Discovery and Validation of Survival-Specific Genes in Papillary Renal Cell Carcinoma Using a Customized Next-Generation Sequencing Gene Panel
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Feature Selection and Machine Learning for Discovering Survival-Specific Genes in PRCC
2.3. Patients
2.4. Samples
2.5. NGS Gene Panel Design for PRCC
2.6. Targeted Library Preparation
2.7. Bioinformatics Analysis
2.8. Datasets
2.9. Data Pre-Processing
2.10. Gene Set Enrichment Analysis (GSEA)
2.11. Statistical Analysis
3. Results
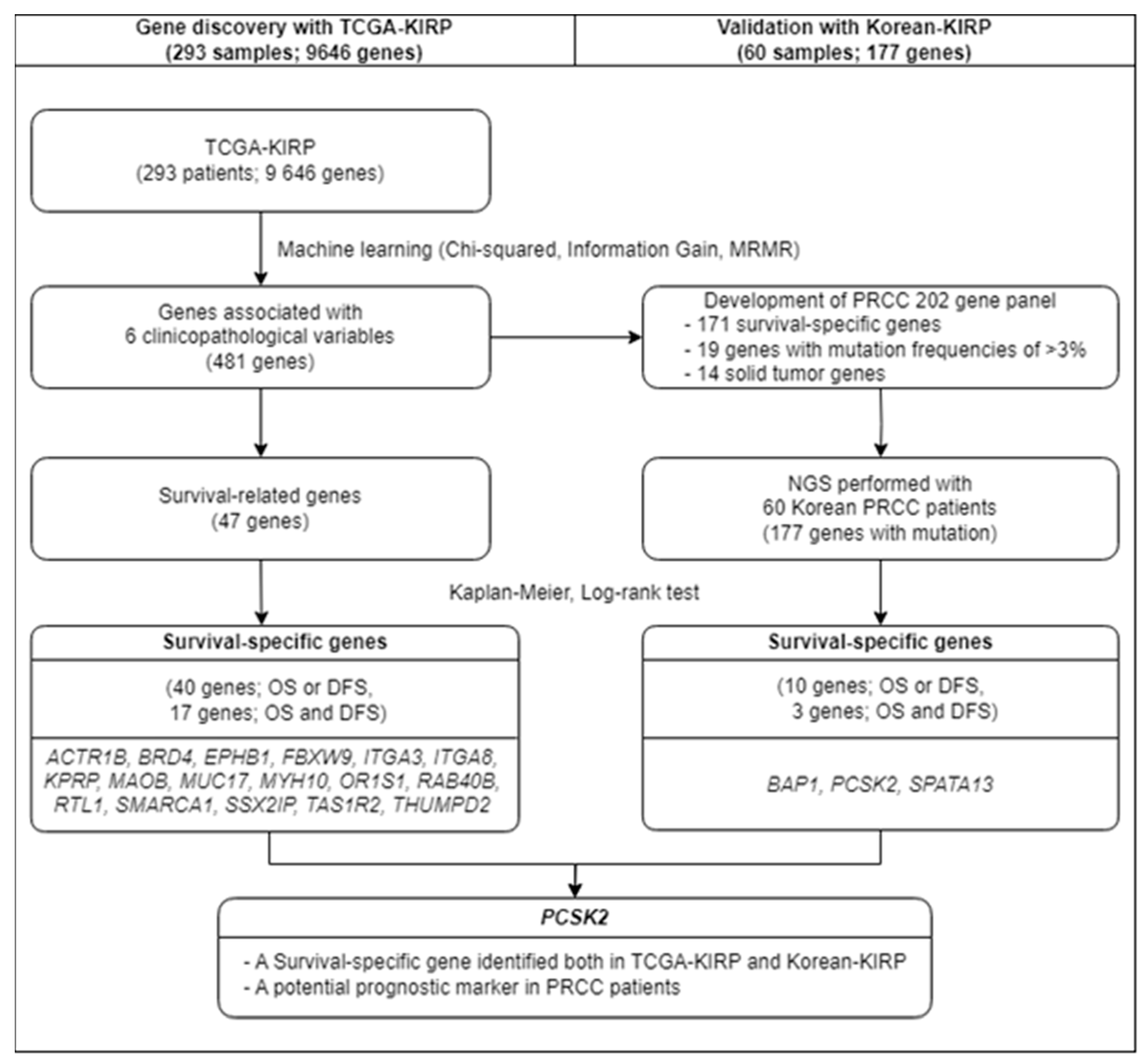
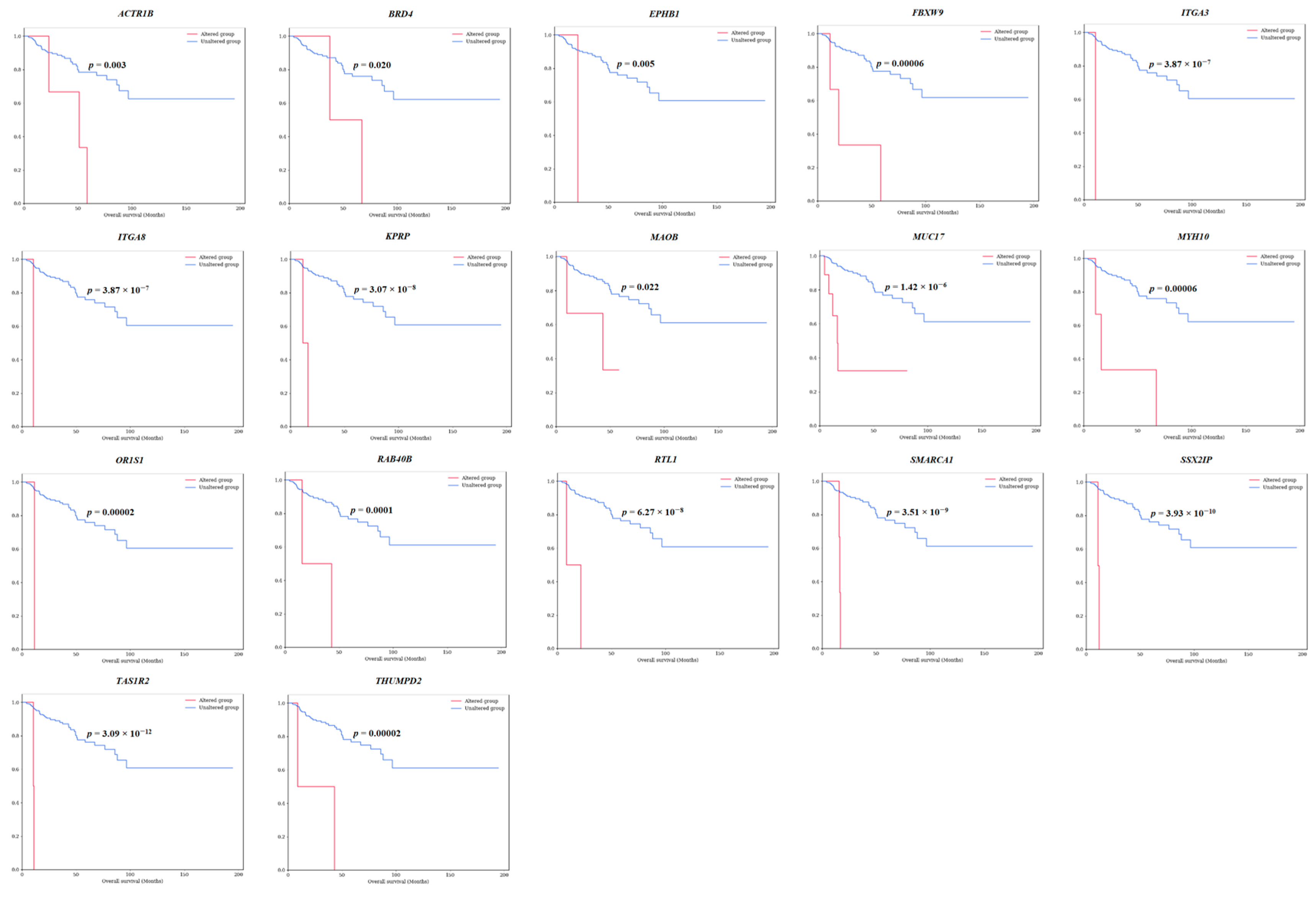
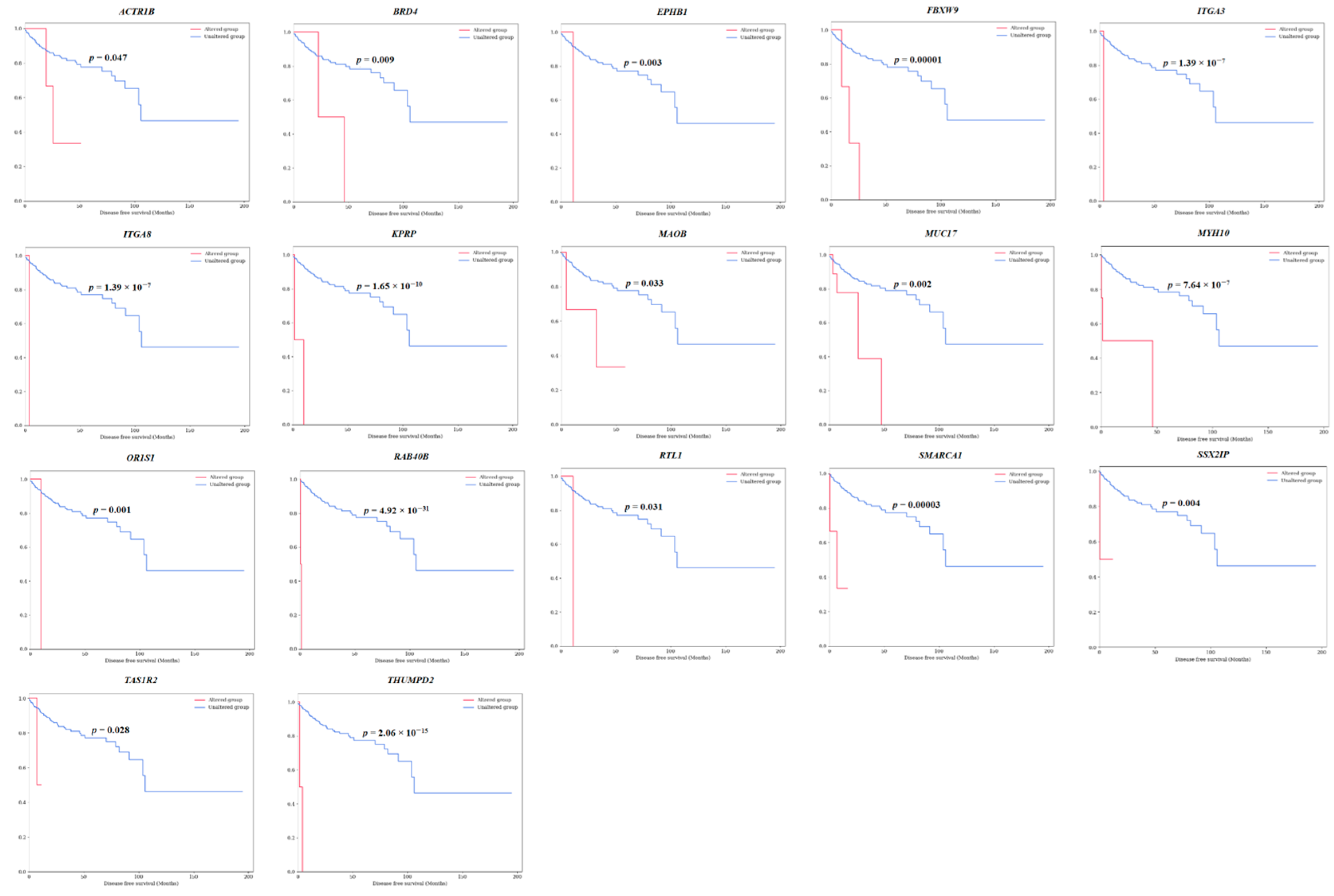
3.1. Discovery of Survival-Specific Genes in TCGA-KIRP Database by Machine Learning
3.1.1. Verification of Survival-Specific Genes in Korean-KIRP Patients through NGS Analysis
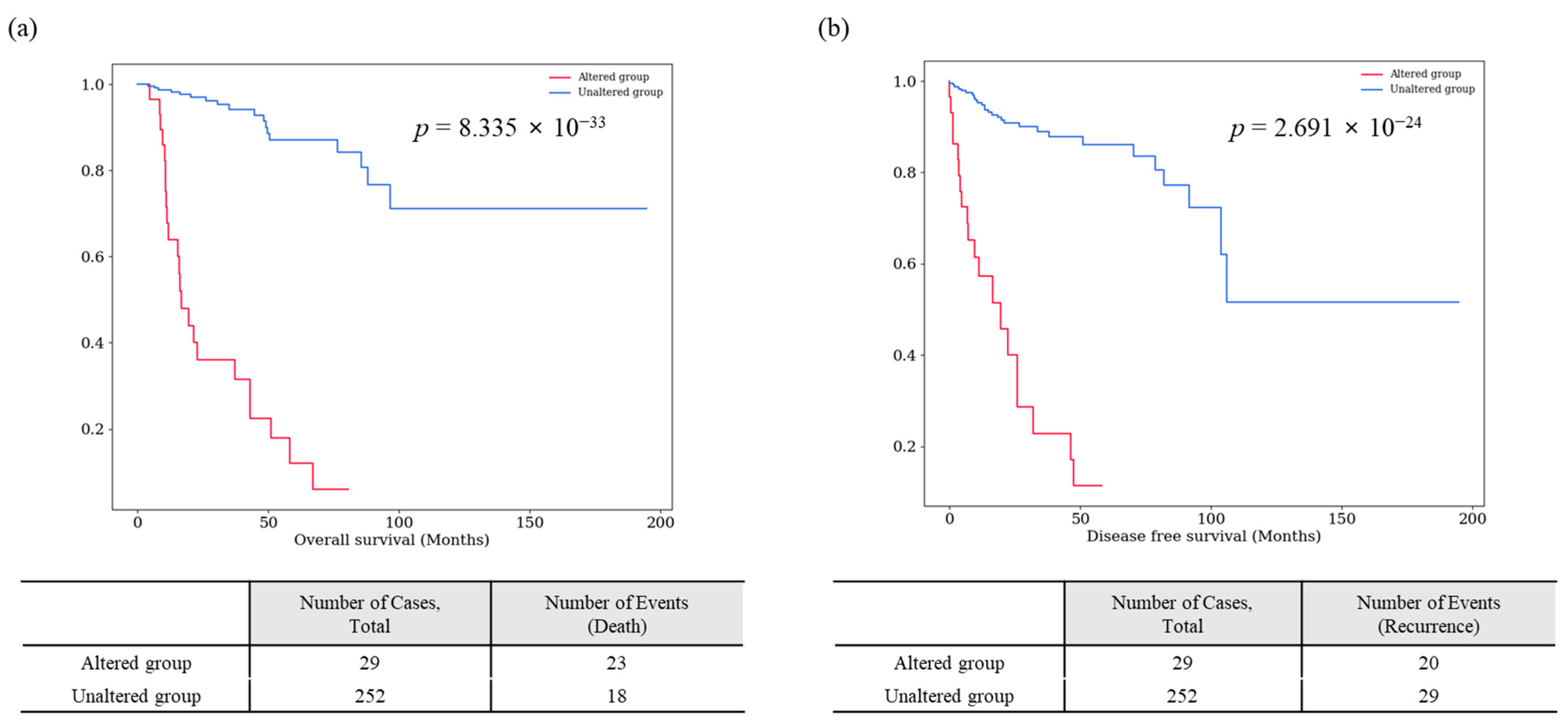
3.1.2. A Survival-Specific Gene Commonly Identified in Both TCGA-KIRP and Korean-KIRP Databases
3.1.3. Clinicopathological Significance of Survival-Specific Genes in Korean-KIRP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linehan, W.M.; Spellman, P.T.; Ricketts, C.J.; Creighton, C.J.; Fei, S.S.; Davis, C.; Wheeler, D.A.; Murray, B.A.; Schmidt, L.; Vocke, C.D.; et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N. Engl. J. Med. 2016, 374, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.K.; Lee, S.E.; Hong, S.K.; Jeong, C.W.; Park, Y.H.; Kang, S.H.; Kim, Y.J.; Hong, S.H.; Choi, W.S.; Byun, S.S. Characteristics and prognostic value of papillary histologic subtype in nonmetastatic renal cell carcinoma in Korea: A multicenter study. Urol. J. 2014, 11, 1884–1890. [Google Scholar] [PubMed]
- Akhtar, M.; Al-Bozom, I.A.; Al Hussain, T. Papillary Renal Cell Carcinoma (PRCC): An Update. Adv. Anat. Pathol. 2019, 26, 124–132. [Google Scholar] [CrossRef]
- Tickoo, S.K.; dePeralta-Venturina, M.N.; Harik, L.R.; Worcester, H.D.; Salama, M.E.; Young, A.N.; Moch, H.; Amin, M.B. Spectrum of epithelial neoplasms in end-stage renal disease: An experience from 66 tumor-bearing kidneys with emphasis on histologic patterns distinct from those in sporadic adult renal neoplasia. Am. J. Surg. Pathol. 2006, 30, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Bukavina, L.; Bensalah, K.; Bray, F.; Carlo, M.; Challacombe, B.; Karam, J.A.; Kassouf, W.; Mitchell, T.; Montironi, R.; O’Brien, T.; et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eur. Urol. 2022, 82, 529–542. [Google Scholar] [CrossRef]
- Petejova, N.; Martinek, A. Renal cell carcinoma: Review of etiology, pathophysiology and risk factors. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc. Czech. Repub. 2016, 160, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Aveta, A.; Cilio, S.; Contieri, R.; Spena, G.; Napolitano, L.; Manfredi, C.; Franco, A.; Crocerossa, F.; Cerrato, C.; Ferro, M.; et al. Urinary MicroRNAs as Biomarkers of Urological Cancers: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 10846. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.K.; Ali, S.M.; Yakirevich, E.; Geynisman, D.M.; Karam, J.A.; Elvin, J.A.; Frampton, G.M.; Huang, X.; Lin, D.I.; Rosenzweig, M.; et al. Characterization of Clinical Cases of Advanced Papillary Renal Cell Carcinoma via Comprehensive Genomic Profiling. Eur. Urol. 2018, 73, 71–78. [Google Scholar] [CrossRef]
- Kovac, M.; Navas, C.; Horswell, S.; Salm, M.; Bardella, C.; Rowan, A.; Stares, M.; Castro-Giner, F.; Fisher, R.; de Bruin, E.C.; et al. Recurrent chromosomal gains and heterogeneous driver mutations characterise papillary renal cancer evolution. Nat. Commun. 2015, 6, 6336. [Google Scholar] [CrossRef]
- Hwang, J.; Kim, H.; Han, J.; Lee, J.; Hong, S.; Kim, S.; Yoon, S.K.; Choi, K.; Yang, J.; Park, U.; et al. Identification of Survival-Specific Genes in Clear Cell Renal Cell Carcinoma Using a Customized Next-Generation Sequencing Gene Panel. J. Pers. Med. 2022, 12, 113. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome. Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Kim, Y.; Han, B.G. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, e20. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Kim, Y.J.; Han, S.; Hwang, M.Y.; Shin, D.M.; Park, M.Y.; Lu, Y.; Yoon, K.; Jang, H.M.; Kim, Y.K.; et al. The Korea Biobank Array: Design and Identification of Coding Variants Associated with Blood Biochemical Traits. Sci. Rep. 2019, 9, 1382. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Moch, H.; Amin, M.B.; Berney, D.M.; Compérat, E.M.; Gill, A.J.; Hartmann, A.; Menon, S.; Raspollini, M.R.; Rubin, M.A.; Srigley, J.R.; et al. The 2022 World Health Organization Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2022, 82, 458–468. [Google Scholar] [CrossRef]
- Ma, C.; Luo, H. A more novel and robust gene signature predicts outcome in patients with esophageal squamous cell carcinoma. Clin. Res. Hepatol. Gastroenterol. 2022, 46, 102033. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Wu, J.Y.; Zhang, J.D.; Sun, Z.J.; Zheng, X.; Yu, B.Z.; Cao, H.Y.; Zhang, F.L.; Gao, Z.H.; Wang, W. A promising Prognostic risk model for advanced renal cell carcinoma (RCC) with immune-related genes. BMC Cancer 2022, 22, 691. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, D.; Duan, Y.; Yan, L.; Fan, Y.; Fang, Z.; Liu, Z. A five-gene signature predicts overall survival of patients with papillary renal cell carcinoma. PLoS ONE 2019, 14, e0211491. [Google Scholar] [CrossRef] [PubMed]
- Butler, S.N.; Blanck, G. Immunoscoring by correlating MHC class II and TCR expression: High level immune functions represented by the KIRP dataset of TCGA. Cell Tissue Res. 2016, 363, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Lasorsa, F.; Rutigliano, M.; Milella, M.; Ferro, M.; Pandolfo, S.D.; Crocetto, F.; Autorino, R.; Battaglia, M.; Ditonno, P.; Lucarelli, G. Cancer Stem Cells in Renal Cell Carcinoma: Origins and Biomarkers. Int. J. Mol. Sci. 2023, 24, 13179. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, A.; Saitoh, M.; Miyazawa, K. Dual Roles for Epithelial Splicing Regulatory Proteins 1 (ESRP1) and 2 (ESRP2) in Cancer Progression. Adv. Exp. Med. Biol. 2017, 925, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Polyak, K. BET Bromodomain Proteins as Cancer Therapeutic Targets. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, T.; Yoshino, H.; Sugita, S.; Miyamoto, K.; Yonemori, M.; Osako, Y.; Meguro-Horike, M.; Horike, S.I.; Nakagawa, M.; Enokida, H. Bromodomain protein BRD4 inhibitor JQ1 regulates potential prognostic molecules in advanced renal cell carcinoma. Oncotarget 2018, 9, 23003–23017. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Hildebrandt, M.A.; Pu, X.; Huang, M.; Lin, J.; Matin, S.F.; Tamboli, P.; Wood, C.G.; Wu, X. Role of inflammatory related gene expression in clear cell renal cell carcinoma development and clinical outcomes. J. Urol. 2011, 186, 2071–2077. [Google Scholar] [CrossRef]
- Shi, B.; Vinyals, A.; Alia, P.; Broceño, C.; Chen, F.; Adrover, M.; Gelpi, C.; Price, J.E.; Fabra, A. Differential expression of MHC class II molecules in highly metastatic breast cancer cells is mediated by the regulation of the CIITA transcription Implication of CIITA in tumor and metastasis development. Int. J. Biochem. Cell Biol. 2006, 38, 544–562. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, J.; Lei, X.; Yang, L.; Li, W.; Zheng, L.; Zhang, S.; Ding, Y.; Shi, J.; Zhang, L.; et al. CD1C is associated with breast cancer prognosis and immune infiltrates. BMC Cancer 2023, 23, 129. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Li, Y.; Lyu, Y.; Tan, L.; Zheng, X.; Jiang, H.; Wen, H.; Feng, C. Development of a Phagocytosis-Dependent Gene Signature to Predict Prognosis and Response to Checkpoint Inhibition in Clear-Cell Renal Cell Carcinoma. Front. Immunol. 2022, 13, 853088. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Ma, H.M.; Huang, H.B.; Li, Y.W.; Zhang, P.; Huang, J.J.; Cheng, L.; Li, G.R. Overexpression of COL5A1 promotes tumor progression and metastasis and correlates with poor survival of patients with clear cell renal cell carcinoma. Cancer Manag. Res. 2019, 11, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Boguslawska, J.; Kedzierska, H.; Poplawski, P.; Rybicka, B.; Tanski, Z.; Piekielko-Witkowska, A. Expression of Genes Involved in Cellular Adhesion and Extracellular Matrix Remodeling Correlates with Poor Survival of Patients with Renal Cancer. J. Urol. 2016, 195, 1892–1902. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Shao, T.; Bao, G.; Gao, Z.; Zhang, Y.; Ding, H.; Zhang, W.; Liu, F.; Guo, C. Identification of potential core genes in metastatic renal cell carcinoma using bioinformatics analysis. Am. J. Transl. Res. 2019, 11, 6812–6825. [Google Scholar] [PubMed]
- Huang, T.; OuYang, X.; Li, J.; Shi, B.; Shan, Z.; Shi, Z.; Yang, Z. Pan-cancer analysis of FBXW family with potential implications in prognosis and immune infiltration. Front. Immunol. 2022, 13, 1084339. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Liang, Z.; Fan, Z.; Cao, B.; Wang, N.; Wu, R.; Sun, H. A Comprehensive Analysis Revealing FBXW9 as a Potential Prognostic and Immunological Biomarker in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 5262. [Google Scholar] [CrossRef] [PubMed]
- Howell, M.C.; Green, R.; Khalil, R.; Foran, E.; Quarni, W.; Nair, R.; Stevens, S.; Grinchuk, A.; Hanna, A.; Mohapatra, S.; et al. Lung cancer cells survive epidermal growth factor receptor tyrosine kinase inhibitor exposure through upregulation of cholesterol synthesis. FASEB Bioadv. 2020, 2, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wan, F.; Zhang, H.; Shi, G.; Ye, D. ITGA2B and ITGA8 are predictive of prognosis in clear cell renal cell carcinoma patients. Tumour Biol. 2016, 37, 253–262. [Google Scholar] [CrossRef]
- Li, C.; Zhou, W.; Zhu, J.; Shen, Q.; Wang, G.; Chen, L.; Zhao, T. Identification of an Immune-Related Gene Signature Associated with Prognosis and Tumor Microenvironment in Esophageal Cancer. Biomed. Res. Int. 2022, 2022, 7413535. [Google Scholar] [CrossRef]
- Brugarolas, J. PBRM1 and BAP1 as novel targets for renal cell carcinoma. Cancer J. 2013, 19, 324–332. [Google Scholar] [CrossRef]
- Jin, S.; Wu, J.; Zhu, Y.; Gu, W.; Wan, F.; Xiao, W.; Dai, B.; Zhang, H.; Shi, G.; Shen, Y.; et al. Comprehensive Analysis of BAP1 Somatic Mutation in Clear Cell Renal Cell Carcinoma to Explore Potential Mechanisms in Silico. J. Cancer 2018, 9, 4108–4116. [Google Scholar] [CrossRef]
- Gong, Z.; Wu, X.; Guo, Q.; Du, H.; Zhang, F.; Kong, Y. Comprehensive Analysis of HMCN1 Somatic Mutation in Clear Cell Renal Cell Carcinoma. Genes 2022, 13, 1282. [Google Scholar] [CrossRef]
- Ricketts, C.J.; Linehan, W.M. Gender Specific Mutation Incidence and Survival Associations in Clear Cell Renal Cell Carcinoma (CCRCC). PLoS ONE 2015, 10, e0140257. [Google Scholar] [CrossRef]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 3698. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.C.; Hines, K.M.; Forsythe, J.G.; Erdogan, B.; Shi, M.; Hill, S.; Rose, K.L.; McLean, J.A.; Webb, D.J. Phosphorylation of serine 106 in Asef2 regulates cell migration and adhesion turnover. J. Proteome Res. 2014, 13, 3303–3313. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Marsaud, A.; Dadone, B.; Ambrosetti, D.; Baudoin, C.; Chamorey, E.; Rouleau, E.; Lefol, C.; Roussel, J.F.; Fabas, T.; Cristofari, G.; et al. Dismantling papillary renal cell carcinoma classification: The heterogeneity of genetic profiles suggests several independent diseases. Genes Chromosomes Cancer 2015, 54, 369–382. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, J.; Park, W.S.; Hong, D.; Chung, J. A retrospective single-centered, comprehensive targeted genetic sequencing analysis of prognostic survival using tissues from Korean patients with metastatic renal cell carcinoma after targeted therapy. Investig. Clin. Urol. 2022, 63, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, J.; Baselga, J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J. Clin. Oncol. 2003, 21, 2787–2799. [Google Scholar] [CrossRef]
- Dorđević, G.; Matušan Ilijaš, K.; Hadžisejdić, I.; Maričić, A.; Grahovac, B.; Jonjić, N. EGFR protein overexpression correlates with chromosome 7 polysomy and poor prognostic parameters in clear cell renal cell carcinoma. J. Biomed. Sci. 2012, 19, 40. [Google Scholar] [CrossRef]
- Wang, S.; Yu, Z.H.; Chai, K.Q. Identification of EGFR as a Novel Key Gene in Clear Cell Renal Cell Carcinoma (ccRCC) through Bioinformatics Analysis and Meta-Analysis. Biomed. Res. Int. 2019, 2019, 6480865. [Google Scholar] [CrossRef] [PubMed]
- D’Avella, C.; Abbosh, P.; Pal, S.K.; Geynisman, D.M. Mutations in renal cell carcinoma. Urol. Oncol. 2020, 38, 763–773. [Google Scholar] [CrossRef]
- Zhou, Y.; Qiu, J.; Liu, S.; Wang, P.; Ma, D.; Zhang, G.; Cao, Y.; Hu, L.; Wang, Z.; Wu, J.; et al. CFDP1 promotes hepatocellular carcinoma progression through activating NEDD4/PTEN/PI3K/AKT signaling pathway. Cancer Med. 2023, 12, 425–444. [Google Scholar] [CrossRef] [PubMed]
- Formicola, D.; Lasorsa, V.A.; Cantalupo, S.; Testori, A.; Cardinale, A.; Avitabile, M.; Diskin, S.; Iolascon, A.; Capasso, M. CFDP1 is a neuroblastoma susceptibility gene that regulates transcription factors of the noradrenergic cell identity. HGG Adv. 2023, 4, 100158. [Google Scholar] [CrossRef] [PubMed]
- Wehde, B.L.; Rädler, P.D.; Shrestha, H.; Johnson, S.J.; Triplett, A.A.; Wagner, K.U. Janus Kinase 1 Plays a Critical Role in Mammary Cancer Progression. Cell Rep. 2018, 25, 2192–2207.e2195. [Google Scholar] [CrossRef]
- Behbahani, T.E.; Thierse, C.; Baumann, C.; Holl, D.; Bastian, P.J.; von Ruecker, A.; Müller, S.C.; Ellinger, J.; Hauser, S. Tyrosine kinase expression profile in clear cell renal cell carcinoma. World J. Urol. 2012, 30, 559–565. [Google Scholar] [CrossRef]
- Chen, B.; Lai, J.; Dai, D.; Chen, R.; Li, X.; Liao, N. JAK1 as a prognostic marker and its correlation with immune infiltrates in breast cancer. Aging 2019, 11, 11124–11135. [Google Scholar] [CrossRef]
- Schier, A.F.; Shen, M.M. Nodal signalling in vertebrate development. Nature 2000, 403, 385–389. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, T.; Li, Q.; Wang, J.; Yang, D.; Li, X.; Wang, Q.; Song, X. Nodal activates smad and extracellular signal-regulated kinases 1/2 pathways promoting renal cell carcinoma proliferation. Mol. Med. Rep. 2015, 12, 587–594. [Google Scholar] [CrossRef]
- Wu, Y.; Du, K.; Guan, W.; Wu, D.; Tang, H.; Wang, N.; Qi, J.; Gu, Z.; Yang, J.; Ding, J. A novel definition of microvessel density in renal cell carcinoma: Angiogenesis plus vasculogenic mimicry. Oncol. Lett. 2020, 20, 192. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Ding, L.; Lu, Z.; Wang, R.; Yu, C.; Wang, H.; Zheng, Q.; Wang, X.; Xu, W.; Yu, H.; et al. METTL14-mediated Lnc-LSG1 m6A modification inhibits clear cell renal cell carcinoma metastasis via regulating ESRP2 ubiquitination. Mol. Ther. Nucleic Acids 2022, 27, 547–561. [Google Scholar] [CrossRef]
- Yue, P.J.; Sun, Y.Y.; Li, Y.H.; Xu, Z.M.; Fu, W.N. MYCT1 inhibits the EMT and migration of laryngeal cancer cells via the SP1/miR-629-3p/ESRP2 pathway. Cell Signal 2020, 74, 109709. [Google Scholar] [CrossRef] [PubMed]
- Freytag, M.; Kluth, M.; Bady, E.; Hube-Magg, C.; Makrypidi-Fraune, G.; Heinzer, H.; Höflmayer, D.; Weidemann, S.; Uhlig, R.; Huland, H.; et al. Epithelial splicing regulatory protein 1 and 2 (ESRP1 and ESRP2) upregulation predicts poor prognosis in prostate cancer. BMC Cancer 2020, 20, 1220. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, M.; Wu, W.; Miao, W.; Liu, H. Downregulated ESRP1/2 promotes lung metastasis of bladder carcinoma through altering FGFR2 splicing and macrophage polarization. Front. Immunol. 2023, 14, 1161273. [Google Scholar] [CrossRef]
- Zekri, J.; Baghdadi, M.A.; Meliti, A.; Sobahy, T.M.; Imtiaz, S. Variants of Human Mucin Genes in Clear Cell Renal Cell Carcinoma and their Potential Prognostic and Predictive Values. Gulf J. Oncol. 2023, 1, 35–39. [Google Scholar]
- Milella, M.; Rutigliano, M.; Lasorsa, F.; Ferro, M.; Bianchi, R.; Fallara, G.; Crocetto, F.; Pandolfo, S.D.; Barone, B.; d’Amati, A.; et al. The Role of MUC1 in Renal Cell Carcinoma. Biomolecules 2024, 14, 315. [Google Scholar] [CrossRef]
- Jin, Q.; Cheng, M.; Xia, X.; Han, Y.; Zhang, J.; Cao, P.; Zhou, G. Down-regulation of MYH10 driven by chromosome 17p13.1 deletion promotes hepatocellular carcinoma metastasis through activation of the EGFR pathway. J. Cell Mol. Med. 2021, 25, 11142–11156. [Google Scholar] [CrossRef]
- Liu, L.; Chen, C.; Liu, P.; Li, J.; Pang, Z.; Zhu, J.; Lin, Z.; Zhou, H.; Xie, Y.; Lan, T.; et al. MYH10 Combines with MYH9 to Recruit USP45 by Deubiquitinating Snail and Promotes Serous Ovarian Cancer Carcinogenesis, Progression, and Cisplatin Resistance. Adv. Sci. 2023, 10, e2203423. [Google Scholar] [CrossRef]
- Chang, S.L.; Lee, S.W.; Yang, S.F.; Chien, C.C.; Chan, T.C.; Chen, T.J.; Yang, C.C.; Li, C.F.; Wei, Y.C. Expression and prognostic utility of SSX2IP in patients with nasopharyngeal carcinoma. Apmis 2020, 128, 287–297. [Google Scholar] [CrossRef]
- Li, P.; Lin, Y.; Zhang, Y.; Zhu, Z.; Huo, K. SSX2IP promotes metastasis and chemotherapeutic resistance of hepatocellular carcinoma. J. Transl. Med. 2013, 11, 52. [Google Scholar] [CrossRef]
- Ewens, K.G.; Lalonde, E.; Richards-Yutz, J.; Shields, C.L.; Ganguly, A. Comparison of Germline versus Somatic BAP1 Mutations for Risk of Metastasis in Uveal Melanoma. BMC Cancer 2018, 18, 1172. [Google Scholar] [CrossRef]
- Frasca, F.; Nucera, C.; Pellegriti, G.; Gangemi, P.; Attard, M.; Stella, M.; Loda, M.; Vella, V.; Giordano, C.; Trimarchi, F.; et al. BRAF(V600E) mutation and the biology of papillary thyroid cancer. Endocr. Relat. Cancer 2008, 15, 191–205. [Google Scholar] [CrossRef]
- Shin, D.Y.; Na, I.I.; Kim, C.H.; Park, S.; Baek, H.; Yang, S.H. EGFR mutation and brain metastasis in pulmonary adenocarcinomas. J. Thorac. Oncol. 2014, 9, 195–199. [Google Scholar] [CrossRef]
- Mendhiratta, N.; Muraki, P.; Sisk, A.E., Jr.; Shuch, B. Papillary renal cell carcinoma: Review. Urol. Oncol. 2021, 39, 327–337. [Google Scholar] [CrossRef]
- Jonasch, E.; Walker, C.L.; Rathmell, W.K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 2021, 17, 245–261. [Google Scholar] [CrossRef]

| Variables | Number of Patients (%), N = 293 | |
|---|---|---|
| Sex | Male | 214 (73.0) |
| Female | 78 (26.6) | |
| Not available | 1 (0.4) | |
| Survival | Alive | 248 (84.6) |
| Deceased | 44 (15.0) | |
| Not available | 1 (0.4) | |
| Recurrence | Disease free | 218 (74.4) |
| Recurred/Progressed | 54 (18.4) | |
| Not available | 21 (7.2) | |
| Metastasis | Absent | 209 (71.3) |
| Present | 12 (4.1) | |
| Not available | 72 (24.6) | |
| No. | Gene | Number of Patients with Mutation (%) | Cytoband | Mutation Type | Survival (%) | Metastasis (%) | Overall Survival | Disease Free Survival | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Truncating | Missense | Splice | Inframe | Alive | Deceased | Absent | Present | Not Available | ||||||
| 1 | ACTR1B | 3 (1.02) | 2q11.2 | 1 | 2 | 0 | 0 | 0 | 3 (100) | 2 (67) | 0 | 1 (33) | 0.003 * | 0.047 * |
| 2 | BPNT1 | 2 (0.68) | 1q41 | 1 | 1 | 1 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.011 * | 0.204 |
| 3 | BRD4 | 2 (0.68) | 19p13.12 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 2 (100) | 0 | 0 | 0.020 * | 0.009 * |
| 4 | BZRAP1 | 2 (0.68) | 17q22 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 2 (100) | 0 | 0 | 0.003 * | 0.512 |
| 5 | C15orf27 | 1 (0.34) | 15q24.2 | 0 | 1 | 0 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 | 0.019 * | 0.652 |
| 6 | C16orf72 | 2 (0.68) | 16p13.2 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.002 * | 0.144 |
| 7 | CD1C | 2 (0.68) | 1q23.1 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.017 * | 0.410 |
| 8 | CEP128 | 2 (0.68) | 14q31.1 | 2 | 0 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.017 * | 0.330 |
| 9 | CIITA | 2 (0.68) | 16p13.13 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 1 (50) | 1 (50) | 0 | 0.000002 ** | 0.140 |
| 10 | COL5A1 | 4 (1.37) | 9q34.3 | 2 | 1 | 1 | 0 | 1 (25) | 3 (75) | 3 (75) | 0 | 1 (25) | 0.001 ** | 0.404 |
| 11 | CYP51A1 | 2 (0.68) | 9q34.3 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 0 | 0 | 2 (100) | 0.001 * | 0.139 |
| 12 | DNAAF2 | 2 (0.68) | 14q21.3 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 2 (100) | 0 | 0 | 0.001 * | 0.548 |
| 13 | EPHB1 | 1 (0.34) | 3q22.2 | 0 | 1 | 0 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 | 0.005 * | 0.003 * |
| 14 | ESRP2 | 2 (0.68) | 16q22.1 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 0 | 1 (50) | 1 (50) | 0.016 * | 0.492 |
| 15 | FBXW9 | 3 (1.02) | 19p13.13 | 0 | 3 | 0 | 0 | 0 | 3 (100) | 3 (100) | 0 | 0 | 0.0001 ** | 0.00001 ** |
| 16 | ITGA3 | 1 (0.34) | 17q21.33 | 0 | 1 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 1 (100) | 3.87 × 10−7 ** | 1.42 × 10−7 ** |
| 17 | ITGA4 | 1 (0.34) | 2q31.3 | 0 | 1 | 0 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 | 0.00001 ** | 0.766 |
| 18 | ITGA8 | 1 (0.34) | 10p13 | 0 | 1 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 1 (100) | 3.87 × 10−7 ** | 1.42 × 10−7 ** |
| 19 | KIF5C | 2 (0.68) | 2q23.1-q23.2 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.0001 ** | 0.146 |
| 20 | KPRP | 2 (0.68) | 1q21.3 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 2 (100) | 0 | 0 | 3.07 × 10−8 ** | 1.68 × 10−10 ** |
| 21 | MAOB | 3 (1.02) | Xp11.3 | 0 | 2 | 1 | 0 | 1 (33) | 2 (67) | 2 (67) | 1 (33) | 0 | 0.022 * | 0.033 * |
| 22 | MUC17 | 9 (3.07) | 7q22.1 | 1 | 9 | 0 | 1 | 4 (44) | 5 (56) | 1 (11) | 3 (33) | 5 (56) | 1.42 × 10−6 ** | 0.002 * |
| 23 | MYH10 | 4 (1.37) | 17p13.1 | 1 | 3 | 0 | 0 | 1 (25) | 3 (75) | 2 (50) | 1 (25) | 1 (25) | 0.00006 ** | 6.40 × 10−7 ** |
| 24 | OGFR | 1 (0.34) | 20q13.33 | 0 | 1 | 0 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 | 0.0006 ** | 0.736 |
| 25 | OR1S1 | 1 (0.34) | 11q12.1 | 0 | 1 | 0 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 | 0.00002 ** | 0.0007 ** |
| 26 | PCBP4 | 2 (0.68) | 3p21.2 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.025 * | 0.481 |
| 27 | PCGF2 | 2 (0.68) | 17q12 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.042 * | 0.441 |
| 28 | PCSK2 | 2 (0.68) | 20p12.1 | 0 | 1 | 0 | 1 | 0 | 2 (100) | 2 (100) | 0 | 0 | 0.010 * | 0.301 |
| 29 | PLEKHB2 | 2 (0.68) | 2q21.1 | 1 | 2 | 0 | 0 | 0 | 2 (100) | 2 (100) | 0 | 0 | 0.002 * | 0.175 |
| 30 | PPM1F | 2 (0.68) | 22q11.22 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 1 (50) | 1 (50) | 0 | 0.0002 ** | 0.180 |
| 31 | RAB40B | 2 (0.68) | 17q25.3 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.0001 ** | 5.88 × 10−31 ** |
| 32 | RRP36 | 2 (0.68) | 6p21.1 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 2 (100) | 0 | 0 | 0.006 * | 0.176 |
| 33 | RTL1 | 2 (0.68) | 14q32.2 | 3 | 0 | 0 | 1 | 0 | 2 (100) | 1 (50) | 1 (50) | 0 | 6.27 × 10−8 ** | 0.031 * |
| 34 | RYR1 | 7 (2.39) | 19q13.2 | 1 | 6 | 0 | 0 | 3 (43) | 4 (57) | 6 (86) | 1 (14) | 0 | 0.001 ** | 0.051 |
| 35 | SMARCA1 | 3 (1.02) | Xq25-q26.1 | 1 | 2 | 0 | 0 | 0 | 3 (100) | 1 (33) | 0 | 2 (67) | 3.51 × 10−9 ** | 2.64 × 10−5 ** |
| 36 | SNX7 | 2 (0.68) | 1p21.3 | 1 | 1 | 0 | 0 | 0 | 2 (100) | 0 | 1 (50) | 1 (50) | 0.0004 ** | 0.197 |
| 37 | SSX2IP | 2 (0.68) | 1p22.3 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 0 | 2 (100) | 0 | 3.93 × 10−10 ** | 0.006 * |
| 38 | TAS1R2 | 2 (0.68) | 1p36.13 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 2 (100) | 0 | 0 | 3.09 × 10−12 ** | 0.028 * |
| 39 | THUMPD2 | 2 (0.68) | 2p22.1; 2p22-p21 | 2 | 0 | 1 | 0 | 0 | 2 (100) | 1 (50) | 1 (50) | 0 | 0.00002 ** | 2.15 × 10−15 ** |
| 40 | VPS13D | 2 (0.68) | 1p36.22-p36.21 | 0 | 2 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 | 1 (50) | 0.042 * | 0.441 |
| Patients with Metastasis | Age | Sex | TNM Stage | Survival | Tumor Type | Survival-Specific Genes | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| T | N | M | ||||||||
| TCGA-B9-4114-01 | 49 | M | 2 | 0 | 1 | Alive | NA | |||
| TCGA-G7-A8LB-01 | 70 | M | 2 | NA | 1 | Alive | NA | |||
| TCGA-BQ-5894-01 | 42 | M | 3 | 1 | 1 | Alive | II | |||
| TCGA-4A-A93X-01 | 58 | M | 3 | 1 | 1 | Alive | NA | |||
| TCGA-F9-A8NY-01 | 38 | F | 4 | 1 | 1 | Alive | II | |||
| TCGA-A4-A57E-01 | 59 | M | 2 | 0 | 1 | Deceased | NA | MUC17 | RTL1 | RYR1 |
| TCGA-2Z-A9J7-01 | 63 | M | 2 | 0 | 1 | Deceased | NA | MUC17 | ||
| TCGA-AL-3466-01 | 41 | M | 3 | 1 | 1 | Deceased | NA | MAOB | ||
| TCGA-BQ-5877-01 | 60 | M | 3 | 1 | 1 | Deceased | NA | PPM1F | THUMPD2 | |
| TCGA-SX-A7SM-01 | 60 | M | 3 | 1 | 1 | Deceased | II | ESRP2 | MUC17 | SSX2IP |
| TCGA-BQ-5893-01 | 61 | M | 3 | 1 | 1 | Deceased | NA | CIITA | SNX7 | |
| TCGA-BQ-5889-01 | 63 | M | 3 | 1 | 1 | Deceased | NA | MYH10 | SSX2IP | |
| Variables | Patients (%) N = 60 | Survival (%) | p-Value | |||
|---|---|---|---|---|---|---|
| Alive | Deceased | |||||
| Age | <70 | <50 | 11 (18.3) | 9 (24.3) | 2 (5.4) | 1.000 |
| 50–59 | 11 (18.3) | 10 (27.0) | 1 (2.7) | |||
| 60–69 | 15 (25) | 15 (40.5) | 0 (0) | |||
| ≥70 | 70–79 | 15 (25) | 15 (65.2) | 0 (0) | ||
| 80–89 | 7 (11.7) | 7 (30.4) | 0 (0) | |||
| ≥90 | 1 (1.7) | 0 (0) | 1 (4.3) | |||
| Sex | Male | 45 (75) | 42 (93.3) | 3 (6.7) | 1.000 | |
| Female | 15 (25) | 14 (93.3) | 1 (6.7) | |||
| Tumor type | I | 15 (25) | 15 (100.0) | 0 (0) | 0.564 | |
| II | 45 (75) | 41 (91.1) | 4 (8.9) | |||
| Nuclear Grade | I | 0 (0) | 0 (0) | 0 (0) | 0.133 | |
| II | 25 (41.7) | 25 (100.0) | 0 (0) | |||
| III | 34 (56.7) | 31 (88.6) | 3 (8.6) | |||
| IV | 1 (1.7) | 0 (0) | 1 (2.9) | |||
| Tumor size | ≤7.0 cm | 52 (86.7) | 51 (98.1) | 1 (1.9) | 0.006 | |
| >7.0 cm | 8 (13.3) | 5 (62.5) | 3 (37.5) | |||
| T stage | T1 | 47 (78.3) | 46 (97.9) | 1 (2.1) | 0.029 | |
| T2 | 5 (8.3) | 3 (23.1) | 2 (15.4) | |||
| T3 | 8 (13.3) | 7 (53.8) | 1 (7.7) | |||
| N stage | N0 | 57 (95) | 54 (94.7) | 3 (5.3) | 0.190 | |
| N1 | 3 (5) | 2 (66.7) | 1 (33.3) | |||
| M stage | M0 | 52 (86.7) | 52 (100.0) | 0 (0) | 0.0001 | |
| M1 | 8 (13.3) | 4 (50.0) | 4 (50.0) | |||
| Recurrence | Disease free | 52 (86.7) | 52 (100.0) | 0 (0) | 0.0001 | |
| Recurred/Progressed | 8 (13.3) | 4 (50.0) | 4 (50.0) | |||
| Response to laparoscopic | No evidence of disease | 52 (86.7) | 52 (100.0) | 0 (0) | 0.0001 | |
| radical nephrectomy | Fail | 8 (13.3) | 4 (50.0) | 4 (50.0) | ||
| No. | Gene | Number of Patients with Mutation | Mutation Frequency, % | Cytoband | Mutation Type | Survival (%) | Metastasis (%) | Overall Survival | Disease Free Survival | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Truncating | Missense | Inframe | Alive | Deceased | Absent | Present | |||||||
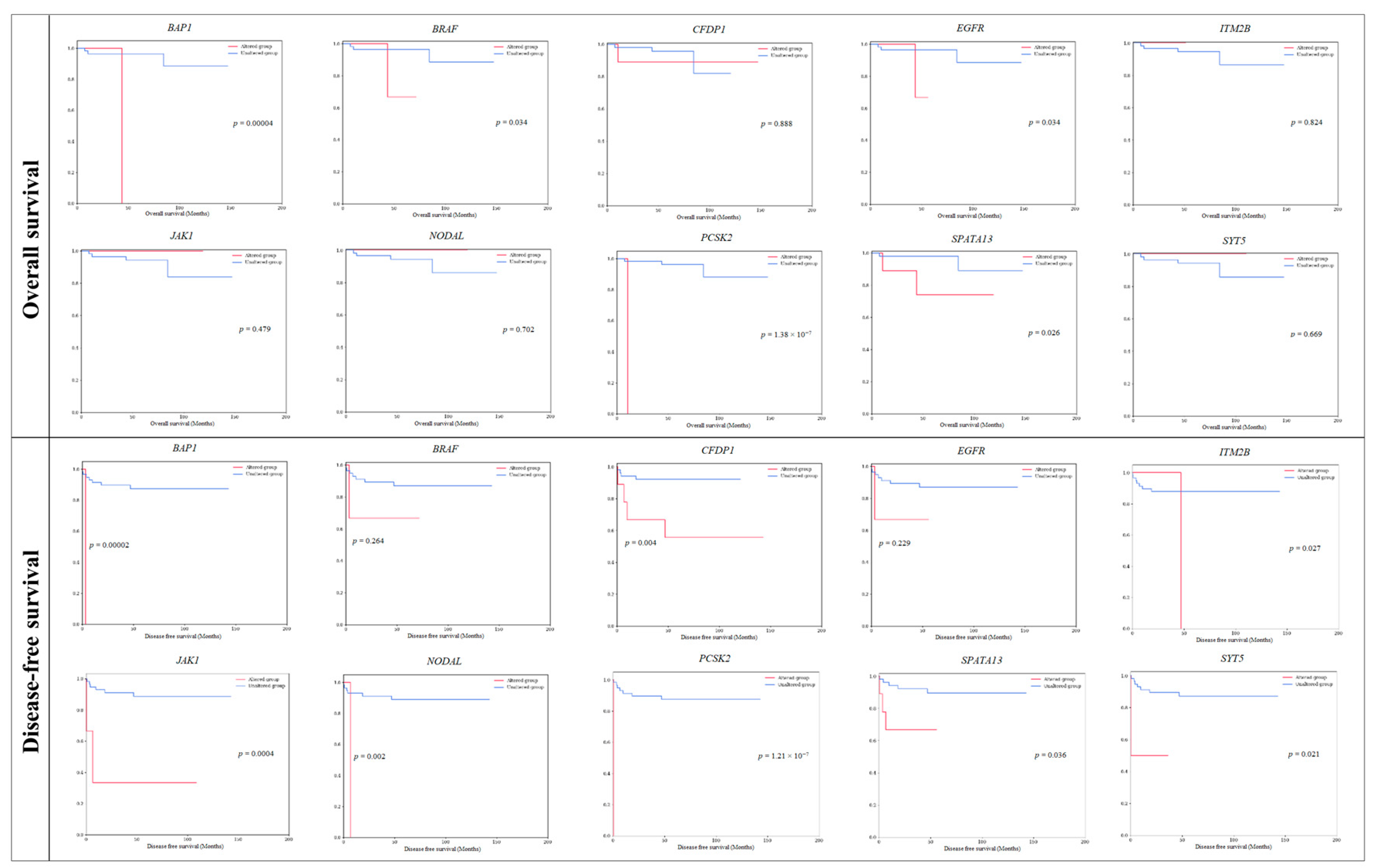
| 1 | BAP1 | 1 | 1.67 | 3p21.1 | 0 | 0 | 1 | 0 | 1 (100) | 0 | 1 (100) | 0.00004 ** | 0.00002 ** |
| 2 | BRAF | 3 | 5.00 | 7q34 | 0 | 3 | 5 | 2 (67) | 1 (33) | 2 (67) | 1 (33) | 0.034 * | 0.264 |
| 3 | CFDP1 | 9 | 15.00 | 16q23.1 | 0 | 15 | 1 | 8 (89) | 1 (11) | 5 (56) | 4 (44) | 0.888 | 0.004 * |
| 4 | EGFR | 3 | 5.00 | 7p11.2 | 0 | 4 | 0 | 2 (67) | 1 (33) | 2 (67) | 1 (33) | 0.034 * | 0.229 |
| 5 | ITM2B | 1 | 1.67 | 13q14.2 | 0 | 1 | 0 | 1 (100) | 0 | 0 | 1 (100) | 0.814 | 0.027 * |
| 6 | JAK1 | 3 | 5.00 | 1p31.3 | 1 | 4 | 1 | 3 (100) | 0 | 1 (33) | 2 (67) | 0.479 | 0.0004 ** |
| 7 | NODAL | 1 | 1.67 | 10q22.1 | 0 | 1 | 0 | 1 (100) | 0 | 0 | 1 (100) | 0.702 | 0.002 * |
| 8 | PCSK2 | 1 | 1.67 | 20p12.1 | 0 | 1 | 0 | 0 | 1 (100) | 0 | 1 (100) | 1.38 × 10−7 ** | 1.21 × 10−7 ** |
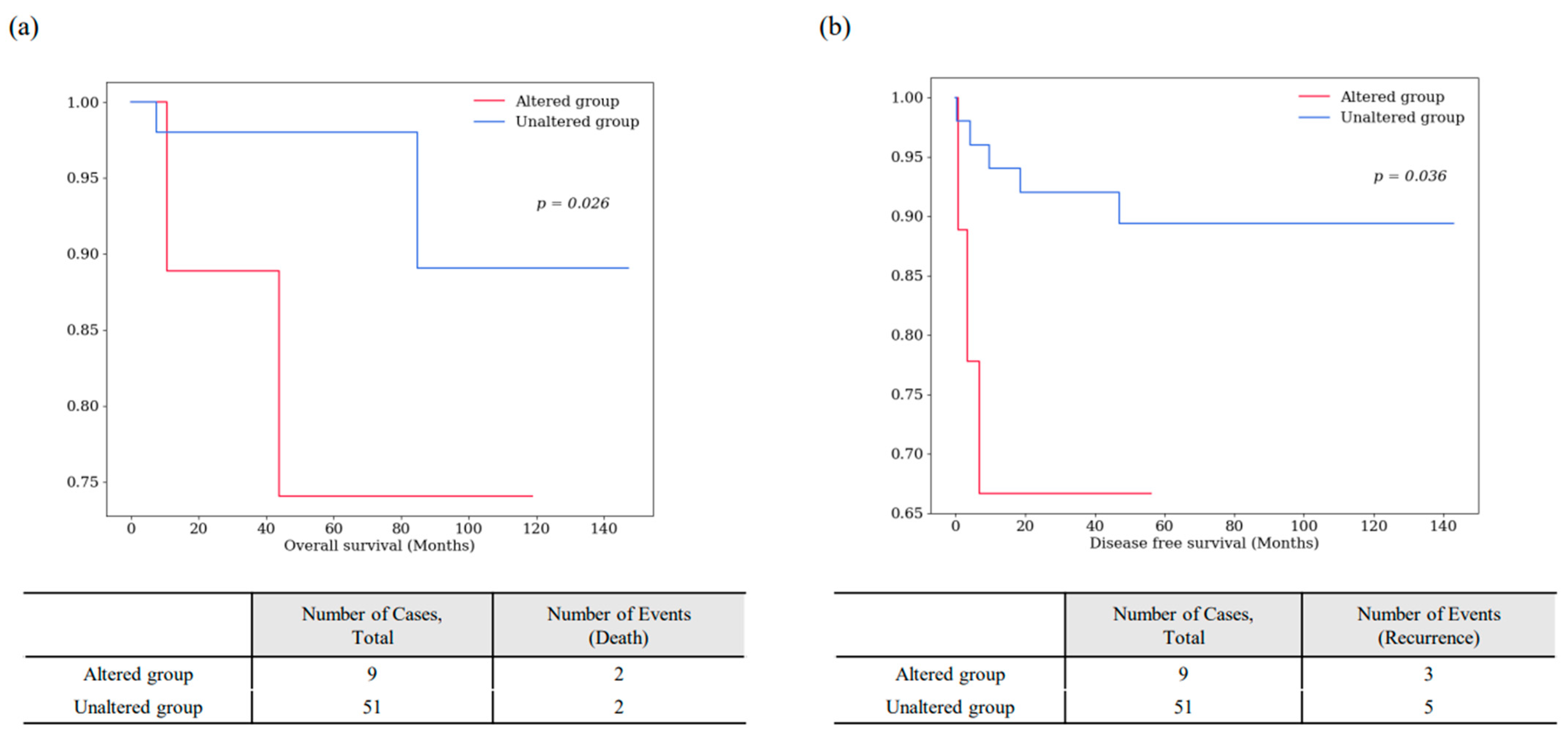
| 9 | SPATA13 | 9 | 15.00 | 13q12.12 | 0 | 9 | 0 | 7 (78) | 2 (22) | 6 (67) | 3 (33) | 0.026 * | 0.036 * |
| 10 | SYT5 | 2 | 3.33 | 19q13.42 | 0 | 2 | 0 | 2 (100) | 0 | 1 (50) | 1 (50) | 0.669 | 0.021 * |
| Patients with Metastasis | Age | Sex | Tumor Type | Nuclear Grade | TNM Stage | Survival | Metastatic Site | Response to LRN | Mutations among 10 Survival-Specific Genes | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | N | M | ||||||||||||
| S_20220322_036 | 80 | M | Ⅱ | III | 3 | 0 | 1 | Alive | Lung | Fail | CFDP1 | ITM2B | ||
| S_20220425_038 | 42 | F | Ⅱ | III | 1 | 0 | 1 | Alive | Adrenal gland, Lung | Fail | JAK1 | SYT5 | ||
| S_20220425_044 | 77 | M | Ⅱ | III | 3 | 0 | 1 | Alive | Bone | Fail | CFDP1 | JAK1 | NODAL | SPATA13 |
| S_20220425_055 | 82 | F | Ⅱ | III | 3 | 1 | 1 | Alive | Lymph node | Fail | CFDP1 | |||
| S_20220425_042 | 49 | M | Ⅱ | III | 2 | 0 | 1 | Deceased | Lung, Lymph node | Fail | ||||
| S_20220322_058 | 42 | M | Ⅱ | III | 1 | 0 | 1 | Deceased | Lung | Fail | ||||
| S_20220425_029 | 51 | M | Ⅱ | III | 2 | 0 | 1 | Deceased | Lung | Fail | BAP1 | BRAF | EGFR | SPATA13 |
| S_20220425_040 | 93 | F | Ⅱ | IV | 3 | 1 | 1 | Deceased | Liver, Lung, Vagina | Fail | CFDP1 | PCSK2 | SPATA13 | |
| Gene | Database | Mutation | Survival Analysis | ||||
|---|---|---|---|---|---|---|---|
| Type | HGVS.c | HGVS.p | Frequency, % | Overall Survival | Disease Free Survival | ||
| PCSK2 | TCGA-KIRP | missense_variant | c.850C>T | p.Leu284Phe | 0.68 | 0.010 * | 0.301 |
| In_Frame_Ins | c.10_12dupGGT | p.Gly4dup | |||||
| Korean-KIRP | missense_variant | c.1879G>T | p.Val627Leu | 1.67 | 1.38× 10−7 ** | 1.21× 10−7 ** | |
| Clinical Variable | Statistic | CFDP1 | JAK1 | SPATA13 |
|---|---|---|---|---|
| Tumor size | OR (CI) | 0.47 (0.06–5.71) | 0.29 (0.01–18.86) | 0.11 (0.02–0.80) |
| p-value | 0.593 | 0.354 | 0.013 * | |
| T stage | OR (CI) | 0.28 (0.05–1.67) | 0.54 (0.03–34.06) | 0.16 (0.02–0.89) |
| p-value | 0.092 | 0.526 | 0.018 * | |
| N stage | OR (CI) | 13.20 (0.62–852.80) | 0.00 (0.00–60.70) | 2.98 (0.05–63.96) |
| p-value | 0.056 | 1.000 | 0.391 | |
| M stage | OR (CI) | 8.83 (1.25–66.35) | 15.50 (0.71–1012.77) | 4.44 (0.55–30.90) |
| p-value | 0.013 * | 0.044 * | 0.090 | |
| Recurrence | OR (CI) | 8.83 (1.25–66.35) | 15.50 (0.71–1012.77) | 4.44 (0.55–30.90) |
| p-value | 0.013 * | 0.044 * | 0.090 | |
| Death | OR (CI) | 1.97 (0.03–28.41) | 0.00 (0.00–40.77) | 6.64 (0.42–105.71) |
| p-value | 0.488 | 1.000 | 0.103 | |
| Response to LRN | OR (CI) | 0.11 (0.02–0.80) | 0.06 (0.00–1.40) | 0.23 (0.03–1.81) |
| p-value | 0.013 * | 0.044 * | 0.090 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, J.; Bang, S.; Choi, M.H.; Hong, S.-H.; Kim, S.W.; Lee, H.E.; Yang, J.H.; Park, U.S.; Choi, Y.J. Discovery and Validation of Survival-Specific Genes in Papillary Renal Cell Carcinoma Using a Customized Next-Generation Sequencing Gene Panel. Cancers 2024, 16, 2006. https://doi.org/10.3390/cancers16112006
Hwang J, Bang S, Choi MH, Hong S-H, Kim SW, Lee HE, Yang JH, Park US, Choi YJ. Discovery and Validation of Survival-Specific Genes in Papillary Renal Cell Carcinoma Using a Customized Next-Generation Sequencing Gene Panel. Cancers. 2024; 16(11):2006. https://doi.org/10.3390/cancers16112006
Chicago/Turabian StyleHwang, Jia, Seokhwan Bang, Moon Hyung Choi, Sung-Hoo Hong, Sae Woong Kim, Hye Eun Lee, Ji Hoon Yang, Un Sang Park, and Yeong Jin Choi. 2024. "Discovery and Validation of Survival-Specific Genes in Papillary Renal Cell Carcinoma Using a Customized Next-Generation Sequencing Gene Panel" Cancers 16, no. 11: 2006. https://doi.org/10.3390/cancers16112006
APA StyleHwang, J., Bang, S., Choi, M. H., Hong, S.-H., Kim, S. W., Lee, H. E., Yang, J. H., Park, U. S., & Choi, Y. J. (2024). Discovery and Validation of Survival-Specific Genes in Papillary Renal Cell Carcinoma Using a Customized Next-Generation Sequencing Gene Panel. Cancers, 16(11), 2006. https://doi.org/10.3390/cancers16112006






