1. Introduction
Hepatocellular carcinoma (HCC) is globally ranked as the third leading cause of cancer-related death, and the incidence of HCC is particularly high in eastern and southeastern Asia [
1]. The main therapeutic options for HCC are liver resection (LR), radiofrequency or microwave ablation, and liver transplant, with the most appropriate treatment depending on tumor burden, liver function, and performance status [
2,
3].
LR in well-selected candidates leads to the best outcomes and can even be an option for patients with more advanced HCC [
3]. Nevertheless, the rate of recurrence after LR is up to 70% at 5 years, and recurrence is associated with poor outcomes; thus, close follow-up after LR is recommended [
4,
5]. At present, follow-up after LR is based on liver sonography combined with measurement of alpha-fetoprotein (AFP) level [
6]. However, this approach is unsatisfactory because the sensitivity of sonography in the detection of recurrent HCC is only 63%—and even lower in patients with cirrhosis—and the specificity of AFP in the detection of recurrent HCC is poor [
7].
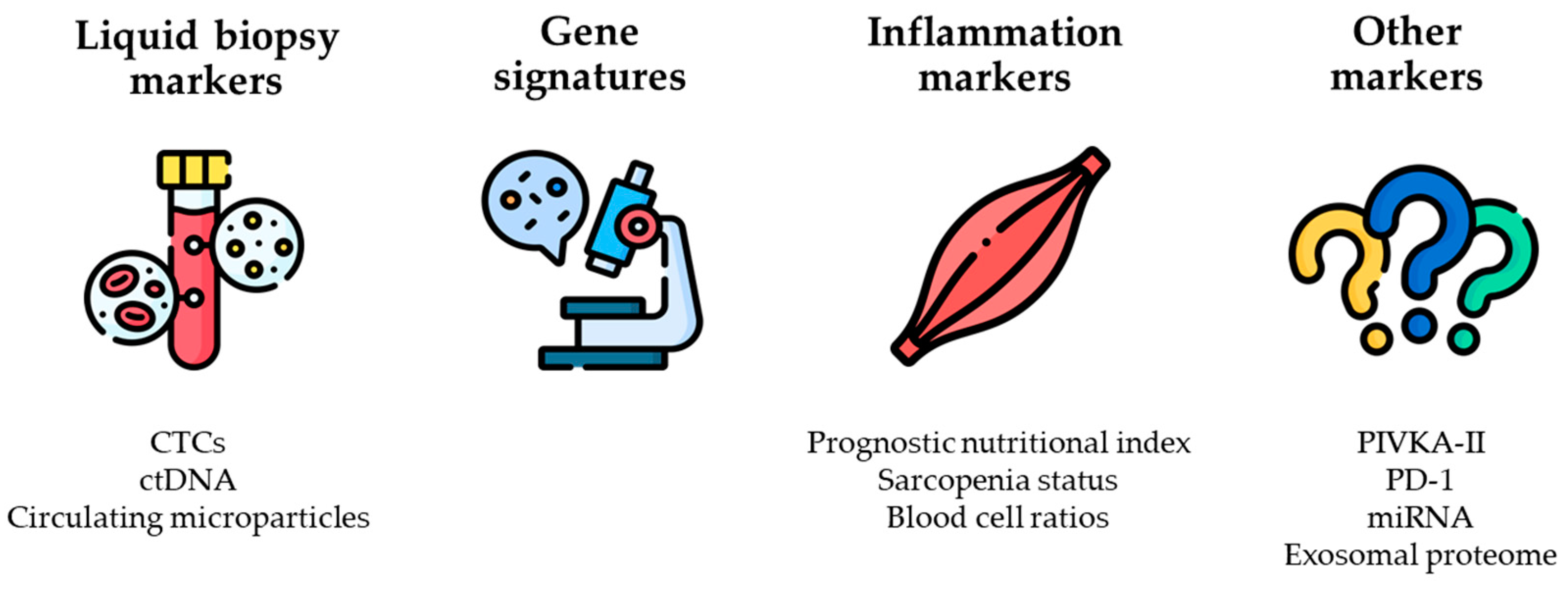
To guide medical and surgical treatments, there is an urgent need to identify novel predictors of recurrence and survival after LR for HCC. In the past decade, many new biomarkers have been reported to correlate with the prognosis of patients with HCC undergoing LR. Specifically, three categories of markers have been identified: (i) markers measured in liquid biopsy specimens, (ii) gene signatures, and (iii) markers associated with inflammation. A few markers not in any of these categories have also been identified, including prothrombin induced by vitamin K absence-II (PIVKA-II), programmed cell death protein-1 (PD-1), microRNAs, and proteins in urinary exosomes (
Figure 1) [
8,
9,
10,
11].
Unfortunately, not all of the aforementioned markers are readily available in clinical practice, and their reproducibility is unclear. The aim of this review is to summarize the use of emerging biomarkers in predicting recurrence and survival after LR for HCC and provide clinicians with a snapshot of the current scientific evidence.
2. Liquid Biopsy Markers
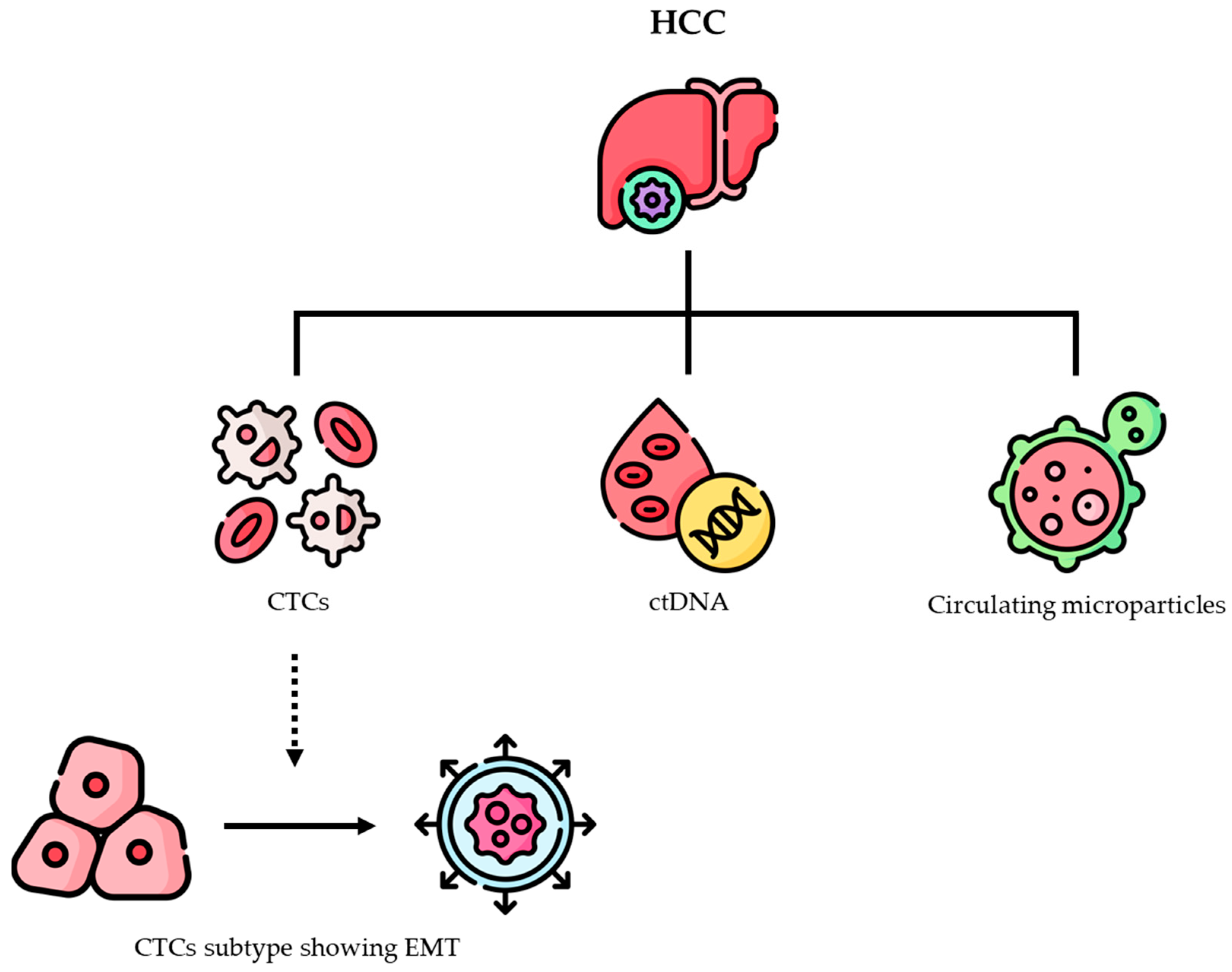
Liquid biopsy, the collection of tumor material from body fluids such as circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and circulating microparticles (CMs) secreted by viable cancer cells, represents a minimally invasive and reproducible tool to acquire molecular data to predict the risk of recurrence and death after LR for HCC [
12,
13] (
Figure 2).
2.1. CTC Level
Epithelial-mesenchymal transition (EMT) is a multistep, reversible process in which epithelial cells de-differentiate and acquire a mesenchymal phenotype. EMT allows tumor cells to enter the bloodstream and become CTCs [
14]. CTSs are defined as nucleated cells positive for epithelial cell adhesion molecule (EpCAM) and cytokeratin and negative for CD45. CTCs are scarce and usually get rapidly damaged by shear stress, killed by the immune system, or apoptosis [
15,
16]. Only a small proportion of CTSs can survive and nestle in peripheral tissue, from which they originate local and distant metastases. Such early hematogenous spread of HCC is associated with increased risk for early recurrence and poor prognosis and should be detected in the perioperative period with high-sensitivity methods [
17].
Detection of CTCs presents technical difficulties and is accomplished using one of three main techniques: (i) use of antibodies against cell surface markers (CellSearch and CTC-Chip assays), (ii) exploitation of cell biophysical properties (e.g., membrane filtration, dielectric mobility, inertial focusing, and dielectric mobility), and (iii) enrichment-free methods (flow cytometry) [
16]. At present, CellSearch is the only U.S. Food and Drug Administration-approved CTC detection tool.
In a 2013 report, Sun et al. [
18] demonstrated that a preoperative CTC count of ≥2 CTCs/7.5 mL predicted tumor recurrence after surgery in 123 HCC patients, especially in patients with AFP levels ≤400 ng/mL or low tumor recurrence risk. Later, the same group divided HCC patients who underwent LR into a retrospective training cohort (144 patients) and a prospective validation cohort (53 patients) and showed that a CTC count ≥3 CTCs/7.5 mL after LR was associated with a higher risk of extrahepatic metastases and reduced survival [
19]. Similar findings were observed in other studies that utilized immunoaffinity technologies for CTC detection [
20,
21,
22].
Fan et al. [
23] performed a prospective study in which multicolor flow cytometry was used to count CTCs in serum samples of 82 HCC patients the day before LR. The authors observed that HCC patients with a CTC percentage (the proportion of examined cells that are CTCs) >0.01% were at higher risk of intrahepatic and extrahepatic recurrence and had a lower overall survival (OS) rate compared with patients with a CTC percentage ≤0.01%. A multivariable analysis showed that CTC percentage >0.01% was an independent predictor of poor disease-free survival (DFS). Hamaoka et al. [
24] used multicolor flow cytometry to detect CTCs in the bloodstream of 85 HCC patients before LR and found a median of 3 CTCs/8 mL per patient. Interestingly, this study proved that CTC count of ≥5 CTCs/8 mL was associated with a higher risk of microscopic portal vein invasion, lower DFS, and lower OS.
Similarly, Ha et al. [
25] performed a prospective study in which 105 HCC patients had blood sampling before and after LR was performed utilizing a tapered slit platform. An increase in CTCs after LR was significantly associated with a higher risk of recurrence, and subgroup analysis showed that an increase in CTCs was also related to lower OS among patients with cirrhosis.
Notably, the ongoing FINDIN-GIBIOREC study (
clinicaltrials.gov ID NCT04800497) is a prospective observational cohort study designed to identify the early presence of CTCs in HCC patients who underwent LR. This multicenter protocol utilizes FACSymphony cell analyzers (BD Biosciences, Freemont, CA, USA) to detect the CTC markers EpCAM, neural cadherin, and CD90 in the perioperative period and integrates information about these markers with findings on computed tomography, magnetic resonance imaging, and AFP level to demonstrate a correlation with recurrence and cancer-related prognosis.
Table 1 summarizes findings from studies assessing the relationship between CTCs and prognosis after LR for HCC.
2.2. CTC Subtype
Epithelial cells usually show apical–basal polarity and are held together laterally by tight junctions and adherens junctions, which maintain the structure of epithelia. EMT allows epithelial cells to lose their polygonal shape and acquire a spindle-shaped mesenchymal morphology [
26]. This transition is associated with the repression of surface epithelial cadherin expression in favor of the expression of mesenchymal markers such as neural cadherin, vimentin, and fibronectin [
27]. EMT and the reverse process, mesenchymal-epithelial transition, are involved in HCC progression and recurrence, giving tumor cells metastatic potential [
28]. Consequently, many authors have focused on markers of EMT and mesenchymal-epithelial transition on CTCs to develop high-sensitivity systems to predict recurrence and OS in patients with HCC [
29,
30,
31,
32,
33,
34,
35].
In particular, Qi et al. [
32] aimed to detect and classify CTCs in the bloodstream before LR. A CTC count ≥ 16 and a mesenchymal-CTC percentage ≥ 2% were found to correlate with ER, multiple intrahepatic recurrences, and lung metastasis. The authors discovered an evident upregulation of the
BCAT1 gene that seemed to regulate EMT. The same group performed a retrospective study using the same technique to classify CTCs in 136 HCC patients undergoing LR with negative margins [
32]. They discovered that the preoperative presence of a high CTC count and the presence of mesenchymal CTC phenotypes were associated with extrahepatic and disseminated intrahepatic recurrence with low DFS. Innovatively, these authors suggested that anatomical resection may be advantageous in patients with low CTC count, and mesenchymal- and epithelial/mesenchymal-negative phenotypes in order to achieve a potentially curative operation. Prognostic benefits and surgical risks should be carefully balanced to plan the optimal resection approach.
Interestingly, CanPatrol was also used by Chen et al. [
35] to count and classify CTCs with EMT markers in HCC patients before and after LR. However, in this study, no correlation was described between mesenchymal CTC count and clinical stage or ER.
Court et al. found that vimentin-positive CTCs can not only help diagnose and stage HCC but also accurately discriminate patients eligible for liver transplant (median, 0 vimentin-positive CTCs) from patients with locally advanced/metastatic disease ineligible for liver transplant (median, 6 vimentin-positive CTCs). Vimentin-positive CTCs can also predict earlier recurrence after curative-intent surgical or locoregional therapy in potentially curable early-stage HCC [
36].
Table 2 summarizes findings from studies assessing the relationship between CTC subtypes and prognosis after LR for HCC.
Orrapin et al. [
37] performed a systematic review of studies that examined CTCs expressing EMT markers (EMT-CTCs), circulating cancer stem cells, or both in patients with HCC. These authors found that most of the studies confirmed a correlation between the presence of EMT-CTCs and circulating cancer stem cells in the bloodstream and other negative prognostic factors: larger tumor size, advanced stage, vascular invasion, distant metastases, and ER.
2.3. Circulating Tumor DNA
ctDNA is the fraction of cell-free DNA (cfDNA) that can be detected in the bloodstream. ctDNA is released from dead tumor cells and from macrophages that have phagocytized tumor cells [
16,
38]; ctDNA can also be actively secreted by tumors as a signaling molecule [
39]. The average concentration of cfDNA in cancer patients is 180 ng/mL and ctDNA usually constitutes approximately 0.01% of total cfDNA [
40]. ctDNA maintains the genetic mutations and epigenetic changes of the originating tumor cells, andit started being considered as a biomarker for tumor diagnosis and monitoring of disease and response to therapy during the last decade [
41].
ctDNA’s short half-life and stability in the bloodstream in theory would allow ctDNA to be a highly sensitive predictor of recurrence and an accurate prognostic factor in HCC patients who underwent LR [
42]. However, the utility of ctDNA in clinical practice is limited. It can be challenging to distinguish ctDNA from cfDNA originating from different body tissues. In addition, ctDNA is thought to be released more easily by less aggressive strains of tumor cells, which means that ctDNA analysis might not reveal information regarding more aggressive tumor cell populations [
16,
43].
Different kits are available for ctDNA extraction, but the current techniques lack standardization. The first approach to ctDNA detection is based on targeting one or a few tumor-specific mutations known from the primary tumor; alternative approaches are to perform a genomewide analysis for copy number aberrations or whole genome sequencing [
39].
Corcoran et al. [
44] illustrated the role of ctDNA in monitoring response to cancer treatment and showed how the ctDNA level decreased after LR in HCC patients, implying the importance of monitoring the trends of ctDNA levels in the blood. Tokuhisa et al. [
45] and An et al. [
46] confirmed that ctDNA level in serum predicted prognosis after LR for HCC in terms of OS, DFS, ER, and development of extrahepatic metastases. Cai et al. [
47] demonstrated how ctDNA might be routinely used as a dynamic biomarker for HCC recurrence after curative-intent hepatectomy. They proved that serial ctDNA sampling performed better in HCC surveillance than did protein biomarkers such as AFP, AFP-L3 (an isoform of AFP), and des-gamma-carboxy prothrombin, identifying microscopic residual tumors and revealing recurrence before it was revealed by imaging.
Liao et al. [
48] utilized the MiSeq system to highlight
TERT,
CTNNB1, and
TP53 gene mutations in ctDNA and correlated their presence with the clinical outcome of HCC patients who underwent LR. The authors suggested that tumor-associated ctDNA mutations correlate with vascular invasion and shorter DFS in HCC patients.
Shen et al. [
49] divided 895 HCC patients into three cohorts and demonstrated that the
TP53 R249S mutation in ctDNA, detected with droplet digital polymerase chain reaction (PCR), represents a promising prognostic biomarker for HCC patients, regardless of whether they have undergone LR. Among patients who had undergone LR (275 patients), patients with the R249S mutation had worse OS and DFS than patients without this mutation.
Another interesting potential ctDNA-related biomarker is the level of methylation or demethylation of ctDNA gene sequences, which seems to provide an early warning of residual HCC and ER after hepatectomy. For instance, Liu et al. [
50] found that
LINE1 hypomethylation and
RASSF1A promoter hypermethylation predict worse OS and ER. Additionally, Chan et al. [
51] demonstrated that residual HCC after LR is related to the extent of ctDNA demethylation. A clinical trial by Xu et al. [
52] showed the importance of HCC methylation markers in surveillance and prognostication for patients with HCC and established a prognostic prediction model based on a 10-marker panel. Furthermore, these authors built a prognostic score (cd-score) that was validated as an independent prognostic risk factor and appeared to be superior to AFP for predicting prognosis.
Table 3 summarizes findings from studies assessing the relationship between ctDNA and ctDNA mutations and the prognosis after LR for HCC.
2.4. Circulating Microparticles
Another interesting biomarker that can be measured in liquid biopsy specimens is CMs, which are extracellular vesicles with a diameter of 100 nm to 1000 nm [
53]. CMs are produced through the budding of the cellular membrane and released in the bloodstream, and they are consequently marked with the same surface antigens as the originating cells. CMs are involved in horizontal communication between cells, can allow for the detection of liver malignancies, and predict a high risk of HCC recurrence after hepatectomy [
54].
Abbate et al. [
55] utilized flow cytometry to identify CMs expressing hepatocyte paraffin 1 (HepPar1) in the bloodstream of HCC patients, and these authors found that CM concentration was higher than in healthy patients. The authors also demonstrated that CM concentration before LR was significantly higher in HCC patients who developed ER than in those with no recurrence, suggesting that CMs expressing HepPar1 are a predictor of HCC recurrence after LR.
Furthermore, several studies have shown that circulating exosomal noncoding RNA (microRNA), often released from necrotic tumor cells, might represent a novel prognostic biomarker in HCC patients. Liu et al. [
56], Shi et al. [
57], and Tian et al. [
58] described several microRNAs (miR-638, miR-125b, miR-21, and miR-10b) whose exosomal levels were associated with negative prognostic factors, such as larger tumor size and number, vascular invasion, advanced TNM stage, and poor prognosis. Reduced concentrations of miR-638 in CMs (detected with real-time PCR) in HCC patients after LR were associated with poorer OS, suggesting exosome-delivered miR-638 as an emerging biomarker to predict HCC prognosis [
56].
Notably, Luo et al. [
59] used quantitative PCR to measure exosomal circular RNA AKT3 (circAKT3) in serum samples of patients with HCC who had undergone LR and found that levels were significantly higher in patients with ER and poor OS.
Table 4 summarizes findings from studies assessing the relationship between CMs and prognosis after LR for HCC.
3. Gene Signatures
Pathological factors, including tumor extension, microvascular invasion, and lymph node involvement, have been widely recognized as predictors of recurrence and survival after LR for HCC [
60], but the molecular mechanisms underlying HCC recurrence remain unclear.
Notably, gene signatures, including autophagy-related gene signatures, are emerging from molecular profiling of HCC [
61,
62,
63]. This seems particularly relevant since the identification of commonly altered pathways has historically been more challenging for HCC than for other solid malignancies.
A gene signature consists of a single gene or a group of genes in a cell having a unique pattern of expression associated with a specific phenotype or outcome [
64]. RNA sequencing and microarray are among the most commonly used technologies for transcriptome profiling and the identification of characteristic molecular gene signatures [
61].
Several genetic mutations and gene expression patterns have been reported to be excellent tools to predict HCC recurrence and OS after LR for HCC [
61,
65,
66,
67,
68,
69,
70,
71]. Notably, He et al. [
67] found that gene rearrangement in HCC was frequently associated with low tumor differentiation, tumor necrosis, microvascular invasion, elevated AFP levels, and mutations in
TP53,
NTRK3, and
BRD9. These authors found that cumulative DFS rates at 1, 2, and 3 years after LR in patients with HCC were 45.1%, 31.9%, and 31.9%, respectively, for patients with gene rearrangement and 72.5%, 57.9%, and 49.0%, respectively, for patients without gene rearrangement. Furthermore, gene rearrangement appeared to be an independent risk factor for lower DFS.
In one of the largest published studies of gene expression and prognosis in patients with HCC, Wang et al. [
70] matched the mRNA expression data of 372 patients with their corresponding clinical characteristics. The authors identified a prognostic signature consisting of seven ferroptosis-related genes (
MAPK9,
SLC1A4,
PCK2,
ACSL3,
STMN1,
CDO1, and
CXCL2) that was independently associated with poor DFS, and they employed this signature to construct a reliable risk model of ER, which was validated in an independent cohort of The Cancer Genome Atlas.
Table 5 summarizes findings from studies assessing the relationship between gene signatures and prognosis after LR for HCC.
Autophagy is a process that leads to the lysosome-mediated degradation of damaged or aging cellular components. Autophagy is pivotal in maintaining tissue homeostasis and is believed to play a role in tumorigenesis, cancer progression, and resistance to systemic therapy. [
63] The prognosis after LR for HCC seems to be associated with autophagy-related genes. In particular, recent evidence showed how underexpression of autophagic genes leads to HCC recurrence and poor OS [
72,
73,
74,
75].
Notably, Lin et al. [
73] assessed the expression of LC3, Beclin-1, and p62 in HCC and adjacent healthy tissue in 535 HCC patients who underwent radical hepatectomy. The authors performed a multivariate analysis and found that both early recurrence and late recurrence were associated with LC3 underexpression in the entire tumor microenvironment. In this study, HCC patients with low LC3 expression had a 5-year recurrence rate of 94.3%. The same research group also proved that a combination of Axl overexpression and LC3 underexpression in HCC patients who underwent LR was associated with the highest risk of recurrence (90% at 5 years) and mortality (83.2% at 5 years) [
74].
Table 6 summarizes findings from studies assessing the relationship between autophagy-related genes and prognosis after LR for HCC.
4. Inflammation Markers
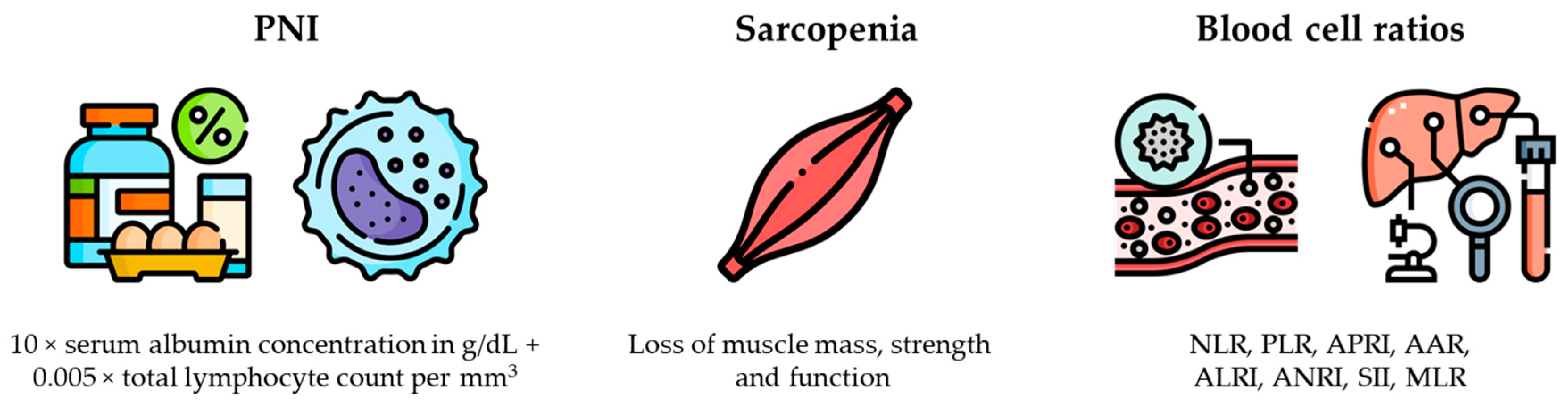
Several recent studies have highlighted a close correlation between inflammatory status, nutritional status, and the prognosis of HCC patients after curative-intent hepatectomy. Specifically, prognostic nutritional index (PNI), sarcopenia status, and several blood cell ratios have been adopted as independent predictors of OS [
76,
77] (
Figure 3).
The role of the immune system in HCC development and progression is difficult to comprehend. Chronic inflammation is common in liver disease and plays a pivotal role in tumor cells’ ability to elude immune surveillance [
78]. To further investigate the role of inflammation in HCC, Wu et al. [
79] analyzed the NF-κB signal pathway in tumor specimens and correlated HCC progression with the inflammatory state of hepatitis. They showed that high levels of TNFα in the tumor microenvironment promote EMT by upregulating the transcriptional regulator Snail, promoting tumor invasion, and indirectly reducing DFS. In contrast, the presence of high levels of tumor-associated macrophages and memory T cells appeared to protect HCC patients from recurrence after hepatectomy, improving DFS and OS [
80].
Several other studies showed that preoperative white blood cell counts and liver function test results were reproducible predictors of DFS and OS. Specifically, neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio, and aspartate aminotransferase-to-platelet count ratio index were found to be independent risk factors for poor outcomes in HCC patients after LR [
81,
82,
83,
84,
85,
86]. In a multicenter study [
87], the aforementioned markers and additional preoperatively measured inflammation markers (aspartate aminotransferase-to-alanine aminotransferase ratio, aspartate aminotransferase-to-lymphocyte ratio index, and aspartate aminotransferase-to-neutrophil ratio index) were used to create an inflammation score system. A multivariate Cox analysis showed that a high inflammation score was an independent predictor of worse DFS and OS in HCC patients who underwent hepatectomy. This principle appeared to be significant for patients with early-stage HCC.
Notable new indexes include the following:
- The systemic immune-inflammation index, which utilizes lymphocyte, neutrophil, and platelet counts [
88] to predict OS. A systemic immune-infiltration index ≥330 was significantly associated with ER after LR and higher CTC blood levels.
- The monocyte-to-lymphocyte ratio, which was shown to be a better predictor than systemic immune-infiltration index, neutrophil-to-lymphocyte ratio, or platelet-to-lymphocyte ratio for ER and DFS [
89]. Furthermore, the monocyte-to-lymphocyte ratio combined with tumor size, tumor differentiation, and the Barcelona Clinic Liver Cancer stage appeared to be more reliable than the monocyte-to-lymphocyte ratio by itself.
Pinato et al. [
90] first demonstrated how PNI was an independent predictor of OS in HCC patients, showing that nutritional status was strictly related to HCC progression. In particular, the 5-year DFS and OS rates in HCC patients with PNI < 45 were significantly lower than those in HCC patients with higher PNI, and low PNI was an independent outcome predictor for patients with early-stage HCC after curative-intent hepatectomy [
91,
92].
A meta-analysis by Man et al. [
93], including 13 articles and 3738 HCC cases, proved that preoperatively low levels of PNI were statistically related to postoperative early recurrence and worse OS. Furthermore, a multicenter retrospective study encompassing both nutritional and inflammatory factors established that detection of sarcopenia on computed tomography and/or a high platelet-to-lymphocyte ratio is independently associated with poor OS in HCC patients undergoing LR [
77].
Table 7 summarizes findings from studies assessing the relationship between inflammation markers and prognosis after LR for HCC.
5. Other Markers
In addition to the markers discussed above, a few other markers have been identified that are associated with prognosis after LR for HCC. These include PIVKA-II, immune checkpoint molecules, microRNAs, and proteins in urinary exosomes.
PIVKA-II is frequently used in combination with AFP to diagnose HCC, but the presence of PIVKA-II was recently proven to be an independent predictor of ER after LR. Wang et al. [
8] observed 751 HCC patients undergoing LR and assessed serum AFP and PIVKA-II levels in the perioperative period and within 2 years after surgery. They demonstrated that PIVKA-II had a higher positivity rate than AFP in patients with ER. Moreover, unlike AFP, on multivariate Cox regression analysis, PIVKA-II was an independent predictor of ER after curative-intent hepatectomy.
The checkpoint molecule programmed cell death protein-1 (PD-1) has been proven to correlate with tumor aggressiveness. Shi et al. [
94] conducted a study that included 56 hepatitis B virus-infected patients with HCC, 54 of whom underwent LR. These authors found high levels of T cells positive for both PD-1 and CD8 in peripheral blood samples of patients with advanced HCC, suggesting a positive correlation with HCC progression. Additionally, circulating PD-1 was found to be promising in predicting shorter DFS [
94]. Nie et al. [
95] utilized flow cytometry to detect programmed cell death-ligand 1 (PD-L1) levels in blood specimens of HCC patients undergoing radical hepatectomy and performed a multivariate Cox regression analysis, which showed that PD-L1 expression was one of the most significant independent predictors of DFS.
Researchers are searching for circulating microRNAs for diagnosis and prediction of prognosis in various liver diseases, including viral hepatitis, cirrhosis, and HCC [
96]. Dysregulated microRNA expression and protein production are crucial to cancer initiation and malignant progression in some cancers. Chen et al. [
97] reported that miR-182 and miR-331-3p might be useful in predicting the TNM stage and progression of HCC and are correlated with postoperative OS. For patients with hepatitis B virus-related HCC who undergo LR, Cho et al. [
98] support circulating levels of miR-26a and miR-29a as predictors of poor DFS and OS. Recently, Wong et al. [
10] combined quantitative PCR and artificial intelligence to integrate circulating microRNA with laboratory tumor characteristics and create a diagnostic and predictive model of DFS in patients undergoing LR. These authors identified a panel of eight microRNAs (which they termed the HCC seek-8 panel) that, in combination with serum biomarkers, was significantly associated with DFS.
Recently, several studies have focused on assessing the role of the exosomal proteome as a potential source of biomarkers for HCC [
11,
99]. Feng et al. [
11] used supramolecular probe-based capture and in situ detection technology to demonstrate how exosomes are efficiently enriched from urine samples with high concentration and purity. The urinary exosome proteomic analysis identified 68 upregulated proteins in HCC patients. Additionally, three previously identified biomarkers, OLFM4, HDGF, and GDF15, were validated as diagnostic biomarkers for HCC, showcasing the ability to diagnose HCC and potentially recognize the ER of HCC in a noninvasive, reproducible, and reliable manner.
Table 8 summarizes findings from studies assessing the relationship between other markers and the prognosis after LR for HCC.
6. Conclusions
During the past decade, researchers have increased their effort to identify new biomarkers able to reliably predict survival and recurrence in HCC patients undergoing surgical resection. While some of the proposed markers have been widely investigated and demonstrated to be reproducible, others seem less reproducible in clinical practice.
Liquid biopsy has been proven to be a powerful tool to predict long-term outcomes of patients with resected HCC, with costs related to its technical implementation representing the main limitation. Inflammation markers, including PNI and several blood cell ratios, seem more affordable on a large scale. While numerous groups have tried to identify patterns of gene expression capable of predicting the survival of patients undergoing curative-intent hepatectomy for HCC, there is still too much heterogeneity in the findings for gene expression to be used in clinical practice.
It is important to note that the great majority of the available studies on this topic come from Eastern Asia, reflecting the incidence of HCC in the world. It would be important to obtain more evidence from Western countries in the future to confirm that findings from Eastern countries hold true for patients in Western countries.
Author Contributions
Conceptualization, E.P. and A.M.D.R.; methodology, E.P. and A.C.; investigation, E.P., A.C. and C.M.; writing—original draft preparation, E.P. and A.C.; writing—review and editing, E.P., F.G. and F.A.; supervision, J.-N.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors thank Stephanie Deming, Senior Scientific Editor at MD Anderson Cancer Center, for copyediting the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Giuliante, F.; Ratti, F.; Panettieri, E.; Mazzaferro, V.; Guglielmi, A.; Ettorre, G.M.; Gruttadauria, S.; Di Benedetto, F.; Cillo, U.; De Carlis, L.; et al. Short and long-term outcomes after minimally invasive liver resection for single small hepatocellular carcinoma: An analysis of 714 patients from the IGoMILS (Italian group of minimally invasive liver surgery) registry. HPB 2023, 25, 674–683. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- Famularo, S.; Donadon, M.; Cipriani, F.; Ardito, F.; Carissimi, F.; Perri, P.; Iaria, M.; Dominioni, T.; Zanello, M.; Conci, S.; et al. Hepatocellular carcinoma surgical and oncological trends in a national multicentric population: The HERCOLES experience. Updates Surg. 2020, 72, 399–411. [Google Scholar] [CrossRef]
- Erridge, S.; Pucher, P.H.; Markar, S.R.; Malietzis, G.; Athanasiou, T.; Darzi, A.; Sodergren, M.H.; Jiao, L.R. Meta-analysis of determinants of survival following treatment of recurrent hepatocellular carcinoma. Br. J. Surg. 2017, 104, 1433–1442. [Google Scholar] [CrossRef]
- Kanwal, F.; Singal, A.G. Surveillance for Hepatocellular Carcinoma: Current Best Practice and Future Direction. Gastroenterology 2019, 157, 54–64. [Google Scholar] [CrossRef]
- Tzartzeva, K.; Obi, J.; Rich, N.E.; Parikh, N.D.; Marrero, J.A.; Yopp, A.; Waljee, A.K.; Singal, A.G. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients with Cirrhosis: A Meta-analysis. Gastroenterology 2018, 154, 1706–1718. [Google Scholar] [CrossRef]
- Wang, M.D.; Sun, L.Y.; Qian, G.J.; Li, C.; Gu, L.H.; Yao, L.Q.; Diao, Y.K.; Pawlik, T.M.; Lau, W.Y.; Huang, D.S.; et al. Prothrombin induced by vitamin K Absence-II versus alpha-fetoprotein in detection of both resectable hepatocellular carcinoma and early recurrence after curative liver resection: A retrospective cohort study. Int. J. Surg. 2022, 105, 106843. [Google Scholar] [CrossRef]
- Wan, H.; Lu, S.; Xu, L.; Yuan, K.; Xiao, Y.; Xie, K.; Wu, H. Immune-Related Biomarkers Improve Performance of Risk Prediction Models for Survival in Patients with Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 925362. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.C.; Wong, M.I.; Lee, V.H.; Man, K.; Ng, K.T.; Cheung, T.T. Prognostic MicroRNA Fingerprints Predict Recurrence of Early-Stage Hepatocellular Carcinoma Following Hepatectomy. J. Cancer 2023, 14, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Jia, S.; Ali, M.M.; Zhang, G.; Li, D.; Tao, W.A.; Hu, L. Proteomic Discovery and Array-Based Validation of Biomarkers from Urinary Exosome by Supramolecular Probe. J. Proteome Res. 2023, 22, 2516–2524. [Google Scholar] [CrossRef] [PubMed]
- Bhan, I.; Haber, D.A.; Chung, R.T.; Ting, D.T. Liquid Biopsy in Hepatocellular Carcinoma. In Hepatocellular Carcinoma: Translational Precision Medicine Approaches; Hoshida, Y., Ed.; Humana Press: Totowa, NJ, USA, 2019; Chapter 7. [Google Scholar]
- Chen, V.L.; Xu, D.; Wicha, M.S.; Lok, A.S.; Parikh, N.D. Utility of Liquid Biopsy Analysis in Detection of Hepatocellular Carcinoma, Determination of Prognosis, and Disease Monitoring: A Systematic Review. Clin. Gastroenterol. Hepatol. 2020, 18, 2879–2902. [Google Scholar] [CrossRef] [PubMed]
- van Zijl, F.; Zulehner, G.; Petz, M.; Schneller, D.; Kornauth, C.; Hau, M.; Machat, G.; Grubinger, M.; Huber, H.; Mikulits, W. Epithelial-mesenchymal transition in hepatocellular carcinoma. Future Oncol. 2009, 5, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J. Oncol. 2010, 2010, 617421. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.C.; Teng, P.C.; Chen, P.J.; Posadas, E.; Tseng, H.R.; Lu, S.C.; Yang, J.D. Detection of Circulating Tumor Cells and Their Implications as a Biomarker for Diagnosis, Prognostication, and Therapeutic Monitoring in Hepatocellular Carcinoma. Hepatology 2021, 73, 422–436. [Google Scholar] [CrossRef] [PubMed]
- Funaki, N.O.; Tanaka, J.; Seto, S.I.; Kasamatsu, T.; Kaido, T.; Imamura, M. Hematogenous spreading of hepatocellular carcinoma cells: Possible participation in recurrence in the liver. Hepatology 1997, 25, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.F.; Xu, Y.; Yang, X.R.; Guo, W.; Zhang, X.; Qiu, S.J.; Shi, R.Y.; Hu, B.; Zhou, J.; Fan, J. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013, 57, 1458–1468. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.F.; Wang, P.X.; Cheng, J.W.; Gong, Z.J.; Huang, A.; Zhou, K.Q.; Hu, B.; Gao, P.T.; Cao, Y.; Qiu, S.J.; et al. Postoperative circulating tumor cells: An early predictor of extrahepatic metastases in patients with hepatocellular carcinoma undergoing curative surgical resection. Cancer Cytopathol. 2020, 128, 733–745. [Google Scholar] [CrossRef] [PubMed]
- von Felden, J.; Schulze, K.; Krech, T.; Ewald, F.; Nashan, B.; Pantel, K.; Lohse, A.W.; Riethdorf, S.; Wege, H. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget 2017, 8, 89978–89987. [Google Scholar] [CrossRef]
- Yu, J.J.; Xiao, W.; Dong, S.L.; Liang, H.F.; Zhang, Z.W.; Zhang, B.X.; Huang, Z.Y.; Chen, Y.F.; Zhang, W.G.; Luo, H.P.; et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMC Cancer 2018, 18, 835. [Google Scholar] [CrossRef]
- Amado, V.; González-Rubio, S.; Zamora, J.; Alejandre, R.; Espejo-Cruz, M.L.; Linares, C.; Sánchez-Frías, M.; García-Jurado, G.; Montero, J.L.; Ciria, R.; et al. Clearance of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma Undergoing Surgical Resection or Liver Transplantation. Cancers 2021, 13, 2476. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.T.; Yang, Z.F.; Ho, D.W.; Ng, M.N.; Yu, W.C.; Wong, J. Prediction of posthepatectomy recurrence of hepatocellular carcinoma by circulating cancer stem cells: A prospective study. Ann. Surg. 2011, 254, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Hamaoka, M.; Kobayashi, T.; Tanaka, Y.; Mashima, H.; Ohdan, H. Clinical significance of glypican-3-positive circulating tumor cells of hepatocellular carcinoma patients: A prospective study. PLoS ONE 2019, 14, e0217586. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.; Kim, T.H.; Shim, J.E.; Yoon, S.; Jun, M.J.; Cho, Y.H.; Lee, H.C. Circulating tumor cells are associated with poor outcomes in early-stage hepatocellular carcinoma: A prospective study. Hepatol. Int. 2019, 13, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, G.; Koudelkova, P.; Dituri, F.; Mikulits, W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016, 65, 798–808. [Google Scholar] [CrossRef]
- Wang, Z.; Luo, L.; Cheng, Y.; He, G.; Peng, B.; Gao, Y.; Jiang, Z.S.; Pan, M. Correlation Between Postoperative Early Recurrence of Hepatocellular Carcinoma and Mesenchymal Circulating Tumor Cells in Peripheral Blood. J. Gastrointest. Surg. 2018, 22, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Ou, H.; Huang, Y.; Xiang, L.; Chen, Z.; Fang, Y.; Lin, Y.; Cui, Z.; Yu, S.; Li, X.; Yang, D. Circulating Tumor Cell Phenotype Indicates Poor Survival and Recurrence After Surgery for Hepatocellular Carcinoma. Dig. Dis. Sci. 2018, 63, 2373–2380. [Google Scholar] [CrossRef]
- Yin, L.C.; Luo, Z.C.; Gao, Y.X.; Li, Y.; Peng, Q.; Gao, Y. Twist Expression in Circulating Hepatocellular Carcinoma Cells Predicts Metastasis and Prognoses. Biomed. Res. Int. 2018, 2018, 3789613. [Google Scholar] [CrossRef]
- Qi, L.N.; Xiang, B.D.; Wu, F.X.; Ye, J.Z.; Zhong, J.H.; Wang, Y.Y.; Chen, Y.Y.; Chen, Z.S.; Ma, L.; Chen, J.; et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4731–4744. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.N.; Ma, L.; Chen, Y.Y.; Chen, Z.S.; Zhong, J.H.; Gong, W.F.; Lu, Y.; Xiang, B.D.; Li, L.Q. Outcomes of anatomical versus non-anatomical resection for hepatocellular carcinoma according to circulating tumour-cell status. Ann. Med. 2020, 52, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xing, W.; Zhang, J.; Hu, J.; Qi, L.; Xiang, B. Circulating Tumor Cells Undergoing the Epithelial-Mesenchymal Transition: Influence on Prognosis in Cytokeratin 19-Positive Hepatocellular Carcinoma. Onco Targets Ther. 2021, 14, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, S.; Li, W.; Yang, R.; Zhang, X.; Ye, Y.; Yu, J.; Ye, L.; Tang, W. Circulating tumor cells undergoing EMT are poorly correlated with clinical stages or predictive of recurrence in hepatocellular carcinoma. Sci. Rep. 2019, 9, 7084. [Google Scholar] [CrossRef] [PubMed]
- Court, C.M.; Hou, S.; Winograd, P.; Segel, N.H.; Li, Q.W.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Ganapathy, E.; Song, M.; et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 2018, 24, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Orrapin, S.; Udomruk, S.; Lapisatepun, W.; Moonmuang, S.; Phanphaisarn, A.; Phinyo, P.; Pruksakorn, D.; Chaiyawat, P. Clinical Implication of Circulating Tumor Cells Expressing Epithelial Mesenchymal Transition (EMT) and Cancer Stem Cell (CSC) Markers and Their Perspective in HCC: A Systematic Review. Cancers 2022, 14, 3373. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Schwarzenbach, H.; Pantel, K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012, 63, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378. [Google Scholar] [CrossRef]
- Lee, H.; Park, C.; Na, W.; Park, K.H.; Shin, S. Precision cell-free DNA extraction for liquid biopsy by integrated microfluidics. NPJ Precis. Oncol. 2020, 4, 3. [Google Scholar] [CrossRef]
- Benesova, L.; Belsanova, B.; Suchanek, S.; Kopeckova, M.; Minarikova, P.; Lipska, L.; Levy, M.; Visokai, V.; Zavoral, M.; Minarik, M. Mutation-based detection and monitoring of cell-free tumor DNA in peripheral blood of cancer patients. Anal. Biochem. 2013, 433, 227–234. [Google Scholar] [CrossRef]
- Marzese, D.M.; Hirose, H.; Hoon, D.S. Diagnostic and prognostic value of circulating tumor-related DNA in cancer patients. Expert. Rev. Mol. Diagn. 2013, 13, 827–844. [Google Scholar] [CrossRef] [PubMed]
- Fiala, C.; Diamandis, E.P. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med. 2018, 16, 166. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef]
- Tokuhisa, Y.; Iizuka, N.; Sakaida, I.; Moribe, T.; Fujita, N.; Miura, T.; Tamatsukuri, S.; Ishitsuka, H.; Uchida, K.; Terai, S.; et al. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br. J. Cance 2007, 97, 1399–1403. [Google Scholar] [CrossRef]
- An, Y.; Guan, Y.; Xu, Y.; Han, Y.; Wu, C.; Bao, C.; Zhou, B.; Wang, H.; Zhang, M.; Liu, W.; et al. The diagnostic and prognostic usage of circulating tumor DNA in operable hepatocellular carcinoma. Am. J. Transl. Res. 2019, 11, 6462–6474. [Google Scholar]
- Cai, Z.; Chen, G.; Zeng, Y.; Dong, X.; Li, Z.; Huang, Y.; Xin, F.; Qiu, L.; Xu, H.; Zhang, W.; et al. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients with Hepatocellular Carcinoma. Clin. Cancer Res. 2019, 25, 5284–5294. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Yang, H.; Xu, H.; Wang, Y.; Ge, P.; Ren, J.; Xu, W.; Lu, X.; Sang, X.; Zhong, S.; et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016, 7, 40481–40490. [Google Scholar] [CrossRef]
- Shen, T.; Li, S.F.; Wang, J.L.; Zhang, T.; Zhang, S.; Chen, H.T.; Xiao, Q.Y.; Ren, W.H.; Liu, C.; Peng, B.; et al. TP53 R249S mutation detected in circulating tumour DNA is associated with Prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020, 40, 2834–2847. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Huang, Y.; Wei, L.; He, J.Y.; Liu, Q.Y.; Yu, X.Q.; Li, Z.L.; Zhang, J.; Li, B.; Sun, C.J.; et al. Combination of LINE-1 hypomethylation and RASSF1A promoter hypermethylation in serum DNA is a non-invasion prognostic biomarker for early recurrence of hepatocellular carcinoma after curative resection. Neoplasma 2017, 64, 795–802. [Google Scholar] [CrossRef]
- Chan, K.C.; Jiang, P.; Chan, C.W.; Sun, K.; Wong, J.; Hui, E.P.; Chan, S.L.; Chan, W.C.; Hui, D.S.; Ng, S.S.; et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 18761–18768. [Google Scholar] [CrossRef]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Julich-Haertel, H.; Urban, S.K.; Krawczyk, M.; Willms, A.; Jankowski, K.; Patkowski, W.; Kruk, B.; Krasnodębski, M.; Ligocka, J.; Schwab, R.; et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 2017, 67, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Abbate, V.; Marcantoni, M.; Giuliante, F.; Vecchio, F.M.; Gatto, I.; Mele, C.; Saviano, A.; Arciuolo, D.; Gaetani, E.; Ferrari, M.C.; et al. HepPar1-Positive Circulating Microparticles Are Increased in Subjects with Hepatocellular Carcinoma and Predict Early Recurrence after Liver Resection. Int. J. Mol. Sci. 2017, 18, 1043. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Hu, J.; Zhou, K.; Chen, F.; Wang, Z.; Liao, B.; Dai, Z.; Cao, Y.; Fan, J.; Zhou, J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017, 10, 3843–3851. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Jiang, Y.; Yang, L.; Yan, S.; Wang, Y.G.; Lu, X.J. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell Biochem. 2018, 119, 4711–4716. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.P.; Wang, C.Y.; Jin, X.H.; Li, M.; Wang, F.W.; Huang, W.J.; Yun, J.P.; Xu, R.H.; Cai, Q.Q.; Xie, D. Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis. Theranostics 2019, 9, 1965–1979. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, F.; Gui, R. High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg. Oncol. 2020, 33, 276–281. [Google Scholar] [CrossRef]
- Marsh, J.W.; Dvorchik, I.; Bonham, C.A.; Iwatsuki, S. Is the pathologic TNM staging system for patients with hepatoma predictive of outcome? Cancer 2000, 88, 538–543. [Google Scholar] [CrossRef]
- Son, J.A.; Ahn, H.R.; You, D.; Baek, G.O.; Yoon, M.G.; Yoon, J.H.; Cho, H.J.; Kim, S.S.; Nam, S.W.; Eun, J.W.; et al. Novel Gene Signatures as Prognostic Biomarkers for Predicting the Recurrence of Hepatocellular Carcinoma. Cancers 2022, 14, 865. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, J.; Huang, G.; Zhu, J.; Jian, H.; Xia, G.; Wei, Q.; Li, Y.; Yu, H. A novel epithelial-mesenchymal transition gene signature for the immune status and prognosis of hepatocellular carcinoma. Hepatol. Int. 2022, 16, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Shi, Z.; Zhou, Y.; Zhang, Z.; Zhou, Y.; Chen, B.; Zhang, Q. Autophagy-Related Signatures as Prognostic Indicators for Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 654449. [Google Scholar] [CrossRef] [PubMed]
- Mallik, S.; Zhao, Z. Identification of gene signatures from RNA-seq data using Pareto-optimal cluster algorithm. BMC Syst. Biol. 2018, 12, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Ashida, R.; Okamura, Y.; Ohshima, K.; Kakuda, Y.; Uesaka, K.; Sugiura, T.; Ito, T.; Yamamoto, Y.; Sugino, T.; Urakami, K.; et al. CYP3A4 Gene Is a Novel Biomarker for Predicting a Poor Prognosis in Hepatocellular Carcinoma. Cancer Genom. Proteom. 2017, 14, 445–453. [Google Scholar]
- Wang, Y.; Tan, P.Y.; Handoko, Y.A.; Sekar, K.; Shi, M.; Xie, C.; Jiang, X.D.; Dong, Q.Z.; Goh, B.K.P.; Ooi, L.L.; et al. NUF2 is a valuable prognostic biomarker to predict early recurrence of hepatocellular carcinoma after surgical resection. Int. J. Cancer 2019, 145, 662–670. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Song, K.; Guan, G.; Huo, J.; Xin, Y.; Li, T.; Liu, C.; Zhu, Q.; Fan, N.; Guo, Y.; et al. The Phenomenon of Gene Rearrangement is Frequently Associated with TP53 Mutations and Poor Disease-Free Survival in Hepatocellular Carcinoma. Pharmacogenomics Pers. Med. 2021, 14, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.S.; An, J.; Kang, H.J.; Oh, B.; Oh, Y.J.; Oh, J.H.; Kim, W.; Sung, C.O.; Shim, J.H.; Yu, E. Prognostic Molecular Indices of Resectable Hepatocellular Carcinoma: Implications of S100P for Early Recurrence. Ann. Surg. Oncol. 2021, 28, 6466–6478. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; He, F.; Xin, Y.; Guan, G.; Huo, J.; Zhu, Q.; Fan, N.; Guo, Y.; Zang, Y.; Wu, L. TSC2 Mutations Were Associated with the Early Recurrence of Patients with HCC Underwent Hepatectomy. Pharmacogenomics Pers. Med. 2021, 14, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, C.; Jiang, Y.; Hu, H.; Fang, J.; Yang, F. A novel ferroptosis-related gene signature for clinically predicting recurrence after hepatectomy of hepatocellular carcinoma patients. Am. J. Cancer Res. 2022, 12, 1995–2011. [Google Scholar]
- Xin, Z.; Li, J.; Zhang, H.; Zhou, Y.; Song, J.; Chen, P.; Bai, L.; Chen, H.; Zhou, J.; Chen, J.; et al. Cancer Genomic Alterations Can Be Potential Biomarkers Predicting Microvascular Invasion and Early Recurrence of Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 783109. [Google Scholar] [CrossRef]
- Ding, Z.B.; Shi, Y.H.; Zhou, J.; Qiu, S.J.; Xu, Y.; Dai, Z.; Shi, G.M.; Wang, X.Y.; Ke, A.W.; Wu, B.; et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008, 68, 9167–9175. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Chen, Y.S.; Lin, C.C.; Lee, P.H.; Lo, G.H.; Hsu, C.C.; Hsieh, P.M.; Koh, K.W.; Chou, T.C.; Dai, C.Y.; et al. Autophagy-related gene LC3 expression in tumor and liver microenvironments significantly predicts recurrence of hepatocellular carcinoma after surgical resection. Clin. Transl. Gastroenterol. 2018, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.C.; Hsieh, P.M.; Chen, Y.S.; Lo, G.H.; Lin, H.Y.; Dai, C.Y.; Huang, J.F.; Chuang, W.L.; Chen, Y.L.; Yu, M.L.; et al. Axl and autophagy LC3 expression in tumors is strongly associated with clinical prognosis of hepatocellular carcinoma patients after curative resection. Cancer Med. 2019, 8, 3453–3463. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, C.; Li, D.; Wang, R.; Li, Y.; Lv, L. Bioinformatics analysis and experimental validation of a novel autophagy-related signature relevant to immune infiltration for recurrence prediction after curative hepatectomy. Aging 2023, 15, 2610–2630. [Google Scholar] [CrossRef]
- Mao, S.; Yu, X.; Sun, J.; Yang, Y.; Shan, Y.; Sun, J.; Mugaanyi, J.; Fan, R.; Wu, S.; Lu, C. Development of nomogram models of inflammatory markers based on clinical database to predict prognosis for hepatocellular carcinoma after surgical resection. BMC Cancer 2022, 22, 249. [Google Scholar] [CrossRef]
- Kim, H.; Choi, H.Z.; Choi, J.M.; Kang, B.M.; Lee, J.W.; Hwang, J.W. Sarcopenia with systemic inflammation can predict survival in patients with hepatocellular carcinoma undergoing curative resection. J. Gastrointest. Oncol. 2022, 13, 744–753. [Google Scholar] [CrossRef]
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491. [Google Scholar] [CrossRef]
- Wu, T.J.; Chang, S.S.; Li, C.W.; Hsu, Y.H.; Chen, T.C.; Lee, W.C.; Yeh, C.T.; Hung, M.C. Severe Hepatitis Promotes Hepatocellular Carcinoma Recurrence via NF-κB Pathway-Mediated Epithelial-Mesenchymal Transition after Resection. Clin. Cancer Res. 2016, 22, 1800–1812. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.W.; Qiu, S.J.; Fan, J.; Gao, Q.; Zhou, J.; Xiao, Y.S.; Xu, Y.; Wang, X.Y.; Sun, J.; Huang, X.W. Tumor-infiltrating macrophages can predict favorable prognosis in hepatocellular carcinoma after resection. J. Cancer Res. Clin. Oncol. 2009, 135, 439–449. [Google Scholar] [CrossRef]
- Mano, Y.; Shirabe, K.; Yamashita, Y.; Harimoto, N.; Tsujita, E.; Takeishi, K.; Aishima, S.; Ikegami, T.; Yoshizumi, T.; Yamanaka, T.; et al. Preoperative neutrophil-to-lymphocyte ratio is a predictor of survival after hepatectomy for hepatocellular carcinoma: A retrospective analysis. Ann. Surg. 2013, 258, 301–305. [Google Scholar] [CrossRef]
- Shen, S.L.; Fu, S.J.; Chen, B.; Kuang, M.; Li, S.Q.; Hua, Y.P.; Liang, L.J.; Guo, P.; Hao, Y.; Peng, B.G. Preoperative aspartate aminotransferase to platelet ratio is an independent prognostic factor for hepatitis B-induced hepatocellular carcinoma after hepatic resection. Ann. Surg. Oncol. 2014, 21, 3802–3809. [Google Scholar] [CrossRef]
- Ji, F.; Liang, Y.; Fu, S.J.; Guo, Z.Y.; Shu, M.; Shen, S.L.; Li, S.Q.; Peng, B.G.; Liang, L.J.; Hua, Y.P. A novel and accurate predictor of survival for patients with hepatocellular carcinoma after surgical resection: The neutrophil to lymphocyte ratio (NLR) combined with the aspartate aminotransferase/platelet count ratio index (APRI). BMC Cancer 2016, 16, 137. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Z.X.; Cao, Y.; Zhang, G.; Chen, W.B.; Jiang, C.P. Preoperative inflammation-based markers predict early and late recurrence of hepatocellular carcinoma after curative hepatectomy. Hepatobiliary Pancreat. Dis. Int. 2016, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.G.; Mao, W.; Park, Y.K.; Xu, W.G.; Kim, B.W.; Wang, H.J. Blood Neutrophil-to-Lymphocyte Ratio Predicts Tumor Recurrence in Patients with Hepatocellular Carcinoma within Milan Criteria after Hepatectomy. Yonsei Med. J. 2016, 57, 1115–1123. [Google Scholar] [CrossRef]
- Wang, D.; Bai, N.; Hu, X.; OuYang, X.W.; Yao, L.; Tao, Y.; Wang, Z. Preoperative inflammatory markers of NLR and PLR as indicators of poor prognosis in resectable HCC. PeerJ 2019, 7, e7132. [Google Scholar] [CrossRef]
- Chen, Q.; Li, F.; Zhong, C.; Zou, Y.; Li, Z.; Gao, Y.; Zou, Q.; Xia, Y.; Wang, K.; Shen, F. Inflammation Score System using Preoperative Inflammatory Markers to Predict Prognosis for Hepatocellular Carcinoma after Hepatectomy: A Cohort Study. J. Cancer 2020, 11, 4947–4956. [Google Scholar] [CrossRef]
- Hu, B.; Yang, X.R.; Xu, Y.; Sun, Y.F.; Sun, C.; Guo, W.; Zhang, X.; Wang, W.M.; Qiu, S.J.; Zhou, J.; et al. Systemic immune-inflammation index predicts prognosis of patients after curative resection for hepatocellular carcinoma. Clin. Cancer Res. 2014, 20, 6212–6222. [Google Scholar] [CrossRef]
- Wu, Y.; Tu, C.; Shao, C. Inflammatory indexes in preoperative blood routine to predict early recurrence of hepatocellular carcinoma after curative hepatectomy. BMC Surg. 2021, 21, 178. [Google Scholar] [CrossRef] [PubMed]
- Pinato, D.J.; North, B.V.; Sharma, R. A novel, externally validated inflammation-based prognostic algorithm in hepatocellular carcinoma: The prognostic nutritional index (PNI). British J. Cancer 2012, 106, 1439–1445. [Google Scholar] [CrossRef]
- Fan, X.; Chen, G.; Li, Y.; Shi, Z.; He, L.; Zhou, D.; Lin, H. The Preoperative Prognostic Nutritional Index in Hepatocellular Carcinoma After Curative Hepatectomy: A Retrospective Cohort Study and Meta-Analysis. J. Investig. Surg. 2021, 34, 826–833. [Google Scholar] [CrossRef]
- Chan, A.W.; Chan, S.L.; Wong, G.L.; Wong, V.W.; Chong, C.C.; Lai, P.B.; Chan, H.L.; To, K.F. Prognostic Nutritional Index (PNI) Predicts Tumor Recurrence of Very Early/Early Stage Hepatocellular Carcinoma After Surgical Resection. Ann. Surg. Oncol. 2015, 22, 4138–4148. [Google Scholar] [CrossRef] [PubMed]
- Man, Z.; Pang, Q.; Zhou, L.; Wang, Y.; Hu, X.; Yang, S.; Jin, H.; Liu, H. Prognostic significance of preoperative prognostic nutritional index in hepatocellular carcinoma: A meta-analysis. HPB 2018, 20, 888–895. [Google Scholar] [CrossRef]
- Shi, F.; Shi, M.; Zeng, Z.; Qi, R.Z.; Liu, Z.W.; Zhang, J.Y.; Yang, Y.P.; Tien, P.; Wang, F.S. PD-1 and PD-L1 upregulation promotes CD8(+) T-cell apoptosis and postoperative recurrence in hepatocellular carcinoma patients. Int. J. Cancer 2011, 128, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; He, T.; Wang, L.; Zhang, L. Expression and Prognostic Value of Tumor-Infiltrating Lymphocytes and PD-L1 in Hepatocellular Carcinoma. Onco Targets Ther. 2021, 14, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Latief, U.; Tung, G.K.; Per, T.S.; Kaur, M.; Thakur, S.; Singh, H.; Jain, S.K. Micro RNAs as Emerging Therapeutic Targets in Liver Diseases. Curr. Protein Pept. Sc. 2022, 23, 369–383. [Google Scholar]
- Chen, L.; Chu, F.; Cao, Y.; Shao, J.; Wang, F. Serum miR-182 and miR-331-3p as diagnostic and prognostic markers in patients with hepatocellular carcinoma. Tumour Biol. 2015, 36, 7439–7447. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Kim, S.S.; Nam, J.S.; Kim, J.K.; Lee, J.H.; Kim, B.; Wang, H.J.; Kim, B.W.; Lee, J.D.; Kang, D.Y.; et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin. Res. Hepatol. Gastroenterol. 2017, 41, 181–189. [Google Scholar] [CrossRef]
- Zhao, L.; Shi, J.; Chang, L.; Wang, Y.; Liu, S.; Li, Y.; Zhang, T.; Zuo, T.; Fu, B.; Wang, G.; et al. Serum-Derived Exosomal Proteins as Potential Candidate Biomarkers for Hepatocellular Carcinoma. ACS Omega 2021, 6, 827–835. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).