SMARCD3 Overexpression Promotes Epithelial–Mesenchymal Transition in Gastric Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Transfection
2.2. Western Blotting
2.3. Proliferation Assay
2.4. Colony Formation Assay
2.5. Wound-Healing Migration Assay
2.6. Invasion Assay
2.7. Kaplan-Meier Survival Curve Analysis
2.8. Microscopic Examination
2.9. Immunocytochemistry
2.10. Statistical Analysis
2.11. Ethics Statement
3. Results
3.1. SMARCD3 Was More Highly Expressed in Gastric Cancer Tissues Than in Normal Tissues, Especially in SRCs
3.2. The High-SMARCD3 Expression Group Had Poor Overall Survival according to Kaplan-Meier Plotter Data
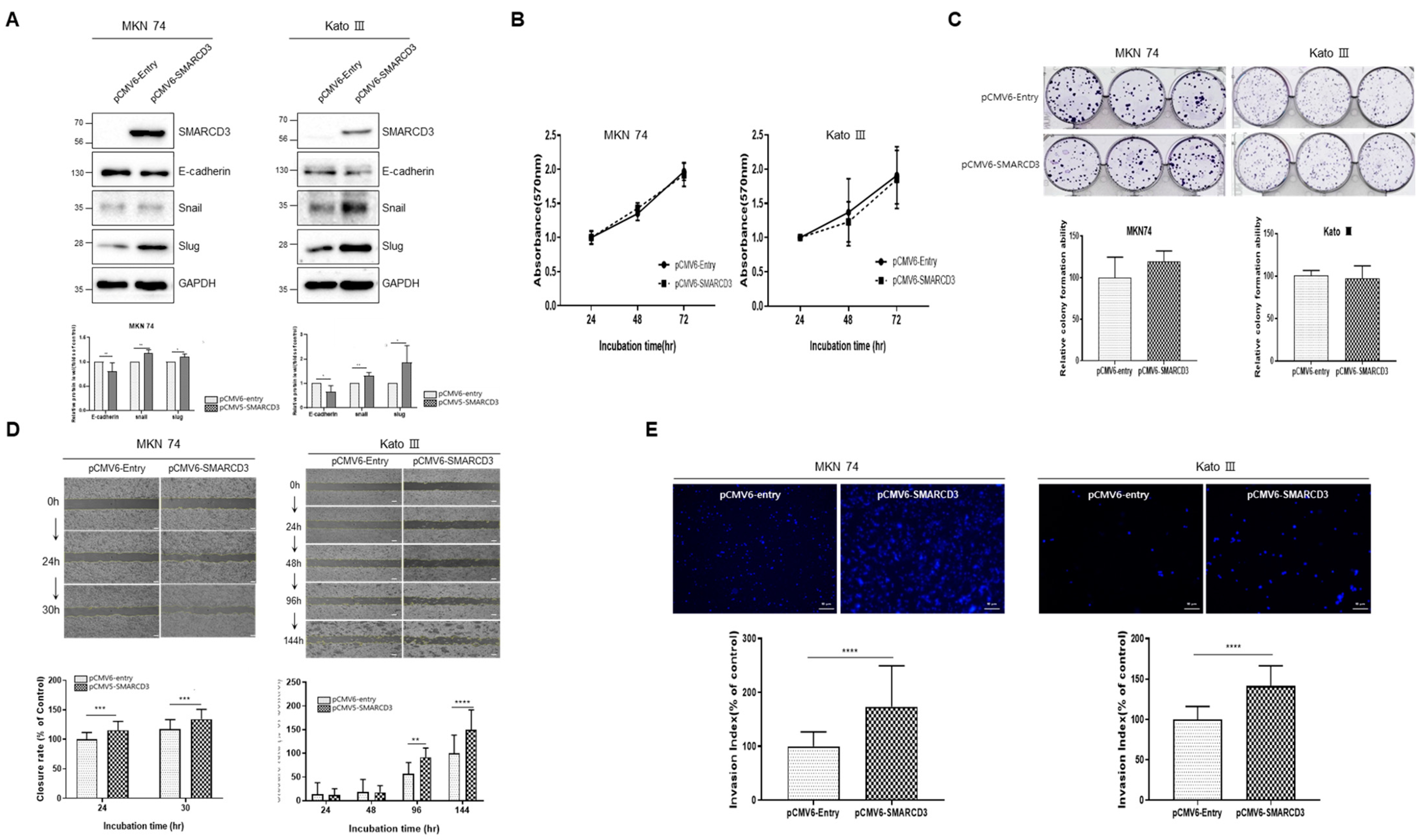
3.3. SMARCD3-Overexpressing Gastric Cancer Cell Lines Exhibit Increased Migration, Invasion, and EMT Markers
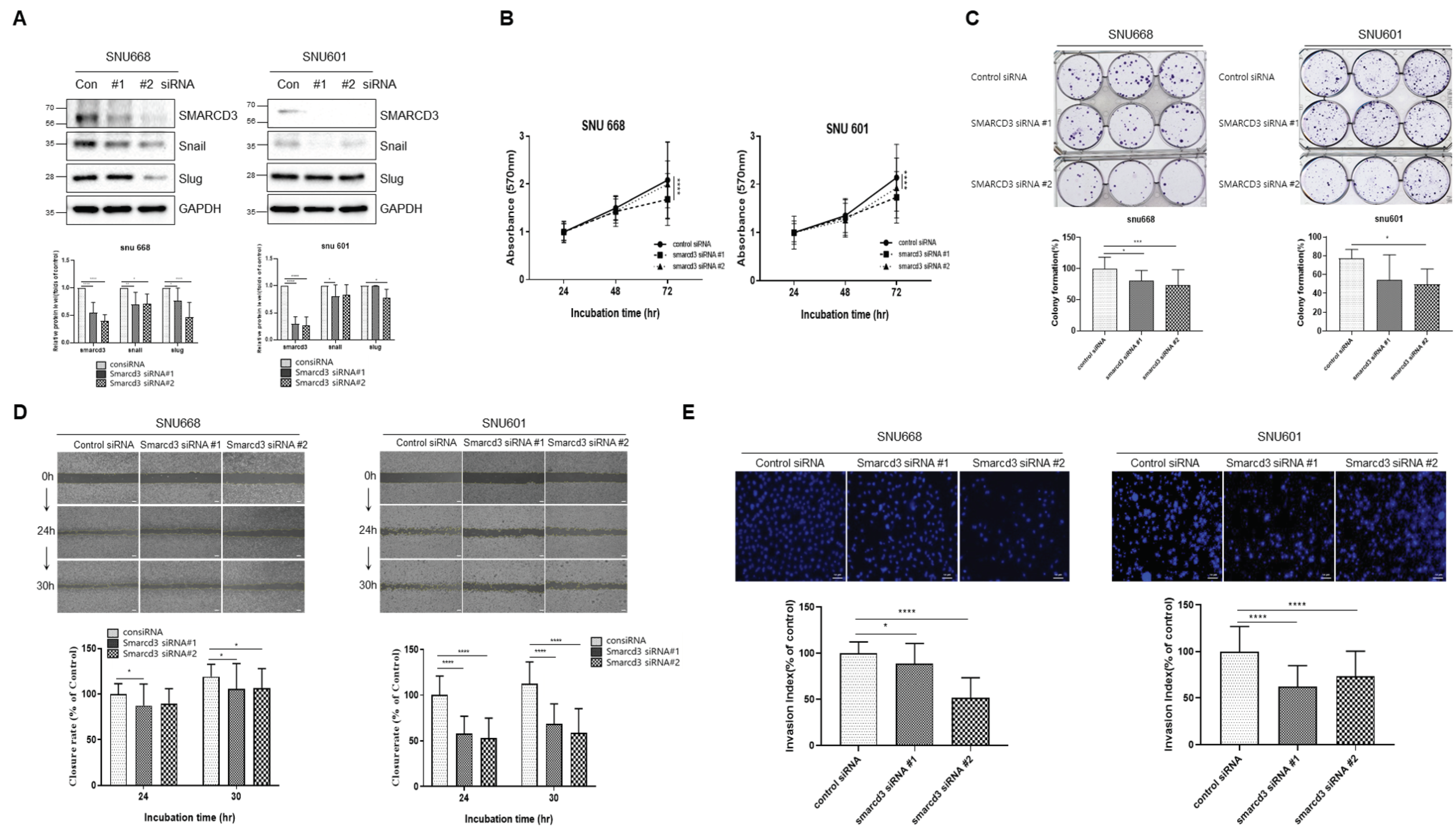
3.4. SMARCD3-Knock-Out Gastric Cancer Cell Lines Exhibited Decreased Proliferation, Migration, Invasion and EMT
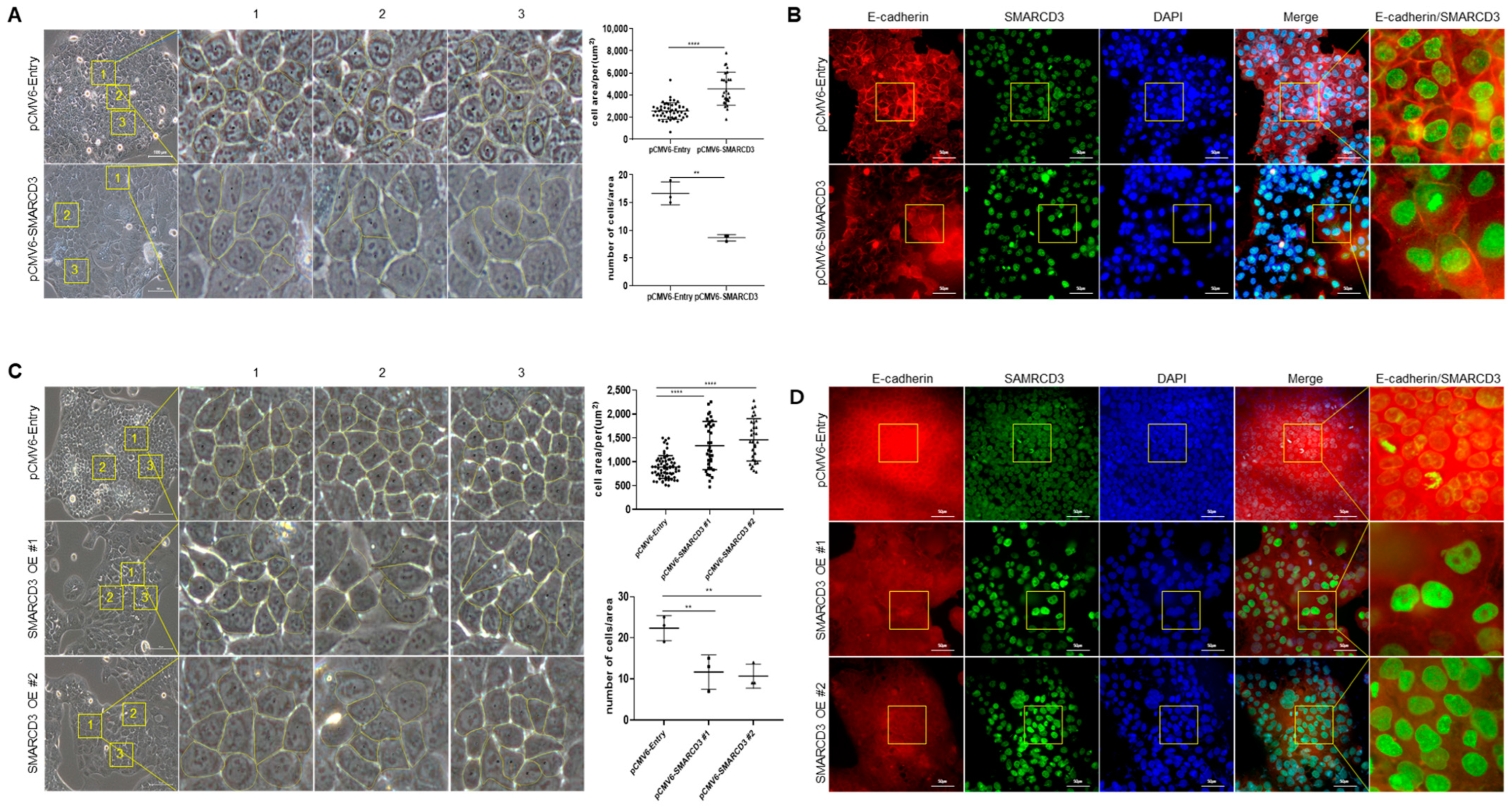
3.5. Macroscopic Changes after SMARCD3 Overexpression Revealed Irregular Shapes and Increased Average Cell Areas
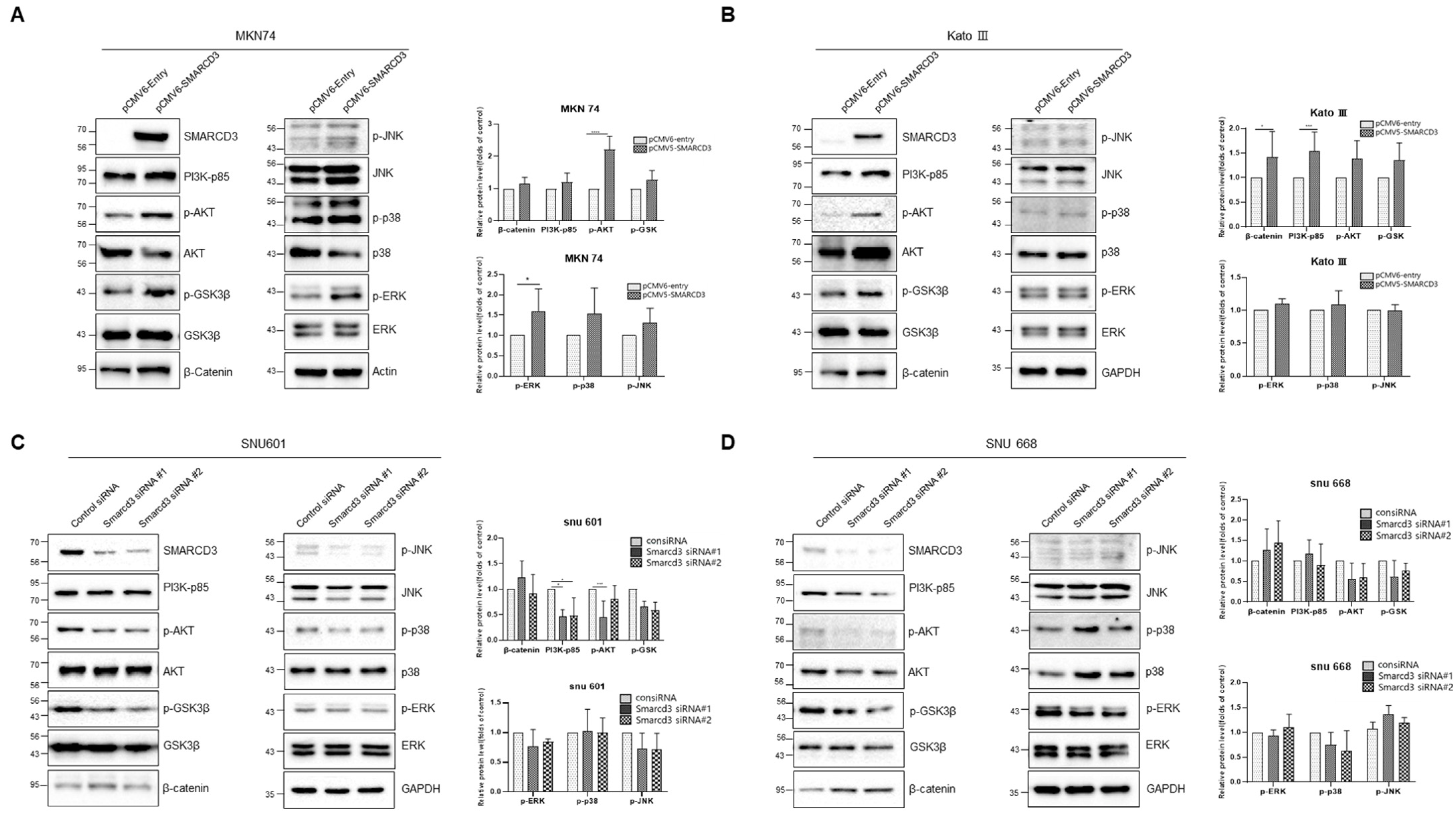
3.6. Investigation of Cancer Signaling Pathways Using SMARCD3 Overexpression and Knockdown
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Park, E.H.; Jung, K.W.; Park, N.J.; Kang, M.J.; Yun, E.H.; Kim, H.J.; Kim, J.E.; Kong, H.J.; Im, J.S.; Seo, H.G. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2021. Cancer Res. Treat. 2024, 56, 357–371. [Google Scholar] [CrossRef]
- Shah, M.A.; Kelsen, D.P. Gastric cancer: A primer on the epidemiology and biology of the disease and an overview of the medical management of advanced disease. J. Natl. Compr. Cancer Netw. 2010, 8, 437–447. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Health Insurance Review & Assessment Service. Results of Gastric Cancer Adequacy Evaluation, 5th ed.; Health Insurance Review & Assessment Service: Gwangju, Republic of Korea, 2021. [Google Scholar]
- Škovierová, H.; Okajčeková, T.; Strnádel, J.; Vidomanová, E.; Halašová, E. Molecular regulation of epithelial-to-mesenchymal transition in tumorigenesis (Review). Int. J. Mol. Med. 2018, 41, 1187–1200. [Google Scholar] [CrossRef]
- Petrova, Y.I.; Schecterson, L.; Gumbiner, B.M. Roles for E-cadherin cell surface regulation in cancer. Mol. Biol. Cell 2016, 27, 3233–3244. [Google Scholar] [CrossRef]
- Padmanaban, V.; Krol, I.; Suhail, Y.; Szczerba, B.M.; Aceto, N.; Bader, J.S.; Ewald, A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019, 573, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Na, T.Y.; Schecterson, L.; Mendonsa, A.M.; Gumbiner, B.M. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc. Natl. Acad. Sci. USA 2020, 117, 5931–5937. [Google Scholar] [CrossRef] [PubMed]
- Navarro, P.; Gómez, M.; Pizarro, A.; Gamallo, C.; Quintanilla, M.; Cano, A. A role for the E-cadherin cell-cell adhesion molecule during tumor progression of mouse epidermal carcinogenesis. J. Cell Biol. 1991, 115, 517–533. [Google Scholar] [CrossRef]
- Qian, X.; Karpova, T.; Sheppard, A.M.; McNally, J.; Lowy, D.R. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004, 23, 1739–1748. [Google Scholar] [CrossRef]
- Kim, N.G.; Koh, E.; Chen, X.; Gumbiner, B.M. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA 2011, 108, 11930–11935. [Google Scholar] [CrossRef] [PubMed]
- Mendonsa, A.M.; Na, T.Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef] [PubMed]
- Tóthová, Z.; Šemeláková, M.; Solárová, Z.; Tomc, J.; Debeljak, N.; Solár, P. The Role of PI3K/AKT and MAPK Signaling Pathways in Erythropoietin Signalization. Int. J. Mol. Sci. 2021, 22, 7682. [Google Scholar] [CrossRef] [PubMed]
- Buttell, A.; Qiu, W. The action and resistance mechanisms of Lenvatinib in liver cancer. Mol. Carcinog. 2023, 62, 1918–1934. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Iglehart, J.D.; Pardee, A.B. Metastatic potential of 21T human breast cancer cells depends on Akt/protein kinase B activation. Cancer Res. 2007, 67, 5293–5299. [Google Scholar] [CrossRef] [PubMed]
- Kitagishi, Y.; Kobayashi, M.; Kikuta, K.; Matsuda, S. Roles of PI3K/AKT/GSK3/mTOR Pathway in Cell Signaling of Mental Illnesses. Depress. Res. Treat. 2012, 2012, 752563. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Hemmings, B.A. PKB/Akt-dependent regulation of cell motility. J. Natl. Cancer Inst. 2013, 105, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, Z.; Lu, N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh. Migr. 2015, 9, 317–324. [Google Scholar] [CrossRef]
- Vijay, G.V.; Zhao, N.; Den Hollander, P.; Toneff, M.J.; Joseph, R.; Pietila, M.; Taube, J.H.; Sarkar, T.R.; Ramirez-Pena, E.; Werden, S.J.; et al. GSK3β regulates epithelial-mesenchymal transition and cancer stem cell properties in triple-negative breast cancer. Breast Cancer Res. BCR 2019, 21, 37. [Google Scholar] [CrossRef]
- Duda, P.; Akula, S.M.; Abrams, S.L.; Steelman, L.S.; Martelli, A.M.; Cocco, L.; Ratti, S.; Candido, S.; Libra, M.; Montalto, G.; et al. Targeting GSK3 and Associated Signaling Pathways Involved in Cancer. Cells 2020, 9(5), 1110. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Fleming-de-Moraes, C.D.; Rocha, M.R.; Tessmann, J.W.; de Araujo, W.M.; Morgado-Diaz, J.A. Crosstalk between PI3K/Akt and Wnt/β-catenin pathways promote colorectal cancer progression regardless of mutational status. Cancer Biol. Ther. 2022, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jordan, N.V.; Prat, A.; Abell, A.N.; Zawistowski, J.S.; Sciaky, N.; Karginova, O.A.; Zhou, B.; Golitz, B.T.; Perou, C.M.; Johnson, G.L. SWI/SNF chromatin-remodeling factor Smarcd3/Baf60c controls epithelial-mesenchymal transition by inducing Wnt5a signaling. Mol. Cell Biol. 2013, 33, 3011–3025. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Wang, H.; Chen, H.; Han, Y. SMARCD3 is a potential prognostic marker and therapeutic target in CAFs. Aging 2020, 12, 20835–20861. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Park, J.H.; Yang, J.W.; Jung, E.J.; Ju, Y.T.; Jeong, C.Y.; Kim, J.Y.; Park, T.; Park, M.; Lee, Y.J.; et al. HTATIP2 Overexpression was Associated With a Good Prognosis in Gastric Cancer. Technol. Cancer Res. Treat. 2024, 23, 15330338231187254. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Poore, B.; Brown, E.E.; Qian, J.; Xie, B.; Asimakidou, E.; Razskazovskiy, V.; Ayrapetian, D.; Sharma, V.; Xia, S.; et al. A neurodevelopmental epigenetic programme mediated by SMARCD3-DAB1-Reelin signalling is hijacked to promote medulloblastoma metastasis. Nat. Cell Biol. 2023, 25, 493–507. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Dong, K.; Chi, S.; Wei, S.; Zhang, J.; Yu, Z.; Zhang, Q.; Zhang, T.; Cheng, S.; Shi, R.; et al. lncRNA UCA1 promotes tumor progression by targeting SMARCD3 in cervical cancer. Mol. Carcinog. 2024, 63, 384–399. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.P.; Gatchalian, J.; McDermott, M.L.; Nakamura, M.; Chambers, K.; Rajbhandari, N.; Lytle, N.K.; Rosenthal, S.B.; Hamilton, M.; Albini, S.; et al. Smarcd3 is an epigenetic modulator of the metabolic landscape in pancreatic ductal adenocarcinoma. Nat. Commun. 2023, 14, 292. [Google Scholar] [CrossRef] [PubMed]
- Tropée, R.; Avalos, B.d.l.P.; Gough, M.; Snell, C.; Duijf, P.H.G.; Dray, E. The chromatin remodeler SMARCD3 regulates cell cycle progression and its expression predicts survival outcome in ER+ breast cancer. bioRxiv 2019, 684217. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, Y.J.; Park, J.; Kim, T.H.; Hong, S.C.; Jung, E.J.; Ju, Y.T.; Jeong, C.Y.; Park, H.J.; Ko, G.H.; et al. PRDX4 overexpression is associated with poor prognosis in gastric cancer. Oncol. Lett. 2020, 19, 3522–3530. [Google Scholar] [CrossRef]
- Hippo, Y.; Taniguchi, H.; Tsutsumi, S.; Machida, N.; Chong, J.M.; Fukayama, M.; Kodama, T.; Aburatani, H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002, 62, 233–240. [Google Scholar] [PubMed]
- Liu, B.; Zhu, Z.; Yan, M.; Li, J.; Zhang, J.; Li, C. Gene Expression Omnibus GSE54129. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse54129 (accessed on 20 March 2024).
- Tropée, R.; de la Peña Avalos, B.; Gough, M.; Snell, C.; Duijf, P.H.G.; Dray, E. The SWI/SNF subunit SMARCD3 regulates cell cycle progression and predicts survival outcome in ER+ breast cancer. Breast Cancer Res. Treat. 2021, 185, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.H.; Park, M.; Park, S.Y.; Park, J.; Kim, T.H.; Lee, Y.J.; Jung, E.J.; Ju, Y.T.; Jeong, C.Y.; Kim, J.Y.; et al. Transcriptome Analysis and the Prognostic Role of NUDC in Diffuse and Intestinal Gastric Cancer. Technol. Cancer Res. Treat. 2021, 20, 15330338211019501. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Fu, D.; Hu, Z.; Xu, X.; Dai, X.; Liu, Z. Key signal transduction pathways and crosstalk in cancer: Biological and therapeutic opportunities. Transl. Oncol. 2022, 26, 101510. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.Y.; Park, J.-H.; Yang, J.W.; Jung, E.-J.; Ju, Y.-T.; Jeong, C.-Y.; Kim, J.-Y.; Park, T.; Kim, T.-H.; Park, M.; et al. SMARCD3 Overexpression Promotes Epithelial–Mesenchymal Transition in Gastric Cancer. Cancers 2024, 16, 2282. https://doi.org/10.3390/cancers16122282
Park SY, Park J-H, Yang JW, Jung E-J, Ju Y-T, Jeong C-Y, Kim J-Y, Park T, Kim T-H, Park M, et al. SMARCD3 Overexpression Promotes Epithelial–Mesenchymal Transition in Gastric Cancer. Cancers. 2024; 16(12):2282. https://doi.org/10.3390/cancers16122282
Chicago/Turabian StylePark, Sun Yi, Ji-Ho Park, Jung Wook Yang, Eun-Jung Jung, Young-Tae Ju, Chi-Young Jeong, Ju-Yeon Kim, Taejin Park, Tae-Han Kim, Miyeong Park, and et al. 2024. "SMARCD3 Overexpression Promotes Epithelial–Mesenchymal Transition in Gastric Cancer" Cancers 16, no. 12: 2282. https://doi.org/10.3390/cancers16122282
APA StylePark, S. Y., Park, J.-H., Yang, J. W., Jung, E.-J., Ju, Y.-T., Jeong, C.-Y., Kim, J.-Y., Park, T., Kim, T.-H., Park, M., Lee, Y.-J., & Jeong, S.-H. (2024). SMARCD3 Overexpression Promotes Epithelial–Mesenchymal Transition in Gastric Cancer. Cancers, 16(12), 2282. https://doi.org/10.3390/cancers16122282






