ARID1A Mutations in Gastric Cancer: A Review with Focus on Clinicopathological Features, Molecular Background and Diagnostic Interpretation
Abstract
Simple Summary
Abstract
1. Introduction
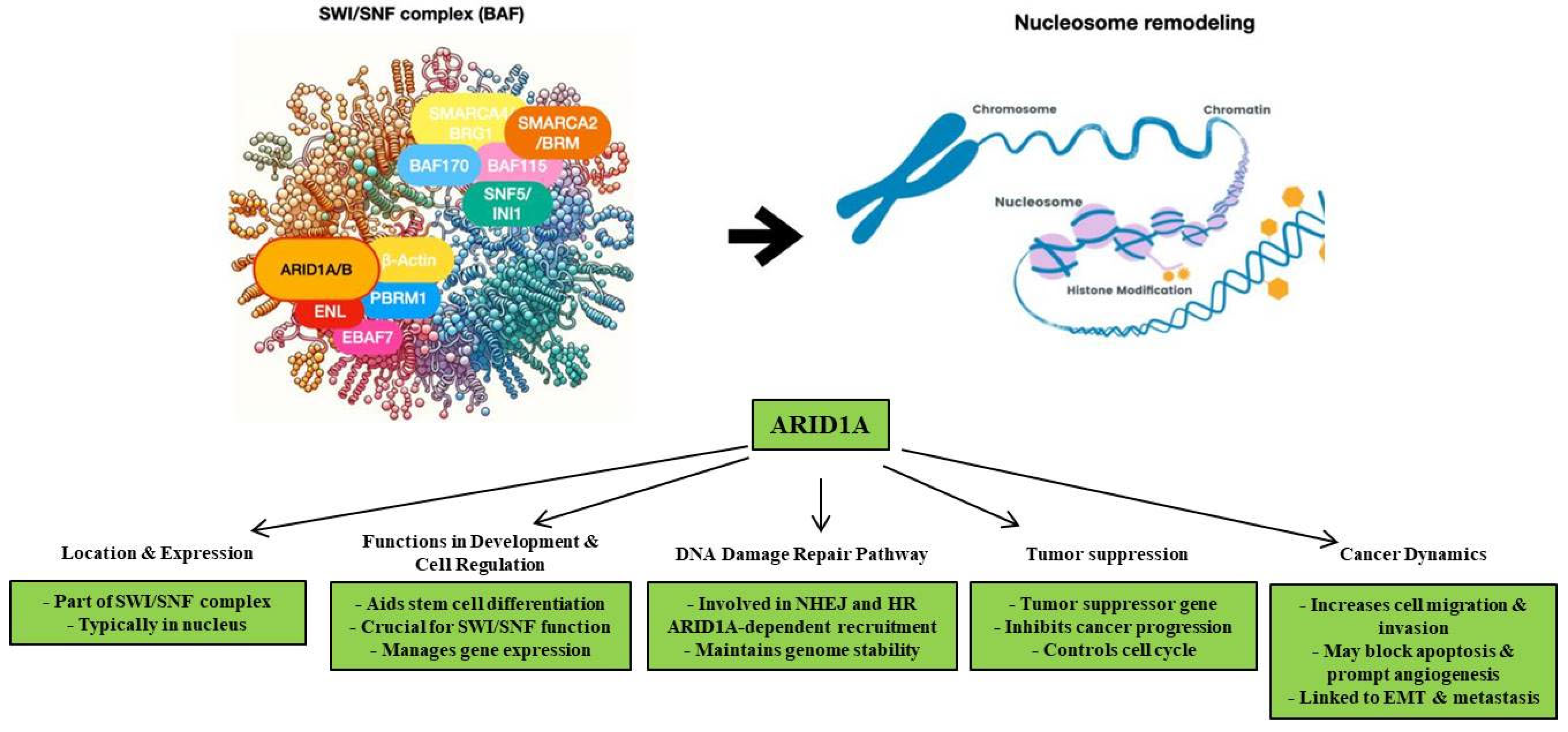
2. Biological Functions of ARID1A
3. ARID1A Mutations in Gastric Cancer
4. Clinical and Prognostic Significance of ARID1A Mutation in Gastric Cancer
5. Molecular Pathways Involved in ARID1A Mutation
6. Therapeutic Approach in ARID1A-Deficient Gastric Cancer
7. ARID1A Immunohistochemistry in Gastric Cancer
8. ARID1A Loss in Precursor Lesions
9. Role of ARID1A in Development and Progression of Tumors Other Than Gastric Cancer
- Hepatocellular Carcinoma (HCC): ARID1A is one of the most frequently mutated genes in hepatocellular carcinoma, with mutations occurring in 10% to 17% of cases. ARID1A mutations affect several pathways critical for tumor growth [16,79,80]. Low ARID1A expression correlates with shorter patient survival, suggesting its involvement in HCC development and metastasis [79,80,81].
- Endometrial Cancer: The rate of ARID1A mutation in low-grade endometrioid adenocarcinomas is 47%, while in high-grade endometrioid adenocarcinomas, serous adenocarcinomas, and carcinosarcomas, it is 60%, 11%, and 24%, respectively [16,82,83,84]. Moreover, in 14–22% of uterine endometrial clear cell carcinoma, ARID1A expression is also found to be downregulated [16,83,84]. Notably, ARID1A mutations have been reported to occur also in preneoplastic lesions, indicating its role in early cancer development. [16,83,84].
- Colorectal Cancer: ARID1A mutations are detected in 10% of colorectal cancers and are strictly related to mismatch repair deficiency [16,90,91]. In detail, ARID1A downregulation has been reported to influence the proliferation of colorectal cancer cells and their resistance to chemotherapy [16,90,91]. Moreover, ARID1A loss has been shown to promote epithelial–mesenchymal transition (EMT) in colon cancer, contributing to metastasis [16,90,91].
- Pancreatic Cancer: Recent comprehensive sequencing analyses of pancreatic cancer have demonstrated ARID1A mutations in 6% of cases [16,92,93]. ARID1A may represent a tumor suppressor gene in pancreatic carcinogenesis, as its expression levels correlate with tumor differentiation and stage, although not with lymph node or distant metastasis, sex, or age [16,92,93]. In mouse models, ARID1A deficiency has been shown to accelerate tumor progression, leading to high-stage disease [16,92,93].
- Breast Cancer: ARID1A not only exerts antitumor effects such as inhibiting cancer cell migration and invasion in breast cancer but also enhances the sensitivity of breast cancer cells to chemotherapy [16,94,95,96,97]. Moreover, it has been shown to influence the activity of estrogen receptor α+ [16,94,95,96,97]. This receptor, when activated, induces an oncogenic signal which regulates tumor cell proliferation in breast cancer [77,92,93,94,95]. Therefore, wild-type ARID1A has been shown to correlate with improved clinical outcomes in ER+ breast cancer patients [77,92,93,94,95]. By contrast, ARID1A inactivating mutations are more frequently detected in treatment-resistant and metastatic tumors [16,94,95,96,97].
10. Clinical Utility of ARID1A in GC: Limitations, Challenge and Future Directions
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- GBD US Health Disparities Collaborators. The burden of stomach cancer mortality by county, race, and ethnicity in the USA, 2000–2019: A systematic analysis of health disparities. Lancet Reg. Health Am. 2023, 24, 100547. [Google Scholar]
- Dicken, B.J.; Bigam, D.L.; Cass, C.; Mackey, J.R.; Joy, A.A.; Hamilton, S.M. Gastric adenocarcinoma: Review and considerations for future directions. Ann. Surg. 2005, 241, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Gullo, I.; Grillo, F.; Mastracci, L.; Vanoli, A.; Carneiro, F.; Saragoni, L.; Limarzi, F.; Ferro, J.; Parente, P.; Fassan, M. Precancerous lesions of the stomach, gastric cancer and hereditary gastric cancer syndromes. Pathologica 2020, 112, 166–185. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Zhao, H.P.; Yu, Y.; Wang, J.H.; Guo, L.; Liu, J.Y.; Pu, J.; Lv, J. Updates on global epidemiology, risk and prognostic factors of gastric cancer. World J. Gastroenterol. 2023, 29, 2452–2468. [Google Scholar] [CrossRef] [PubMed]
- Nshizirungu, J.P.; Bennis, S.; Mellouki, I.; Sekal, M.; Benajah, D.A.; Lahmidani, N.; El Bouhaddouti, H.; Ibn Majdoub, K.; Ibrahimi, S.A.; Celeiro, S.P.; et al. Reproduction of the Cancer Genome Atlas (TCGA) and Asian Cancer Research Group (ACRG) Gastric Cancer Molecular Classifications and Their Association with Clinicopathological Characteristics and Overall Survival in Moroccan Patients. Dis. Markers 2021, 2021, 9980410. [Google Scholar] [CrossRef] [PubMed]
- Sohn, B.H.; Hwang, J.E.; Jang, H.J.; Lee, H.S.; Oh, S.C.; Shim, J.J.; Lee, K.W.; Kim, E.H.; Yim, S.Y.; Lee, S.H.; et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin. Cancer Res. 2017, 23, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Rodriquenz, M.G.; Roviello, G.; D’Angelo, A.; Lavacchi, D.; Roviello, F.; Polom, K. MSI and EBV Positive Gastric Cancer’s Subgroups and Their Link with Novel Immunotherapy. J. Clin. Med. 2020, 9, 1427. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.; Camargo, M.C.; Leite, M.; Fuentes-Pananá, E.M.; Rabkin, C.S.; Machado, J.C. Pathogenesis of Gastric Cancer: Genetics and Molecular Classification. Curr. Top. Microbiol. Immunol. 2017, 400, 277–304. [Google Scholar] [PubMed]
- Garattini, S.K.; Basile, D.; Cattaneo, M.; Fanotto, V.; Ongaro, E.; Bonotto, M.; Negri, F.V.; Berenato, R.; Ermacora, P.; Cardellino, G.G.; et al. Molecular classifications of gastric cancers: Novel insights and possible future applications. World J. Gastrointest. Oncol. 2017, 9, 194–208. [Google Scholar] [CrossRef]
- Bonelli, P.; Borrelli, A.; Tuccillo, F.M.; Silvestro, L.; Palaia, R.; Buonaguro, F.M. Precision medicine in gastric cancer. World J. Gastrointest. Oncol. 2019, 11, 804–829. [Google Scholar] [CrossRef]
- Liu, N.; Wu, Y.; Cheng, W.; Wu, Y.; Wang, L.; Zhuang, L. Identification of novel prognostic biomarkers by integrating multi-omics data in gastric cancer. BMC Cancer 2021, 21, 460. [Google Scholar] [CrossRef]
- Siciliano, M.C.; Tornambè, S.; Cevenini, G.; Sorrentino, E.; Granai, M.; Giovannoni, G.; Marrelli, D.; Biviano, I.; Roviello, F.; Yoshiyama, H.; et al. EBV persistence in gastric cancer cases conventionally classified as EBER-ISH negative. Infect. Agent Cancer 2022, 17, 57. [Google Scholar] [CrossRef] [PubMed]
- Fanaian, N.K.; Cohen, C.; Waldrop, S.; Wang, J.; Shehata, B.M. Epstein-Barr virus (EBV)-encoded RNA: Automated in-situ hybridization (ISH) compared with manual ISH and immunohistochemistry for detection of EBV in pediatric lymphoproliferative disorders. Pediatr. Dev. Pathol. 2009, 12, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tang, C. The Role of ARID1A in Tumors: Tumor Initiation or Tumor Suppression? Front. Oncol. 2021, 11, 745187. [Google Scholar] [CrossRef]
- Huang, S.C.; Ng, K.F.; Chang, I.Y.; Chang, C.J.; Chao, Y.C.; Chang, S.C.; Chen, M.C.; Yeh, T.S.; Chen, T.C. The clinicopathological significance of SWI/SNF alterations in gastric cancer is associated with the molecular subtypes. PLoS ONE 2021, 16, e0245356. [Google Scholar] [CrossRef]
- Li, J.J.; Lee, C.S. The Role of the AT-Rich Interaction Domain 1A Gene (ARID1A) in Human Carcinogenesis. Genes 2023, 15, 5. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Zhao, J.; Wu, Y.; Zhang, N.; Shen, W. ARID1A mutations in cancer development: Mechanism and therapy. Carcinogenesis 2023, 44, 197–208. [Google Scholar] [CrossRef]
- Guan, B.; Gao, M.; Wu, C.H.; Wang, T.L.; Shih, I.e.M. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia 2012, 14, 986–993. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Garcia, E.; García-Puga, M.; Arevalo, S.; Matheu, A. Towards precision medicine: Linking genetic and cellular heterogeneity in gastric cancer. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794628. [Google Scholar] [CrossRef]
- Zhou, H.; Tan, S.; Li, H.; Lin, X. Expression and significance of EBV, ARID1A and PIK3CA in gastric carcinoma. Mol. Med. Rep. 2019, 19, 2125–2136. [Google Scholar] [CrossRef]
- Wu, J.N.; Roberts, C.W. ARID1A mutations in cancer: Another epigenetic tumor suppressor? Cancer Discov. 2013, 3, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Maeda, D.; Hino, R.; Otake, Y.; Isogai, M.; Ushiku, A.S.; Matsusaka, K.; Kunita, A.; Ushiku, T.; Uozaki, H.; et al. ARID1A expression loss in gastric cancer: Pathway-dependent roles with and without Epstein-Barr virus infection and microsatellite instability. Virchows Arch. 2012, 461, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Anabel Sinberger, L.; Zahavi, T.; Sonnenblick, A.; Salmon-Divon, M. Coexistent ARID1A-PIK3CA mutations are associated with immune-related pathways in luminal breast cancer. Sci. Rep. 2023, 13, 20911. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Chandler, R.L.; Damrauer, J.S.; Raab, J.R.; Schisler, J.C.; Wilkerson, M.D.; Didion, J.P.; Starmer, J.; Serber, D.; Yee, D.; Xiong, J.; et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat. Commun. 2015, 6, 6118. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, S.; Klar, M.; Matsuzaki, S.; Roman, L.D.; Sood, A.K.; Matsuo, K. Uterine carcinosarcoma: Contemporary clinical summary, molecular updates, and future research opportunity. Gynecol. Oncol. 2021, 160, 586–601. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, H.; Heo, Y.J.; Kang, S.Y.; Ahn, S.; Lee, J.; Kim, K.M. PIK3CA mutation subtype delineates distinct immune profiles in gastric carcinoma. J. Pathol. 2023, 260, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, C.K.; Noh, S.; Cheong, J.H.; Noh, S.H.; Kim, H. Prognostic Significance of ARID1A Expression Patterns Varies with Molecular Subtype in Advanced Gastric Cancer. Gut Liver 2023, 17, 753–765. [Google Scholar] [CrossRef]
- Ibarrola-Villava, M.; Llorca-Cardeñosa, M.J.; Tarazona, N.; Mongort, C.; Fleitas, T.; Perez-Fidalgo, J.A.; Roselló, S.; Navarro, S.; Ribas, G.; Cervantes, A. Deregulation of ARID1A, CDH1, cMET and PIK3CA and target-related microRNA expression in gastric cancer. Oncotarget 2015, 6, 26935–26945. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Sy, K.; Kalloger, S.E.; Li-Chang, H.; Woods, R.; Kumar, A.; Streutker, C.J.; Hafezi-Bakhtiari, S.; Zhou, C.; Lim, H.J.; et al. ARID1A/BAF250a as a prognostic marker for gastric carcinoma: A study of 2 cohorts. Hum. Pathol. 2014, 45, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Qadir, J.; Majid, S.; Khan, M.S.; Rashid, F.; Wani, M.D.; Bhat, S.A. Implication of ARID1A Undercurrents and PDL1, TP53 Overexpression in Advanced Gastric Cancer. Pathol. Oncol. Res. 2021, 27, 1609826. [Google Scholar] [CrossRef] [PubMed]
- Blanchet, A.; Bourgmayer, A.; Kurtz, J.E.; Mellitzer, G.; Gaiddon, C. Isoforms of the p53 Family and Gastric Cancer: A Ménage à Trois for an Unfinished Affair. Cancers 2021, 13, 916. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Reske, J.J.; Wilson, M.R.; Holladay, J.; Siwicki, R.A.; Skalski, H.; Harkins, S.; Adams, M.; Risinger, J.I.; Hostetter, G.; Lin, K.; et al. Co-existing TP53 and ARID1A mutations promote aggressive endometrial tumorigenesis. PLoS Genet. 2021, 17, e1009986. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.N.; Teng, Q.X.; Tian, Q.; Chen, W.; Xie, Y.; Wu, K.; Zeng, Q.; Zeng, L.; Pan, Y.; Chen, Z.S.; et al. Signaling pathways and therapeutic interventions in gastric cancer. Signal. Transduct. Target. Ther. 2022, 7, 358. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Chen, Y.B.; Pan, K.; Wang, W.; Chen, S.P.; Chen, J.G.; Zhao, J.J.; Lv, L.; Pan, Q.Z.; Li, Y.Q.; et al. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS ONE 2012, 7, e40364. [Google Scholar] [CrossRef]
- Yang, L.; Wei, S.; Zhao, R.; Wu, Y.; Qiu, H.; Xiong, H. Loss of ARID1A expression predicts poor survival prognosis in gastric cancer: A systematic meta-analysis from 14 studies. Sci. Rep. 2016, 6, 28919. [Google Scholar] [CrossRef]
- Inada, R.; Sekine, S.; Taniguchi, H.; Tsuda, H.; Katai, H.; Fujiwara, T.; Kushima, R. ARID1A expression in gastric adenocarcinoma: Clinicopathological significance and correlation with DNA mismatch repair status. World J. Gastroenterol. 2015, 21, 2159–2168. [Google Scholar] [CrossRef] [PubMed]
- Fontana, B.; Gallerani, G.; Salamon, I.; Pace, I.; Roncarati, R.; Ferracin, M. ARID1A in cancer: Friend or foe? Front. Oncol. 2023, 13, 1136248. [Google Scholar] [CrossRef]
- Zhang, Q.; Yan, H.B.; Wang, J.; Cui, S.-J.; Wang, X.-Q.; Jiang, Y.-H.; Feng, L.; Yang, P.-Y.; Liu, F. Chromatin remodeling gene AT-rich interactive domain-containing protein 1A suppresses gastric cancer cell proliferation by targeting PIK3CA and PDK1. Oncotarget 2016, 7, 46127–46141. [Google Scholar] [CrossRef]
- Guan, B.; Wang, T.L.; Shih Ie, M. ARID1A, a factor that promotes formation of SWI/ SNF-mediated chromatin remodeling, is a Tumor suppressor in gynecologic cancers. Cancer Res. 2011, 71, 6718–6727. [Google Scholar] [CrossRef] [PubMed]
- Bosse, T.; Ter Haar, N.T.; Seeber, L.M.; Hes, F.J.; Vasen, H.F.; Nout, R.A.; Creutzberg, C.L.; Morreau, H.; Smit, V.T. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod. Pathol. 2013, 26, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Loe, A.K.H.; Francis, R.; Seo, J.; Du, L.; Wang, Y.; Kim, J.-E.; Hakim, S.W.; Kim, J.-E.; He, H.H.; Guo, H.; et al. Uncovering the dosage-dependent roles of Arid1a in gastric tumorigenesis for combinatorial drug therapy. J. Exp. Med. 2021, 218, 25. [Google Scholar] [CrossRef] [PubMed]
- Setia, N.; Agoston, A.T.; Han, H.S.; Mullen, J.T.; Duda, D.G.; Clark, J.W.; Deshpande, V.; Mino-Kenudson, M.; Srivastava, A.; Lennerz, J.K.; et al. A protein and mRNA expression-based classification of gastric cancer. Mod. Pathol. 2016, 29, 772–784. [Google Scholar] [CrossRef]
- Kim, Y.B.; Ahn, J.M.; Bae, W.J.; Sung, C.O.; Lee, D. Functional loss of ARID1A is tightly associated with high PD-L1 expression in gastric cancer. Int. J. Cancer 2019, 145, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Okamura, R.; Kato, S.; Lee, S.; Jimenez, R.E.; Sicklick, J.K.; Kurzrock, R. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J. Immunother. Cancer. 2020, 8, e000438. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Duan, R.; Cong, L.; Song, Y. The effects of ARID1A mutation in gastric cancer and its significance for treatment. Cancer Cell Int. 2023, 23, 296. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, G.; Ding, Y.; Huang, Y.; Liu, S.; Zhou, L.; Wei, W.; Wang, J.; Hu, G. Combined treatment with PI3K inhibitor BKM120 and PARP inhibitor olaparib is effective in inhibiting the gastric cancer cells with ARID1A deficiency. Oncol. Rep. 2018, 40, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Bitler, B.G.; Aird, K.M.; Garipov, A.; Li, H.; Amatangelo, M.; Kossenkov, A.V.; Schultz, D.C.; Liu, Q.; Shih, I.-M.; Conejo-Garcia, J.R.; et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 2015, 21, 231–238. [Google Scholar] [CrossRef]
- Yamada, L.; Saito, M.; Thar Min, A.K.; Saito, K.; Ashizawa, M.; Kase, K.; Nakajima, S.; Onozawa, H.; Okayama, H.; Endo, H.; et al. Selective sensitivity of EZH2 inhibitors based on synthetic lethality in ARID1A-deficient gastric cancer. Gastric Cancer 2021, 24, 60–71. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, R.; Bitler, B.G. Arid1a controls tissue regeneration. Stem. Cell Investig. 2016, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chuang, J.C.; Kanchwala, M.; Wu, L.; Celen, C.; Li, L.; Liang, H.; Zhang, S.; Maples, T.; Nguyen, L.H.; et al. Suppression of the SWI/SNF Component Arid1a Promotes Mammalian Regeneration. Cell Stem Cell. 2016, 18, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.C.; Wang, T.L.; Shih, I.e.M. The emerging roles of ARID1A in tumor suppression. Cancer Biol. Ther. 2014, 15, 655–664. [Google Scholar] [CrossRef] [PubMed Central]
- Watanabe, R.; Ui, A.; Kanno, S.; Ogiwara, H.; Nagase, T.; Kohno, T.; Yasui, A. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014, 74, 2465–2475. [Google Scholar] [CrossRef] [PubMed]
- Tomihara, H.; Carbone, F.; Perelli, L.; Huang, J.K.; Soeung, M.; Rose, J.L.; Robinson, F.S.; Lissanu Deribe, Y.; Feng, N.; Takeda, M.; et al. Loss of ARID1A Promotes Epithelial-Mesenchymal Transition and Sensitizes Pancreatic Tumors to Proteotoxic Stress. Cancer Res. 2021, 81, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Somsuan, K.; Peerapen, P.; Boonmark, W.; Plumworasawat, S.; Samol, R.; Sakulsak, N.; Thongboonkerd, V. ARID1A knockdown triggers epithelial-mesenchymal transition and carcinogenesis features of renal cells: Role in renal cell carcinoma. FASEB J. 2019, 33, 12226–12239. [Google Scholar] [CrossRef]
- Li, B.; Zhang, F.; Niu, Q.; Liu, J.; Yu, Y.; Wang, P.; Zhang, S.; Zhang, H.; Wang, Z. A molecular classification of gastric cancer associated with distinct clinical outcomes and validated by an XGBoost-based prediction model. Mol. Ther. Nucleic Acids 2022, 31, 224–240. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Q.; Sun, S.; Ye, J.; Li, Z.; Cui, Z.; Liu, Q.; Zhang, Y.; Xiong, S.; Zhang, S. Prognostic and immune infiltration significance of ARID1A in TCGA molecular subtypes of gastric adenocarcinoma. Cancer Med. 2023, 12, 16716–16733. [Google Scholar] [CrossRef]
- Kase, K.; Saito, M.; Nakajima, S.; Takayanagi, D.; Saito, K.; Yamada, L.; Ashizawa, M.; Nakano, H.; Hanayama, H.; Onozawa, H.; et al. ARID1A deficiency in EBV-positive gastric cancer is partially regulated by EBV-encoded miRNAs, but not by DNA promotor hypermethylation. Carcinogenesis 2021, 42, 21–30. [Google Scholar] [CrossRef]
- Kumar, V.; Ramnarayanan, K.; Sundar, R.; Padmanabhan, N.; Srivastava, S.; Koiwa, M.; Yasuda, T.; Koh, V.; Huang, K.K.; Tay, S.T.; et al. Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer. Cancer Discov. 2022, 12, 670–691. [Google Scholar] [CrossRef]
- Lee, D.; Yu, E.J.; Ham, I.-H.; Hur, H.; Kim, Y.-S. AKT inhibition is an effective treatment strategy in ARID1A-deficient gastric cancer cells. Onco. Targets Ther. 2017, 10, 4153–4159. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.S.; Doi, T.; Jang, R.W.; Muro, K.; Satoh, T.; Machado, M.; Sun, W.; Jalal, S.I.; Shah, M.A.; Metges, J.-P.; et al. Safety and Efficacy of Pembrolizumab Monotherapy in patients with previously treated Advanced gastric and gastroesophageal Junction Cancer: Phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. 2018, 4, e180013. [Google Scholar] [CrossRef] [PubMed]
- Janjigian, Y.Y.; Shitara, K.; Moehler, M.; Garrido, M.; Salman, P.; Shen, L.; Wyrwicz, L.; Yamaguchi, K.; Skoczylas, T.; Bragagnoli, A.C.; et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet 2021, 398, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Lemery, S.; Keegan, P.; Pazdur, R. First FDA approval Agnostic of cancer site—When a biomarker defines the indication. N. Engl. J. Med. 2017, 377, 1409–1412. [Google Scholar] [CrossRef] [PubMed]
- Angelico, G.; Broggi, G.; Tinnirello, G.; Puzzo, L.; Vecchio, G.M.; Salvatorelli, L.; Memeo, L.; Santoro, A.; Farina, J.; Mulé, A.; et al. Tumor Infiltrating Lymphocytes (TILS) and PD-L1 Expression in Breast Cancer: A Review of Current Evidence and Prognostic Implications from Pathologist’s Perspective. Cancers 2023, 15, 4479. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.A.; Ramos, M.F.K.P.; Faraj, S.F.; Dias, A.R.; Yagi, O.K.; Zilberstein, B.; Cecconello, I.; Alves, V.A.F.; de Mello, E.S.; Ribeiro, U., Jr. Clinicopathological and prognostic features of Epstein-Barr virus infection, microsatellite instability, and PD-L1 expression in gastric cancer. J. Surg. Oncol. 2018, 117, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, M.; Jiang, Z.; Wang, X. ARID1A mutations are associated with increased immune activity in gastrointestinal cancer. Cells 2019, 8, 678. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Chui, M.H.; Rahmanto, Y.S.; Yu, Z.-C.; Shamanna, R.A.; Bellani, M.A.; Gaillard, S.; Ayhan, A.; Viswanathan, A.; Seidman, M.M.; et al. Loss of ARID1A in Tumor cells renders selective vulnerability to combined ionizing radiation and PARP inhibitor therapy. Clin. Cancer Res. 2019, 25, 5584–5594. [Google Scholar] [CrossRef] [PubMed]
- Noh, K.H.; Kang, T.H.; Kim, J.H.; I Pai, S.; Lin, K.Y.; Hung, C.-F.; Wu, T.-C.; Kim, T.W. Activation of akt as a mechanism for Tumor immune evasion. Mol. Ther. 2009, 17, 439–447. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Massi, D.; Teng, M.W.L.; Mandala, M. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin. Cancer Biol. 2018, 48, 91–103. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Tse, K.; Zeng, T.; Senz, J.; McConechy, M.K.; Anglesio, M.S.; Kalloger, S.E.; et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010, 363, 1532–1543. [Google Scholar] [CrossRef]
- Bin Guan, B.; Mao, T.-L.; Panuganti, P.K.; Kuhn, E.; Kurman, R.J.; Maeda, D.; Chen, E.; Jeng, Y.-M.; Wang, T.-L.; Shih, I.-M. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am. J. Surg. Pathol. 2011, 35, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhou, Y.; Weiser, M.R.; Gönen, M.; Zhang, L.; Samdani, T.; Bacares, R.; DeLair, D.; Ivelja, S.; Vakiani, E.; et al. Immunohistochemical detection of ARID1A in colorectal carcinoma: Loss of staining is associated with sporadic microsatellite unstable tumors with medullary histology and high TNM stage. Hum. Pathol. 2014, 45, 2430–2436. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.M.; Kipp, B.R.; Halling, K.C.; Kerr, S.E.; Smith, D.I.; Distad, T.J.; Clayton, A.C.; Medeiros, F. Molecular characterization of endometrial cancer: A correlative study assessing microsatellite instability, MLH1 hypermethylation, DNA mismatch repair protein expression, and PTEN, PIK3CA, KRAS, and BRAF mutation analysis. Int. J. Gynecol. Pathol. 2012, 31, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Kohno, T.; Kono, K. Heterogeneity of ARID1A expression in gastric cancer may affect patient survival and therapeutic efficacy. Hum. Pathol. 2020, 101, 80–81. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Qin, W.; Yang, L.; Wang, L.; Wang, Y.; Shen, J.; Xiong, W.; Yu, S.; Song, S.; Ajani, J.A.; et al. Genetic alterations and expression characteristics of ARID1A impact tumor immune contexture and survival in early-onset gastric cancer. Am. J. Cancer Res. 2020, 10, 3947–3972. [Google Scholar]
- Abe, H.; Kunita, A.; Otake, Y.; Kanda, T.; Kaneda, A.; Ushiku, T.; Fukayama, M. Virus-host interactions in carcinogenesis of Epstein-Barr virus-associated gastric carcinoma: Potential roles of lost ARID1A expression in its early stage. PLoS ONE 2021, 16, e0256440. [Google Scholar] [CrossRef]
- Abe, H.; Rokutan, H.; Totoki, Y.; Nakamura, H.; Shibata, T.; Ushiku, T.; Fukayama, M. Lost expression of AT-rich interaction domain 1A in the gastric mucosa-A constituent of field cancerization in the stomach. Pathol. Int. 2023, 73, 234–245. [Google Scholar] [CrossRef]
- Fujimoto, A.; Totoki, Y.; Abe, T.; Boroevich, K.A.; Hosoda, F.; Nguyen, H.H.; Aoki, M.; Hosono, N.; Kubo, M.; Miya, F.; et al. Whole-Genome Sequencing of Liver Cancers Identifies Etiological Influences on Mutation Patterns and Recurrent Mutations in Chromatin Regulators. Nat. Genet. 2012, 44, 760–764. [Google Scholar] [CrossRef]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.B.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated Analysis of Somatic Mutations and Focal Copy—Number Changes Identifies Key Genes and Pathways in Hepatocellular Carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef]
- Huang, J.; Deng, Q.; Wang, Q.; Li, K.Y.; Dai, J.H.; Li, N.; Zhu, Z.-D.; Zhou, B.; Liu, X.-Y.; Liu, R.-F.; et al. Exome Sequencing of Hepatitis B Virusassociated Hepatocellular Carcinoma. Nat. Genet. 2012, 44, 1117–1121. [Google Scholar] [CrossRef]
- Santoro, A.; Angelico, G.; Travaglino, A.; Inzani, F.; Arciuolo, D.; Valente, M.; D’Alessandris, N.; Scaglione, G.; Fiorentino, V.; Raffone, A.; et al. New Pathological and Clinical Insights in Endometrial Cancer in View of the Updated ESGO/ESTRO/ESP Guidelines. Cancers 2021, 13, 2623. [Google Scholar] [CrossRef] [PubMed]
- McConechy, M.K.; Ding, J.; Cheang, M.C.; Wiegand, K.; Senz, J.; Tone, A.; Yang, W.; Prentice, L.; Tse, K.; Zeng, T.; et al. Use of Mutation Profiles to Refine the Classification of Endometrial Carcinomas. J. Pathol. 2012, 228, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Hoang, L.N.; McConechy, M.K.; Meng, B.; McIntyre, J.B.; Ewanowich, C.; Gilks, C.B.; Huntsman, D.G.; Köbel, M.; Lee, C. Targeted Mutation Analysis of Endometrial Clear Cell Carcinoma. Histopathology 2015, 66, 664–674. [Google Scholar] [CrossRef]
- Alldredge, J.K.; Eskander, R.N. EZH2 Inhibition in ARID1A Mutated Clear Cell and Endometrioid Ovarian and Endometrioid Endometrial Cancers. Gynecol. Oncol. Res. Pract. 2017, 4, 17. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Hennessy, B.T.; Leung, S.; Wang, Y.; Ju, Z.; McGahren, M.; Kalloger, S.E.; Finlayson, S.; Stemke-Hale, K.; Lu, Y.; et al. A Functional Proteogenomic Analysis of Endometrioid and Clear Cell Carcinomas Using Reverse Phase Protein Array and Mutation Analysis: Protein Expression Is Histotype-Specific and Loss of ARID1A/BAF250a Is Associated with AKT Phosphorylation. BMC Cancer 2014, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Prat, J.; D’Angelo, E.; Espinosa, I. Ovarian carcinomas: At least five different diseases with distinct histological features and molecular genetics. Hum. Pathol. 2018, 80, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Sasagawa, T.; Jinno-Oue, A.; Nagamatsu, T.; Morita, K.; Tsuruga, T.; Mori-Uchino, M.; Fujii, T.; Shibuya, M. Production of an Anti-Angiogenic Factor Sflt1 Is Suppressed via Promoter Hypermethylation of FLT1 Gene in Choriocarcinoma Cells. BMC Cancer 2020, 20, 112. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Xu, S.; Li, J.; Li, L.; Tang, C. Role of ARID1A in the Regulation of Human Trophoblast Migration and Invasion. Reprod. Sci. 2021, 29, 2363–2373. [Google Scholar] [CrossRef]
- Xie, C.; Fu, L.; Han, Y.; Li, Q.; Wang, E. Decreased ARID1A Expression Facilitates Cell Proliferation and Inhibits 5-Fluorouracil-Induced Apoptosis in Colorectal Carcinoma. Tumor Biol. 2014, 35, 7921–7927. [Google Scholar] [CrossRef]
- Sen, M.; Wang, X.; Hamdan, F.H.; Rapp, J.; Eggert, J.; Kosinsky, R.L.; Wegwitz, F.; Kutschat, A.P.; Younesi, F.S.; Gaedcke, J.; et al. ARID1A Facilitates KRAS Signaling-Regulated Enhancer Activity in an AP1-Dependent Manner in Colorectal Cancer Cells. Clin. Epigenet. 2019, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203.e13. [Google Scholar] [CrossRef]
- Ferri-Borgogno, S.; Barui, S.; McGee, A.M.; Griffiths, T.; Singh, P.K.; Piett, C.G.; Ghosh, B.; Bhattacharyya, S.; Singhi, A.; Pradhan, K.; et al. Paradoxical Role of AT-Rich Interactive Domain 1A in Restraining Pancreatic Carcinogenesis. Cancers 2020, 12, 2695. [Google Scholar] [CrossRef] [PubMed]
- Takao, C.; Morikawa, A.; Ohkubo, H.; Kito, Y.; Saigo, C.; Sakuratani, T.; Futamura, M.; Takeuchi, T.; Yoshida, K. Downregulation of ARID1A, a Component of the SWI/SNF Chromatin Remodeling Complex, in Breast Cancer. J. Cancer 2017, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gao, X.; Zhou, K.; Jiang, T.; Gao, S.; Liu, P.; Zuo, X.; Shi, X. Role of ARID1A in Epithelial–Mesenchymal Transition in Breast Cancer Its Effect on Cell Sensitivity to, 5.−.F.U. Int. J. Mol. Med. 2020, 46, 1683–1694. [Google Scholar] [PubMed]
- Blanchard, Z.; Vahrenkamp, J.M.; Berrett, K.C.; Arnesen, S.; Gertz, J. Estrogen-Independent Molecular Actions of Mutant Estrogen Receptor 1 in Endometrial Cancer. Genome Res. 2019, 29, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, S.; Hossan, T.; Alawi, M.; Najafova, Z.; Indenbirken, D.; Bedi, U.; Taipaleenmäki, H.; Ben-Batalla, I.; Scheller, M.; Loges, S.; et al. Bromodomain Protein BRD4 Is Required for Estrogen Receptor-Dependent Enhancer Activation and Gene Transcription. Cell Rep. 2014, 8, 460–469. [Google Scholar] [CrossRef]
- Mullen, J.; Kato, S.; Sicklick, J.K.; Kurzrock, R. Targeting ARID1A mutations in cancer. Cancer Treat. Rev. 2021, 100, 102287. [Google Scholar] [CrossRef]

| Study | ARID1A Expression Status | Sample Size | OS | PFS | Prognostic Significance |
|---|---|---|---|---|---|
| Zhou et al. [20] | Lower than normal tissue | Not specified | Not specified | Reduced | Not specified |
| Wang et al. [29] | Loss. | 272 primary GCs | Associated with poor prognosis | Not specified | Independent risk factor for poor prognosis |
| Ibarrola–Villava et al. [30] | Loss | Not specified | Higher than those with positive expression | Not specified | Challenges the association with poor prognosis |
| Wiegand et al. [31] | Loss | 173 GCs | No clear relationship observed | Not specified | Conflicting findings |
| References | ||
|---|---|---|
| Frequency of ARID1A mutations in GC |
| Qadir et al. [32] Blanchet et al. [33] Reske et al. [34] Lei et al. [35] |
| Prognostic role of ARID1A | loss of ARID1A expression is associated with both reduced progression-free survival (PFS) and overall survival (OS) | Wang et al. [36] Yang et al. [37] Inada et al. [38] Kim et al. [29] Fontana et al. [39] |
| Interaction of ARID1A with other gene pathways |
| Zhang et al. [40] Guan et al. [41] Bosse et al. [42] Loe et al. [43] |
| Immune-related biomarkers related to ARID1A loss |
| Setia et al. [44] Kim et al [45] Carrasco et al. [46] |
| Therapeutic strategies in ARID1A-deficient GC |
| Lu et al. [47] Yang et al. [48] Bitler et al. [49] Yamada et al. [50] |
| Biomarker | Therapeutic Approach | Clinical Evidence | References/ Clinical Trials |
|---|---|---|---|
| PD-L1 Expression | Correlates with response to PD-1/PD-L1 inhibitors | KEYNOTE-059: Pembrolizumab effective in GC with PD-L1 CPS ≥ 1 CHECKMATE-649: Nivolumab + chemotherapy improved OS in GC/EGJ with PD-L1 CPS ≥ 5 | NCT02335411 NCT02872116 |
| Tumor Mutation Burden (TMB) | Predicts effectiveness of immunotherapy across tumor types | Pembrolizumab FDA approved for metastatic/unresectable solid tumors with dMMR or MSI-H biomarkers | Li et al. [67] Lemery et al. [65] |
| Mismatch Repair Deficiency | Significantly responds to immunotherapy | KEYNOTE-016, 164, 012, 028, and 158 trials | NCT01876511 NCT02460198 NCT01848834 NCT02054806 NCT02628067 |
| Tumor-Infiltrating Lymphocytes | Potential biomarker for PD-1/PD-L1 immunotherapy success | Recognized for predicting PD-1/PD-L1 immunotherapy success | Angelico et al. [65] |
| ARID1A Expression | Correlates with PD-L1 expression, TMB, dMMR/MSI-H, and TILs | Associated with upregulation of PD-L1 via PI3K/AKT/mTOR pathway—Bioinformatics suggest ARID1A-mutated GC may benefit from immunotherapy | Kim et al. [66] Li et al. [67] |
| EZH2 Overexpression | Influences tumor-infiltrating lymphocytes and immunosuppression | Targeting EZH2 may enhance existing immunotherapies in ARID1A-mutated cancers | Lu et al. [47] Bitler et al. [49] |
| Staining Pattern | Interpretation | References |
|---|---|---|
| Diffuse nuclear staining | Positive: no ARID1A mutations | Guan et al. [72] Ye et al. [73] Bosse et al. [74] Saito et al. [75] |
| Complete nuclear loss of ARID1A expression | Negative: associated with mutations in ARID1A | |
| Heterogeneous ARID1A staining | Negative: associated with mutations in ARID1A | |
| Neoplastic cell subpopulation showing abrupt absence of nuclear staining | Negative: associated with mutations in ARID1A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angelico, G.; Attanasio, G.; Colarossi, L.; Colarossi, C.; Montalbano, M.; Aiello, E.; Di Vendra, F.; Mare, M.; Orsi, N.; Memeo, L. ARID1A Mutations in Gastric Cancer: A Review with Focus on Clinicopathological Features, Molecular Background and Diagnostic Interpretation. Cancers 2024, 16, 2062. https://doi.org/10.3390/cancers16112062
Angelico G, Attanasio G, Colarossi L, Colarossi C, Montalbano M, Aiello E, Di Vendra F, Mare M, Orsi N, Memeo L. ARID1A Mutations in Gastric Cancer: A Review with Focus on Clinicopathological Features, Molecular Background and Diagnostic Interpretation. Cancers. 2024; 16(11):2062. https://doi.org/10.3390/cancers16112062
Chicago/Turabian StyleAngelico, Giuseppe, Giulio Attanasio, Lorenzo Colarossi, Cristina Colarossi, Matteo Montalbano, Eleonora Aiello, Federica Di Vendra, Marzia Mare, Nicolas Orsi, and Lorenzo Memeo. 2024. "ARID1A Mutations in Gastric Cancer: A Review with Focus on Clinicopathological Features, Molecular Background and Diagnostic Interpretation" Cancers 16, no. 11: 2062. https://doi.org/10.3390/cancers16112062
APA StyleAngelico, G., Attanasio, G., Colarossi, L., Colarossi, C., Montalbano, M., Aiello, E., Di Vendra, F., Mare, M., Orsi, N., & Memeo, L. (2024). ARID1A Mutations in Gastric Cancer: A Review with Focus on Clinicopathological Features, Molecular Background and Diagnostic Interpretation. Cancers, 16(11), 2062. https://doi.org/10.3390/cancers16112062









