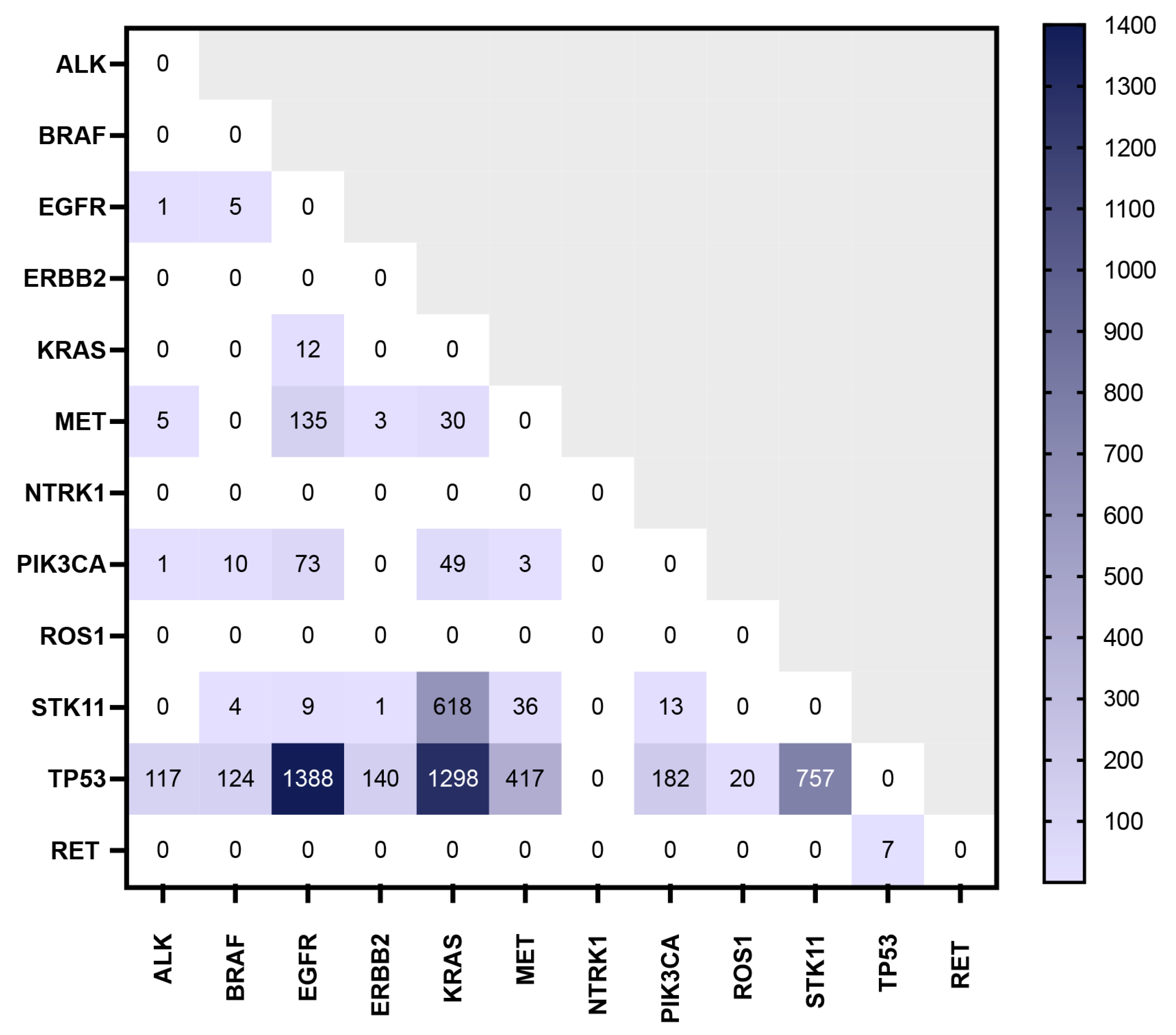
3.4. Co-Occurrence of Mutations of Interest
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- US Department of Health and Human Services, Centers for Disease Control and Prevention, Division of Cancer Prevention and Control. An Update on Cancer Deaths in the United States; US Department of Health and Human Services, Centers for Disease Control and Prevention, Division of Cancer Prevention and Control: Atlanta, GA, USA, 2022.
- National Cancer Institute. SEER Cancer Stat Facts: Lung and Bronchus Cancer; National Cancer Institute: Bethesda, MD, USA, 2024. Available online: https://seer.cancer.gov/statfacts/html/lungb.html (accessed on 9 January 2024).
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Sacher, A.G.; Dahlberg, S.E.; Heng, J.; Mach, S.; Janne, P.A.; Oxnard, G.R. Association between Younger Age and Targetable Genomic Alterations and Prognosis in Non-Small-Cell Lung Cancer. JAMA Oncol. 2016, 2, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. J. Exp. Clin. Cancer Res. 2020, 39, 95. [Google Scholar] [CrossRef] [PubMed]
- Bonanno, L.; Dal Maso, A.; Pavan, A.; Zulato, E.; Calvetti, L.; Pasello, G.; Guarneri, V.; Conte, P.; Indraccolo, S. Liquid biopsy and non-small cell lung cancer: Are we looking at the tip of the iceberg? Br. J. Cancer 2022, 127, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Kasi, P.M.; Lee, J.K.; Pasquina, L.W.; Decker, B.; Vanden Borre, P.; Pavlick, D.C.; Allen, J.M.; Parachoniak, C.; Quintanilha, J.C.F.; Graf, R.P.; et al. Circulating Tumor DNA Enables Sensitive Detection of Actionable Gene Fusions and Rearrangements Across Cancer Types. Clin. Cancer Res. 2024, 30, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Hancock, B.A.; Solzak, J.P.; Radovich, M. Abstract 2280: Co-detection of circulating tumor DNA and RNA in the plasma of patients with breast cancer increases the detectable number of mutated molecules. Cancer Res. 2019, 79, 2280. [Google Scholar] [CrossRef]
- Swanton, C.; Govindan, R. Clinical Implications of Genomic Discoveries in Lung Cancer. N. Engl. J. Med. 2016, 374, 1864–1873. [Google Scholar] [CrossRef] [PubMed]
- Dang, A.H.; Tran, V.U.; Tran, T.T.; Thi Pham, H.A.; Le, D.T.; Nguyen, L.; Nguyen, N.V.; Thi Nguyen, T.H.; Nguyen, C.V.; Le, H.T.; et al. Actionable Mutation Profiles of Non-Small Cell Lung Cancer patients from Vietnamese population. Sci. Rep. 2020, 10, 2707. [Google Scholar] [CrossRef]
- Xiao, D.; Pan, H.; Li, F.; Wu, K.; Zhang, X.; He, J. Analysis of ultra-deep targeted sequencing reveals mutation burden is associated with gender and clinical outcome in lung adenocarcinoma. Oncotarget 2016, 7, 22857–22864. [Google Scholar] [CrossRef]
- Hsu, C.L.; Chen, K.Y.; Shih, J.Y.; Ho, C.C.; Yang, C.H.; Yu, C.J.; Yang, P.C. Advanced non-small cell lung cancer in patients aged 45 years or younger: Outcomes and prognostic factors. BMC Cancer 2012, 12, 241. [Google Scholar] [CrossRef]
- Wakelee, H.A.; Wang, W.; Schiller, J.H.; Langer, C.J.; Sandler, A.B.; Belani, C.P.; Johnson, D.H.; Eastern Cooperative Oncology Group. Survival differences by sex for patients with advanced non-small cell lung cancer on Eastern Cooperative Oncology Group trial 1594. J. Thorac. Oncol. 2006, 1, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Kasi, P.M.; Fehringer, G.; Taniguchi, H.; Starling, N.; Nakamura, Y.; Kotani, D.; Powles, T.; Li, B.T.; Pusztai, L.; Aushev, V.N.; et al. Impact of Circulating Tumor DNA-Based Detection of Molecular Residual Disease on the Conduct and Design of Clinical Trials for Solid Tumors. JCO Precis. Oncol. 2022, 6, e2100181. [Google Scholar] [CrossRef]
- Deveson, I.W.; Gong, B.; Lai, K.; LoCoco, J.S.; Richmond, T.A.; Schageman, J.; Zhang, Z.; Novoradovskaya, N.; Willey, J.C.; Jones, W.; et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat. Biotechnol. 2021, 39, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Politi, K.; Herbst, R.S. Lung cancer in the era of precision medicine. Clin. Cancer Res. 2015, 21, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef]
- Suh, J.H.; Johnson, A.; Albacker, L.; Wang, K.; Chmielecki, J.; Frampton, G.; Gay, L.; Elvin, J.A.; Vergilio, J.A.; Ali, S.; et al. Comprehensive Genomic Profiling Facilitates Implementation of the National Comprehensive Cancer Network Guidelines for Lung Cancer Biomarker Testing and Identifies Patients Who May Benefit from Enrollment in Mechanism-Driven Clinical Trials. Oncologist 2016, 21, 684–691. [Google Scholar] [CrossRef]
- Pavan, A.; Bragadin, A.B.; Calvetti, L.; Ferro, A.; Zulato, E.; Attili, I.; Nardo, G.; Dal Maso, A.; Frega, S.; Menin, A.G.; et al. Role of next generation sequencing-based liquid biopsy in advanced non-small cell lung cancer patients treated with immune checkpoint inhibitors: Impact of STK11, KRAS and TP53 mutations and co-mutations on outcome. Transl. Lung Cancer Res. 2021, 10, 202–220. [Google Scholar] [CrossRef]
- Jiao, X.D.; Qin, B.D.; You, P.; Cai, J.; Zang, Y.S. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 2018, 123, 70–75. [Google Scholar] [CrossRef]
- Shigematsu, H.; Lin, L.; Takahashi, T.; Nomura, M.; Suzuki, M.; Wistuba, I.I.; Fong, K.M.; Lee, H.; Toyooka, S.; Shimizu, N.; et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 2005, 97, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Martella, C.; Felicioni, L.; Barassi, F.; Salvatore, S.; Chella, A.; Camplese, P.P.; Iarussi, T.; Mucilli, F.; Mezzetti, A.; et al. EGFR mutations in non-small-cell lung cancer: Analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J. Clin. Oncol. 2005, 23, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yang, G.; Li, W.; Li, J.; Hao, X.; Xing, P.; Yang, Y.; Wang, Y. EGFR Exon 18 Mutations in Advanced Non-Small Cell Lung Cancer: A Real-World Study on Diverse Treatment Patterns and Clinical Outcomes. Front. Oncol. 2021, 11, 713483. [Google Scholar] [CrossRef]
- Xu, H.; Yang, G.; Liu, R.; Yang, Y.; Li, W.; Li, J.; Hao, X.; Xing, P.; Wang, Y. EGFR uncommon alterations in advanced non-small cell lung cancer and structural insights into sensitivity to diverse tyrosine kinase inhibitors. Front. Pharmacol. 2022, 13, 976731. [Google Scholar] [CrossRef] [PubMed]
- Imianitov, E.; Demidova, I.; Gordiev, M.; Filipenko, M.; Kekeeva, T.; Moliaka, Y.; Gervas, P.; Kozhemyako, V.; Vodolazhsky, D.I.; Aleksakhina, S.; et al. EGFR analysis of 21,039 patients with NSCLC: Age-related gradual increase of the L858R mutation frequency in adenocarcinomas and high occurrence of ex19del/L858R mutations in squamous cell carcinomas from females and/or nonsmokers. J. Clin. Oncol. 2017, 35, 9040. [Google Scholar] [CrossRef]
- Dogan, S.; Shen, R.; Ang, D.C.; Johnson, M.L.; D’Angelo, S.P.; Paik, P.K.; Brzostowski, E.B.; Riely, G.J.; Kris, M.G.; Zakowski, M.F.; et al. Molecular Epidemiology of EGFR and KRAS Mutations in 3026 Lung Adenocarcinomas: Higher Susceptibility of Women to Smoking-Related KRAS-Mutant Cancers. Clin. Cancer Res. 2012, 18, 6169–6177. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.H.; Yang, C.T.; Shih, J.Y.; Huang, M.S.; Su, W.C.; Lai, R.S.; Wang, C.C.; Hsiao, S.H.; Lin, Y.C.; Ho, C.L.; et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas with G719X/L861Q/S768I Mutations. J. Thorac. Oncol. 2015, 10, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.M.; Rosas, G.; Belmar-López, C.; Raez, L.E.; Rolfo, C.D.; Schwarz, L.J.; Infante-Huaytalla, U.; Paez, K.J.; García, L.R.; Alvarado, H.; et al. Influence of Sex in the Molecular Characteristics and Outcomes of Malignant Tumors. Front. Oncol. 2021, 11, 752918. [Google Scholar] [CrossRef] [PubMed]
- Judd, J.; Abdel Karim, N.; Khan, H.; Naqash, A.R.; Baca, Y.; Xiu, J.; VanderWalde, A.M.; Mamdani, H.; Raez, L.E.; Nagasaka, M.; et al. Characterization of KRAS Mutation Subtypes in Non–small Cell Lung Cancer. Mol. Cancer Ther. 2021, 20, 2577–2584. [Google Scholar] [CrossRef]
- Stapelfeld, C.; Dammann, C.; Maser, E. Sex-specificity in lung cancer risk. Int. J. Cancer 2020, 146, 2376–2382. [Google Scholar] [CrossRef]
- Nassar, A.H.; Adib, E.; Kwiatkowski, D.J. Distribution of KRASG12C Somatic Mutations across Race, Sex, and Cancer Type. New Engl. J. Med. 2021, 384, 185–187. [Google Scholar] [CrossRef]
- Araujo, L.H.; Souza, B.M.; Leite, L.R.; Parma, S.A.F.; Lopes, N.P.; Malta, F.S.V.; Freire, M.C.M. Molecular profile of KRAS G12C-mutant colorectal and non-small-cell lung cancer. BMC Cancer 2021, 21, 193. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Alessi, J.V.; Elkrief, A.; Wang, X.; Cortellini, A.; Li, Y.Y.; Vaz, V.R.; Gupta, H.; Pecci, F.; Barrichello, A.; et al. Dissecting the clinicopathologic, genomic, and immunophenotypic correlates of KRASG12D-mutated non-small-cell lung cancer. Ann. Oncol. 2022, 33, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Bergethon, K.; Shaw, A.T.; Ou, S.H.; Katayama, R.; Lovly, C.M.; McDonald, N.T.; Massion, P.P.; Siwak-Tapp, C.; Gonzalez, A.; Fang, R.; et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012, 30, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Takeuchi, K.; Togashi, Y.; Hatano, S.; Ninomiya, H.; Motoi, N.; Mun, M.Y.; Sakao, Y.; Okumura, S.; Nakagawa, K.; et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod. Pathol. 2009, 22, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y. Molecular diagnostic characteristics based on the next generation sequencing in lung cancer and its relationship with the expression of PD-L1. Pathol. Res. Pr. 2020, 216, 152797. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, O.; Ohashi, R.; Yoshioka, Y.; Inagaki, A.; Tajima, M.; Koinuma, Y.; Iwakami, S.; Iwase, A.; Sasaki, S.; Tominaga, S.; et al. High prevalence of gene abnormalities in young patients with lung cancer. J. Thorac. Dis. 2013, 5, 27–30. [Google Scholar] [CrossRef] [PubMed]
- VandenBussche, C.J.; Illei, P.B.; Lin, M.T.; Ettinger, D.S.; Maleki, Z. Molecular alterations in non-small cell lung carcinomas of the young. Hum. Pathol. 2014, 45, 2379–2387. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.X.; Yang, H.; Deng, Q.; Gu, X.; He, P.; Lin, Y.; Zhao, M.; Jiang, J.; Chen, H.; Lin, Y.; et al. Oncogenic driver mutations in patients with non-small-cell lung cancer at various clinical stages. Ann. Oncol. 2013, 24, 1319–1325. [Google Scholar] [CrossRef]
- Wu, S.-G.; Kuo, Y.-W.; Chang, Y.-L.; Shih, J.-Y.; Chen, Y.-H.; Tsai, M.-F.; Yu, C.-J.; Yang, C.-H.; Yang, P.-C. EML4-ALK Translocation Predicts Better Outcome in Lung Adenocarcinoma Patients with Wild-Type EGFR. J. Thorac. Oncol. 2012, 7, 98–104. [Google Scholar] [CrossRef]
- Bi, H.; Ren, D.; Ding, X.; Yin, X.; Cui, S.; Guo, C.; Wang, H. Clinical characteristics of patients with ROS1 gene rearrangement in non-small cell lung cancer: A meta-analysis. Transl. Cancer Res. 2020, 9, 4383–4392. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Felicioni, L.; Malatesta, S.; Sciarrotta, M.G.; Guetti, L.; Chella, A.; Viola, P.; Pullara, C.; Mucilli, F.; Buttitta, F. Clinical Features and Outcome of Patients with Non–Small-Cell Lung Cancer Harboring BRAF Mutations. J. Clin. Oncol. 2011, 29, 3574–3579. [Google Scholar] [CrossRef] [PubMed]
- Marin-Acevedo, J.A.; Hicks, J.K.; Thapa, R.; Chen, D.-T.; Kimbrough, E.; Gray, J.E. Assessment of BRAF class I/II/III mutations, demographics, and treatment outcomes in NSCLC. J. Clin. Oncol. 2021, 39, e21016. [Google Scholar] [CrossRef]
- Farago, A.F.; Taylor, M.S.; Doebele, R.C.; Zhu, V.W.; Kummar, S.; Spira, A.I.; Boyle, T.A.; Haura, E.B.; Arcila, M.E.; Benayed, R.; et al. Clinicopathologic Features of Non–Small-Cell Lung Cancer Harboring an NTRK Gene Fusion. JCO Precis. Oncol. 2018, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Westphalen, C.B.; Krebs, M.G.; Le Tourneau, C.; Sokol, E.S.; Maund, S.L.; Wilson, T.R.; Jin, D.X.; Newberg, J.Y.; Fabrizio, D.; Veronese, L.; et al. Genomic context of NTRK1/2/3 fusion-positive tumours from a large real-world population. NPJ Precis. Oncol. 2021, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H.I.; Sokol, E.S.; Trabucco, S.E.; Jin, D.X.; Frampton, G.M.; Graziano, S.L.; Elvin, J.A.; Vergilio, J.A.; Killian, J.K.; Ngo, N.; et al. 1549P—NTRK1-3 genomic fusions in non-small cell lung cancer (NSCLC) determined by comprehensive genomic profiling. Ann. Oncol. 2019, 30, v638. [Google Scholar] [CrossRef]
- Guinee, D.G., Jr.; Travis, W.D.; Trivers, G.E.; De Benedetti, V.M.; Cawley, H.; Welsh, J.A.; Bennett, W.P.; Jett, J.; Colby, T.V.; Tazelaar, H.; et al. Gender comparisons in human lung cancer: Analysis of p53 mutations, anti-p53 serum antibodies and C-erbB-2 expression. Carcinogenesis 1995, 16, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Feldman, R.; Sukari, A.; Kim, C.; Mamdani, H.; Spira, A.I.; Bepler, G.; Kim, E.S.; Raez, L.E.; Pai, S.G.; et al. Characterization of ERBB2 alterations in non-small cell lung cancer. J. Clin. Oncol. 2020, 38, e21553. [Google Scholar] [CrossRef]
- Wang, R.; Hu, H.; Pan, Y.; Li, Y.; Ye, T.; Li, C.; Luo, X.; Wang, L.; Li, H.; Zhang, Y.; et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J. Clin. Oncol. 2012, 30, 4352–4359. [Google Scholar] [CrossRef]
- Hess, L.M.; Han, Y.; Zhu, Y.E.; Bhandari, N.R.; Sireci, A. Characteristics and outcomes of patients with RET-fusion positive non-small lung cancer in real-world practice in the United States. BMC Cancer 2021, 21, 28. [Google Scholar] [CrossRef]
- Champagnac, A.; Bringuier, P.-P.; Barritault, M.; Isaac, S.; Watkin, E.; Forest, F.; Maury, J.-M.; Girard, N.; Brevet, M. Frequency of MET exon 14 skipping mutations in non-small cell lung cancer according to technical approach in routine diagnosis: Results from a real-life cohort of 2369 patients. J. Thorac. Dis. 2020, 12, 2172–2178. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.G.; Ho, A.T.N.; Altibi, A.M.A.; Nakazawa, T.; Katoh, R.; Kondo, T. Clinicopathological implications of MET exon 14 mutations in non-small cell lung cancer—A systematic review and meta-analysis. Lung Cancer 2018, 123, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Schubart, C.; Stöhr, R.; Tögel, L.; Fuchs, F.; Sirbu, H.; Seitz, G.; Seggewiss-Bernhardt, R.; Leistner, R.; Sterlacci, W.; Vieth, M.; et al. MET Amplification in Non-Small Cell Lung Cancer (NSCLC)-A Consecutive Evaluation Using Next-Generation Sequencing (NGS) in a Real-World Setting. Cancers 2021, 13, 5023. [Google Scholar] [CrossRef] [PubMed]
- Pécuchet, N.; Laurent-Puig, P.; Mansuet-Lupo, A.; Legras, A.; Alifano, M.; Pallier, K.; Didelot, A.; Gibault, L.; Danel, C.; Just, P.A.; et al. Different prognostic impact of STK11 mutations in non-squamous non-small-cell lung cancer. Oncotarget 2017, 8, 23831–23840. [Google Scholar] [CrossRef] [PubMed]
- Pons-Tostivint, E.; Lugat, A.; Fontenau, J.F.; Denis, M.G.; Bennouna, J. STK11/LKB1 Modulation of the Immune Response in Lung Cancer: From Biology to Therapeutic Impact. Cells 2021, 10, 3129. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Liu, L.; Chen, H.; Wang, Y.; Xu, Y.; Mao, H.; Li, J.; Mills, G.B.; Shu, Y.; Li, L.; et al. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer Cell 2016, 29, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Gu, L.; Zhong, D. TP53 Mutation Mapping in Advanced Non-Small Cell Lung Cancer: A Real-World Retrospective Cohort Study. Curr. Oncol. 2022, 29, 7411–7419. [Google Scholar] [CrossRef]
- Jiang, W.; Cheng, H.; Yu, L.; Zhang, J.; Wang, Y.; Liang, Y.; Lou, F.; Wang, H.; Cao, S. Mutation patterns and evolutionary action score of TP53 enable identification of a patient population with poor prognosis in advanced non-small cell lung cancer. Cancer Med. 2023, 12, 6649–6658. [Google Scholar] [CrossRef]
- Shire, N.J.; Klein, A.B.; Golozar, A.; Collins, J.M.; Fraeman, K.H.; Nordstrom, B.L.; McEwen, R.; Hembrough, T.; Rizvi, N.A. STK11 (LKB1) mutations in metastatic NSCLC: Prognostic value in the real world. PLoS ONE 2020, 15, e0238358. [Google Scholar] [CrossRef]
- Skoulidis, F.; Goldberg, M.E.; Greenawalt, D.M.; Hellmann, M.D.; Awad, M.M.; Gainor, J.F.; Schrock, A.B.; Hartmaier, R.J.; Trabucco, S.E.; Gay, L.; et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018, 8, 822–835. [Google Scholar] [CrossRef]
- Pao, W.; Girard, N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011, 12, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Seegobin, K.; Heng, F.; Zhou, K.; Chen, R.; Qin, H.; Manochakian, R.; Zhao, Y.; Lou, Y. Genomic landscape of lung adenocarcinomas in different races. Front. Oncol. 2022, 12, 946625. [Google Scholar] [CrossRef] [PubMed]
- Cornelius, M.E.; Loretan, C.G.; Jamal, A.; Davis Lynn, B.C.; Mayer, M.; Alcantara, I.C.; Neff, L. Tobacco Product Use Among Adults—United States, 2021. Morb. Mortal. Wkly. Rep. 2023, 72, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Govindan, R.; Ding, L.; Griffith, M.; Subramanian, J.; Dees, N.D.; Kanchi, K.L.; Maher, C.A.; Fulton, R.; Fulton, L.; Wallis, J.; et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012, 150, 1121–1134. [Google Scholar] [CrossRef] [PubMed]
- Kuśnierczyk, P. Genetic differences between smokers and never-smokers with lung cancer. Front. Immunol. 2023, 14, 1063716. [Google Scholar] [CrossRef]

| ctDNA+ Population n = 30,790 | |||
|---|---|---|---|
| Characteristics | <70 Years n = 15,495 (50.32%) | ≥70 Years n = 15,237 (49.49%) | Age Unknown n = 58 (0.19%) |
| Median Age (Range) | 61 (18–69) | 77 (70–102) | N/A |
| Sex | |||
| Female | 8416 (50.23%) | 8307 (49.58%) | 33 (0.20%) |
| Male | 7079 (50.44%) | 6930 (49.38%) | 25 (0.18%) |
| Histologic Subtype | |||
| Adenocarcinoma | 14,530 (50.39%) | 14,247 (49.41%) | 58 (0.20%) |
| NSCLC NOS | 965 (49.36%) | 990 (50.64%) | 0 (0.00%) |
| Alteration Detected | Total Patients (n = 30,790) | p-Value | ||
|---|---|---|---|---|
| Females | Males | |||
| 16,756 (54.42%) | 14,034 (45.58%) | |||
| EGFR | Exon 19 deletion | 1600 (9.55%) | 822 (5.86%) | <0.0001 |
| Exon 20 insertion | 209 (1.25%) | 133 (0.95%) | 0.0139 | |
| G719X | 161 (0.96%) | 83 (0.59%) | 0.0003 | |
| L858R | 1141 (6.81%) | 556 (3.96%) | <0.0001 | |
| T790M | 181 (1.08%) | 80 (0.57%) | <0.0001 | |
| S768I | 79 (0.47%) | 35 (0.25%) | 0.0013 | |
| L861Q | 98 (0.58%) | 48 (0.34%) | 0.002 | |
| KRAS | G12C | 1190 (7.10%) | 900 (6.41%) | 0.0169 |
| G12D | 413 (2.46%) | 357 (2.54%) | 0.6604 | |
| G12V | 520 (3.10%) | 395 (2.81%) | 0.1383 | |
| ALK | Fusion | 231 (1.38%) | 172 (1.23%) | 0.366 |
| ROS1 | Fusion | 25 (0.15%) | 29 (0.21%) | 0.274 |
| BRAF | V600E | 157 (0.94%) | 143 (1.02%) | 0.4848 |
| NTRK | NTRK 1 Fusion | 1 (0.01%) | 0 (0.00%) | >0.9999 |
| ERBB2 | Exon 20 insertions | 235 (1.40%) | 140 (1.00%) | 0.0012 |
| RET | Fusion | 13 (0.08%) | 6 (0.04%) | 0.2554 |
| MET | Exon 14 skipping | 188 (1.12%) | 153 (1.09%) | 0.827 |
| Amplification medium | 131 (0.78%) | 157 (1.12%) | 0.0024 | |
| Amplification high | 100 (0.60%) | 153 (1.09%) | <0.0001 | |
| PIK3CA | Mutant | 222 (1.32%) | 177 (1.26%) | 0.649 |
| STK11 | Mutant | 855 (5.10%) | 1068 (7.61%) | <0.0001 |
| TP53 | Mutant | 6370 (38.02%) | 6506 (46.36%) | <0.0001 |
| Alteration Detected | Total Patients (n = 30,732) | p-Value | ||
|---|---|---|---|---|
| Patients < 70 | Patients ≥ 70 | |||
| 15,495 (50.42%) | 15,237 (49.58%) | |||
| EGFR | Exon 19 deletion | 1571 (10.14%) | 851 (5.59%) | <0.0001 |
| Exon 20 insertion | 228 (1.47%) | 114 (0.75%) | <0.0001 | |
| G719X | 127 (0.82%) | 117 (0.77%) | 0.6528 | |
| L858R | 885 (5.71%) | 812 (5.33%) | 0.1475 | |
| T790M | 161 (1.04%) | 100 (0.66%) | 0.0003 | |
| S768I | 60 (0.39%) | 54 (0.35%) | 0.6405 | |
| L861Q | 50 (0.32%) | 96 (0.63%) | <0.0001 | |
| KRAS | G12C | 1174 (7.58%) | 916 (6.01%) | <0.0001 |
| G12D | 421 (2.72%) | 349 (2.29%) | 0.0176 | |
| G12V | 489 (3.16%) | 426 (2.80%) | 0.0649 | |
| ALK | Fusion | 326 (2.10%) | 77 (0.51%) | <0.0001 |
| ROS1 | Fusion | 40 (0.26%) | 14 (0.09%) | 0.0005 |
| BRAF | V600E | 169 (1.09%) | 131 (0.86%) | 0.0422 |
| NTRK | NTRK 1 Fusion | 1 (0.01%) | 0 (0.00%) | >0.9999 |
| ERBB2 | Exon 20 insertions | 230 (1.48%) | 145 (0.95%) | <0.0001 |
| RET | Fusion | 11 (0.07%) | 8 (0.05%) | 0.6477 |
| MET | Exon 14 skipping | 81 (0.52%) | 260 (1.71%) | <0.0001 |
| Amplification medium | 167 (1.08%) | 121 (0.79%) | 0.0108 | |
| Amplification high | 171 (1.10%) | 82 (0.54%) | <0.0001 | |
| PIK3CA | Mutant | 174 (1.12%) | 225 (1.48%) | 0.0065 |
| STK11 | Mutant | 1145 (7.39%) | 778 (5.11%) | <0.0001 |
| TP53 | Mutant | 6914 (44.62%) | 5962 (39.13%) | <0.0001 |
| Alteration Detected | Total Females (n = 16,723) | p-Value | Total Males (n = 14,009) | p-Value | |||
|---|---|---|---|---|---|---|---|
| Females < 70 | Females ≥ 70 | Males < 70 | Males ≥ 70 | ||||
| 8416 (27.39%) | 8307 (27.03%) | 7079 (23.03%) | 6930 (22.55%) | ||||
| EGFR | Exon 19 deletion | 1016 (12.07%) | 584 (7.03%) | <0.0001 | 555 (7.84%) | 267 (3.85%) | <0.0001 |
| Exon 20 insertion | 136 (1.62%) | 73 (0.88%) | <0.0001 | 92 (1.30%) | 41 (0.59%) | <0.0001 | |
| G719X | 85 (1.01%) | 76 (0.91%) | 0.5795 | 42 (0.59%) | 41 (0.59%) | >0.9999 | |
| L858R | 593 (7.05%) | 548 (6.60%) | 0.2565 | 292 (4.12%) | 264 (3.81%) | 0.3414 | |
| T790M | 113 (1.34%) | 68 (0.82%) | 0.0013 | 19 (0.27%) | 16 (0.23%) | 0.7361 | |
| S768I | 41 (0.49%) | 38 (0.46%) | 0.822 | 48 (0.68%) | 32 (0.46%) | 0.0935 | |
| L861Q | 34 (0.40%) | 64 (0.77%) | 0.0022 | 16 (0.23%) | 32 (0.46%) | 0.0201 | |
| KRAS | G12C | 686 (8.15%) | 504 (6.07%) | <0.0001 | 488 (6.89%) | 412 (5.95%) | 0.0229 |
| G12D | 239 (2.84%) | 174 (2.09%) | 0.002 | 182 (2.57%) | 175 (2.53%) | 0.8724 | |
| G12V | 281 (3.34%) | 239 (2.88%) | 0.0904 | 208 (2.94%) | 187 (2.70%) | 0.4142 | |
| ALK | Fusion | 188 (2.23%) | 43 (0.52%) | <0.0001 | 138 (1.95%) | 34 (0.49%) | <0.0001 |
| ROS1 | Fusion | 18 (0.21%) | 7 (0.08%) | 0.0432 | 22 (0.31%) | 7 (0.10%) | 0.0082 |
| BRAF | V600E | 90 (1.07%) | 67 (0.81%) | 0.0919 | 79 (1.12%) | 64 (0.92%) | 0.2749 |
| NTRK | NTRK 1 Fusion | 1 (0.01%) | 0 (0.00%) | >0.9999 | 0 (0.00%) | 0 (0.00%) | >0.9999 |
| ERBB2 | Exon 20 insertions | 139 (1.65%) | 96 (1.16%) | 0.007 | 91 (1.29%) | 49 (0.71%) | 0.0006 |
| RET | Fusion | 7 (0.08%) | 6 (0.07%) | >0.9999 | 4 (0.06%) | 2 (0.03%) | 0.6875 |
| MET | Exon 14 skipping | 48 (0.57%) | 140 (1.69%) | <0.0001 | 33 (0.47%) | 120 (1.73%) | <0.0001 |
| Amplification medium | 77 (0.91%) | 54 (0.65%) | 0.0539 | 90 (1.27%) | 67 (0.97%) | 0.0921 | |
| Amplification high | 71 (0.84%) | 29 (0.35%) | <0.0001 | 100 (1.41%) | 53 (0.76%) | 0.0002 | |
| PIK3CA | Mutant | 101 (1.20%) | 121 (1.46%) | 0.1561 | 73 (1.03%) | 104 (1.50%) | 0.0153 |
| STK11 | Mutant | 533 (6.33%) | 322 (3.88%) | <0.0001 | 612 (8.65%) | 456 (6.58%) | <0.0001 |
| TP53 | Mutant | 3409 (40.51%) | 2961 (35.64%) | <0.0001 | 3505 (49.51%) | 3001 (43.30%) | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kimbrough, E.O.; Marin-Acevedo, J.A.; Drusbosky, L.M.; Mooradian, A.; Zhao, Y.; Manochakian, R.; Lou, Y. Sex- and Age-Associated Differences in Genomic Alterations among Patients with Advanced Non-Small Cell Lung Cancer (NSCLC). Cancers 2024, 16, 2366. https://doi.org/10.3390/cancers16132366
Kimbrough EO, Marin-Acevedo JA, Drusbosky LM, Mooradian A, Zhao Y, Manochakian R, Lou Y. Sex- and Age-Associated Differences in Genomic Alterations among Patients with Advanced Non-Small Cell Lung Cancer (NSCLC). Cancers. 2024; 16(13):2366. https://doi.org/10.3390/cancers16132366
Chicago/Turabian StyleKimbrough, ErinMarie O., Julian A. Marin-Acevedo, Leylah M. Drusbosky, Ariana Mooradian, Yujie Zhao, Rami Manochakian, and Yanyan Lou. 2024. "Sex- and Age-Associated Differences in Genomic Alterations among Patients with Advanced Non-Small Cell Lung Cancer (NSCLC)" Cancers 16, no. 13: 2366. https://doi.org/10.3390/cancers16132366
APA StyleKimbrough, E. O., Marin-Acevedo, J. A., Drusbosky, L. M., Mooradian, A., Zhao, Y., Manochakian, R., & Lou, Y. (2024). Sex- and Age-Associated Differences in Genomic Alterations among Patients with Advanced Non-Small Cell Lung Cancer (NSCLC). Cancers, 16(13), 2366. https://doi.org/10.3390/cancers16132366






