Simple Summary
This review discusses the importance of advancements in cancer diagnosis and treatment, emphasizing the need for early detection methods. It highlights the role of DNA methylation, an epigenetic change, as a promising marker for detecting cancer. This review explains how DNA methylation patterns differ between cancerous and healthy cells, influencing gene expression and contributing to genomic instability. It also delves into the challenges and methods associated with detecting DNA methylation, particularly in cell-free DNA (cfDNA) from bodily fluids, known as liquid biopsies. Different tests for single-cancer detection, such as colorectal, lung, and bladder cancer, as well as multi-cancer detection tests, are discussed along with their methodologies and clinical applications. Biological factors influencing DNA methylation, such as age, gender, and environmental influences, are also explored. The conclusion emphasizes the potential of cfDNA methylation analysis in cancer diagnostics but acknowledges the current limitations and challenges, highlighting the need for further research and advancements in methodology and understanding.
Abstract
In current clinical practice, effective cancer testing and screening paradigms are limited to specific types of cancer, exhibiting varying efficiency, acceptance, and adherence. Cell-free DNA (cfDNA) methylation profiling holds promise in providing information about the presence of malignity regardless of its type and location while leveraging blood-based liquid biopsies as a method to obtain analytical samples. However, technical difficulties, costs and challenges resulting from biological variations, tumor heterogeneity, and exogenous factors persist. This method exploits the mechanisms behind cfDNA release but faces issues like fragmentation, low concentrations, and high background noise. This review explores cfDNA methylation’s origins, means of detection, and profiling for cancer diagnostics. The critical evaluation of currently available multi-cancer early detection methods (MCEDs) as well as tests targeting single genes, emphasizing their potential and limits to refine strategies for early cancer detection, are explained. The current methodology limitations, workflows, comparisons of clinically approved liquid biopsy-based methylation tests for cancer, their utilization in companion diagnostics as well as the biological limitations of the epigenetics approach are discussed, aiming to help healthcare providers as well as researchers to orient themselves in this increasingly complex and evolving field of diagnostics.
1. Introduction
Cancer is poised to become the primary cause of death worldwide, highlighting the urgency for advancements not only in more impactful treatments [1] but also in new effective diagnostic methods with high clinical utility for early cancer detection [2,3]. The detection of cancer before stage IV could potentially reduce cancer-related deaths by at least 15% within a five-year period [1,4]. The emerging paradigm shift in disease diagnosis involves a growing reliance on molecular characterization, moving beyond conventional clinical and symptom-based assessments. While genetic alterations and transcription signatures were initially proposed as biomarkers, their clinical use is constrained by issues of reproducibility and accuracy. Consequently, attention has shifted towards exploring epigenetic changes as a viable alternative for disease diagnosis [5].
DNA methylation, a prominent epigenetic change in humans, has been heavily studied in cancer [6,7]. It plays crucial roles in gene expression regulation, X-chromosome inactivation, genomic imprinting, and allele-specific expression [8,9,10]. DNA methylation primarily targets cytosine residues in CpG dinucleotides, typically clustered in CpG islands. These islands are primarily located at the 5′ end of genes and represent approximately 60% of the promoter regions of human genes. Approximately 30,000 CpG islands are present in the human genome, and under physiological conditions, 70–80% of them are methylated [11]. The dysregulation of DNA methylation is linked to various pathological conditions including cancer, developmental disorders, and aging. The methylomes of cancer cells differ from healthy cells and may be used to distinguish between different cancer types, as some genes develop tissue-specific DNA methylation patterns. Since these changes can be assessed in various body fluid samples, DNA-methylation-based liquid biopsies are widely recognized as promising less-invasive approaches for prognostic assessment as well as for the identification of premalignant/early cancer [12]. One of the significant advantages of a liquid biopsy is its ability to follow the progression of malignant tumors, both primary and metastatic, and to identify when their recurrence has occurred [13]. In addition, it is cheaper and less invasive, thus increasing patient acceptance [14]. However, DNA methylation detection is associated with certain limitations that must be carefully considered. Cell-type composition, cellular contamination, technical problems, allele- and strand-specific DNA methylation, and random loss of DNA methylation at a specific locus are examples of potential sources of heterogeneity [15,16].
2. DNA Methylation and Cancer
Cancer development can be described as an interplay between genetic and epigenetic alterations, with changes in DNA methylation patterns being among the earliest, most consistent, and prevalent indicators [17]. Numerous theories have been advanced about the reason for aberrant methylation in cancer cells, primarily revolving around two fundamental concepts: differential fitness and targeted selection [18]. The differential fitness theory proposes that abnormal DNA hypermethylation initiates with the indiscriminate dissemination of DNA methylation across the genome, potentially stemming from dysregulation of the DNA methylation apparatus. Such methylation occurrences predominantly manifest within gene promoters responsible for constraining cell viability and growth. The alternative theory suggests that hypermethylation arises from the anomalous localization of DNMTs to specific genomic regions and/or that these regions inherently harbor cis-regulatory elements, rendering them more conducive to de novo DNA methylation [19]. The methylome of malignant cells in comparison with normal cells is characterized by two types of general changes: global hypomethylation or focal hypermethylation at CpG islands. Since DNA methylation affects chromatin structure and function, these changes lead to an abnormal regulation of gene expression and the derepression of imprinted genes and retrotransposons and contribute to the occurrence of somatic mutations, resulting in overall genomic instability [11,12,20,21]. Genes with hypomethylated promoter regions have higher expression levels, whereas the acquisition of methylation in the initially non-methylated promoter region can inhibit the ability of transcription factors to bind, resulting in transcription down-regulation or gene silencing [22]. Different genes are methylated at various stages of cancer development, influencing oncogene activation and tumor-suppressor gene inactivation [11,12,20]. It is well known that focused promoter hypermethylation causes the transcriptional suppression of tumor-suppressor genes in cancer [14]. On the contrary, global methylation levels in benign tissue were found to be considerably higher than in malignant tissue [23], indicating that hypermethylated CpG sites are more prone to mutations than their regularly methylated counterparts [24]. Hypomethylation of repeated DNA sequences or proto-oncogenes has a markedly negative impact on chromosomal stability and cellular function [25]. Moreover, it has been proposed that aneuploidy results from the excessive hypomethylation of centromeric and pericentromeric satellite sequences, which is frequently observed in a variety of malignancies [12]. Meta-analyses of global DNA hypomethylation in a variety of malignancies have revealed a correlation between the degree of hypomethylation and cancer stage, confirming its role in carcinogenesis. On an overall methylation level in malignant cells, colorectal cancer (CRC) is very well studied, where a comparison between CRC cells and normal colorectal epithelial cells revealed that over 10% of protein-coding genes exhibit differential methylation [26]. Several methylation markers have been found to date that are related to a wide spectrum of cancers, including breast cancer (e.g., hypermethylated APC, FOXA1, and RASSF1A) [27], esophageal cancer (e.g., hypermethylated TP53, CDKN2A, CTNNB1, and APC) [28,29], gastric cancer (e.g., hypermethylated p16, RUNX3, RASSF10, APC, and hypomethylated LINE1) [28,29], lung cancer (e.g., hypermethylated DAL-1, EPHB6, HS3ST2, MGMT, and hypomethylated ELMO3) [30], CRC (e.g., hypermethylated SEPT9, MGMT, SDC2, N-Myc, and APC) [30,31], and hepatocellular carcinoma (e.g., hypermethylated HOXA1, EMX1, ECE1, and PFKP) [32].
3. Methylation Detection in Cell-Free DNA
Cell-free DNA (cfDNA) fragments released from cells preserve genetics and epigenetics background information from the original tissue [33]. Apart from primary structure and epigenetic marks, the increased concentration of cfDNA in the body fluids of cancer patients compared to healthy individuals makes it a promising marker of neoplasms [34,35]. Results have clearly shown that it is tumor-type- and progression-specific [36], allowing for the real-time monitoring of disease dynamics and treatment response. Among the most common epigenetic modifications of cfDNA is methylation, which has received increased attention in recent years. The results of experiments indicate that cfDNA isolated from two different tumors is more likely to vary in somatic genetic mutations than in epigenetic information, which remains consistent in many cases, again suggesting the potential of cfDNA methylation [37].
Until now, the detection of epigenetic alterations in cancer cells has been carried out mainly from solid tumors, but due to the invasiveness and complexity of samplings, over time, research is starting to move to liquid biopsy sampling as a reliable alternative. [38,39].
Liquid biopsy represents a method focused on reducing the invasiveness of the biological sampling from the patient. For liquid biopsy body fluids, blood and its individual components, cerebrospinal fluid, urine, or saliva are commonly used to analyze cfDNA or circulating tumor DNA (ctDNA) as well as other cancer-related analytes, e.g., exosomes, miRNAs, and proteins [40].
The principle of ctDNA detection in bodily fluids is based precisely on the methylation profile of the DNA fragment, which is typical for tumor cells [41,42]. Hypermethylation occurs in ctDNA derived from a wide range of tumor types, such as lung [43], breast [43,44], colon [45], or prostate [46] neoplasms. Since DNA fragments from necrotizing and apoptotic tumor cells are directly excreted into the body during the tumor process, methylated ctDNA is considered tumor-specific because of its primary source, which provides additional valuable information in diagnostic applications [47].
3.1. Methods for Analyzing DNA Methylation Status
There are two main approaches for detecting methylated sites of ctDNA regarding the range of the target DNA sequence: (I) whole-genome analysis and (II) methods focused on specific DNA loci. Generally, the manual and technical difficulty is higher for methods analyzing the whole genome, and these methods suddenly require validation at the level of the selected loci [47,48].
There are three categories of methods in terms of the molecular mechanism for DNA-methylation detection: (I) methods based on the detection of sodium bisulfite conversion, (II) methods using the interaction of antibodies against 5mC with methylated sections of DNA, and (III) methods using enzymatic cleavage. Among the above-stated methods, antibody–DNA interaction methods are widely used, e.g., enzyme-linked immunosorbent assay (ELISA) assays, mainly due to their availability and easy handling. There are several currently commercially available kits, preponderantly based on the sandwich ELISA principle, which is rough and, unfortunately, does not provide specific information about methylation. In addition, their further disadvantage is high variability together with cross-reactivity [49], which ultimately indicates the improbable use of ELISA-based assays in the detection of DNA methylation in clinical practice.
Methods based on the selective cleavage of DNA using methylation-sensitive restriction enzymes (MSREs) capable of cleaving unmethylated sections of CpG islands while leaving methylated fragments uncleaved are also often used [50]. With the subsequent use of qPCR, it is a rapid, reproducible, and robust method that can also be used in combination with sequencing, either Sanger, bisulfite, or even next-generation sequencing (NGS), while the last-mentioned method is accompanied by increased complexity and costs.
Last but not least, the method of DNA methylation detection using sodium bisulfite conversion is currently considered a gold standard in DNA methylation screening. The treatment of the DNA sample with sodium bisulfite converts unmethylated cytosine to uracil and then converts uracil to thymine during subsequent methylation-specific PCR (MSP) [51]. Primers used for MSP are designed as methylation-specific, complementary to unconverted 5-methylcytosines, or, conversely, to the converted sequence. It is also applicable with high sensitivity for sequences with a low density of methylation, while a high density of CpG islands is directly related to increasing the specificity of this test [52]. In the case of quantitative methylation-specific PCR (qMSP), in addition to specifically designed primers, methylation-specific fluorescent reporter probes are also used, which allow not only for very sensitive detection but also for the quantification of the DNA methylation level [53].
In clinical practice, genome-wide bisulfite sequencing is practically inapplicable, mainly due to its difficulty and high cost; thus, currently, it serves primarily for research purposes to map epigenome methylation [54]. The analytical boundaries of genomic ctDNA assays are commonly assessed through the measurement of the variant allele frequency that denotes the proportion of sequencing reads containing tumor-specific mutations relative to the total number of sequencing reads aligning with the corresponding genomic loci [55].
A targeted bisulfite sequencing of precise loci is deemed more effective [56], offering a reliable tool for validating data obtained, for example, by array-based methods. Array-based methods enable the screening of methylation on a genome-wide scale, but similar to whole-genome sequencing, this method also has a relatively high error rate, and targeted bisulfite sequencing could serve as one of its validation options in clinical practice [56,57].
Although improvements in amplicon-based NGS have enhanced reliability and sensitivity, its utility is constrained to established hotspots. Additionally, panels are comparatively more affordable than capture NGS [58]. Methylation signatures of ctDNA identified through various bioinformatics models may exhibit inconsistency, resulting in disparities or partiality within their predictive models [59]. Furthermore, each assay developed by different research teams has its criteria for defining positive or negative results, as well as distinct methodologies for ctDNA extraction, analysis, and quantification [60]. An overview of the current laboratory methods for methylation detection and their respective characteristics, including their major advantages and disadvantages, can be found in Supplementary Table S1.
3.2. Clinically Approved Methylation-Based Cancer Screening
In clinical practice, the most frequently used methodology is the bisulfite conversion method, followed by PCR or sequencing; thus, the majority of clinically approved methylation-based screenings are based on this principle [61]. Below, we summarize the currently available tests implementing a methylation and liquid biopsy approach, distinguishing between cancer-specific and multi-cancer detection. A list of the currently available tests, along with their targets, specificity, sensitivity, and source of cfDNA, can be found in Table 1, and their visual overview is shown in Figure 1.
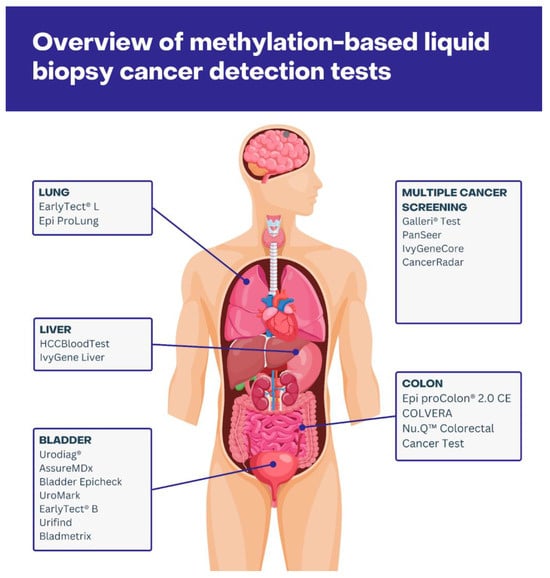
Figure 1.
Visual overview of methylation-based liquid biopsy tests focusing on multi-cancer detection or detection of neoplasm in particular organs.
3.2.1. Single-Cancer Detection Tests
Colorectal Cancer
The majority of commercially available methylation-based tests designed for CRC screening use stool samples as a source of DNA, such as Colovantage® [62] CologuardTM [62,63], and ColoSureTM [64], but only a few blood-based tests are currently available on the market.
The Epi proColon® 2.0 CE (Epigenomics AG, Berlin, Germany) test represents a second-generation screening tool designed to detect the methylation status of the gene Septin9 from plasma-derived cfDNA based on PCR. The test demonstrates an overall sensitivity in detecting CRC of 68.2% across all stages, alongside a specificity of 79.1% [65]. This test is accessible on the European market and in various other regions, including China [66]. Compared with the original 1.0 version of the test, the newest tool boasts enhanced sensitivity and specificity compared to its predecessor, which analyzed samples in duplicate rather than triplicate [67].
Another qPCR-based diagnostic test is COLVERA, designed to identify DNA methylation status in the BCAT1 and IKZF1 genes that exhibit hypermethylation in 95% of CRC tissue. COLVERA has demonstrated enhanced sensitivity for detecting recurrent disease compared to using carcinoembryonic antigen across various clinical populations [68]. The clinical performance of this test varies, as reported in a validation study by Murray et al., with sensitivity and specificity of 73.1% and 89.3%, respectively [69]. Another study was conducted on a cohort of 322 individuals, where the COLVERA test reached an overall sensitivity of 59.3% (95% CI: 38.8–77.6) and specificity of 98.3% (96.1–99.5) [68].
A different approach applies the Nu.Q™ Colorectal Cancer Test, aiming to reduce the number of unnecessary colonoscopies. The test was developed on the principle of global hypomethylation present in cfDNA of a CRC origin [70] and utilizes the ELISA method to analyze the methylation status of nucleosomes present in the serum, with sensitivity at 75% and with a 70% specificity [71,72].
Lung Cancer
EarlyTect® L is a validated epigenetic biomarker test centered on PCDHGA12 methylation status in serum, demonstrating reliable clinical sensitivity and specificity in detecting lung cancer. A study involving 522 participants has confirmed its capability to identify lung cancer, exhibiting a sensitivity of 77.8% and specificity of 92.3%. The test employs standard qPCR instrument systems to analyze epigenetic markers present in 2 mL serum samples [73,74].
Another test able to distinguish malignant lung disease is Epi ProLung. A qPCR method evaluates the DNA methylation levels of SHOX2, PTGER4, and the reference gene ACTB using plasma samples obtained from individuals both with and without malignant disease. The validation process yielded a sensitivity of 78% and a specificity of 96% [75].
Bladder Cancer
Urodiag® employs Mutated-Allele-Specific Oligonucleotide PCR (MASO-PCR) for the simultaneous amplification of SLIT2, SEPTIN9, and HS3ST2 methylated genes [76] for the surveillance of non-muscle-invasive bladder cancer. In a surveillance setting, the test exhibited a sensitivity of 94.5% and a specificity of 75.9% [77].
AssureMDx (MDxHealth) is another qPCR-based assay that integrates the genetic mutation status assessment of FGFR3, TERT, and HRAS with the examination of epigenetic patterns in OTX1, ONECUT2, and TWIST1 within exfoliated urinary cells. By employing a logistic regression analysis of these markers along with patient age, a diagnostic model demonstrated a sensitivity of 97% and specificity of 83% according to the initial findings [78].
The Bladder Epicheck (Nucleix) assay is a commercially available qPCR-based method designed to evaluate DNA methylation patterns in urinary exfoliated cells across 15 genomic regions. In the surveillance context, the test exhibited a sensitivity of 90% and a specificity of 83% [79], while in a larger-scale prospective study, it reached a sensitivity of only 68.2% with a specificity of 88% [79,80].
The UroMark assay presents another promising method for detecting bladder cancer through residual DNA in voided urine. This assay focuses on 150 CpG loci utilizing NGS bisulfite sequencing. It successfully differentiated primary bladder cancer from non-bladder cancer urine samples with a sensitivity and specificity of 98% and 97% [81].
EarlyTect® B is a qPCR-based method for analyzing multiple genomic and epigenomic targets in cfDNA extracted from urine samples [70,82] and is commercially available in East Asia.
Other available tests for bladder cancer screening utilizing exfoliating cells in urine are Urifind (Anchor Dx), providing a 91.7% sensitivity and 77.3% specificity [83], and Bladmetrix (SDU), with a 92.1% sensitivity and 93.3% specificity [84].
Liver Cancer
As imagining and biochemical markers remain substandard methods for the screening of hepatocellular carcinoma (HCC), an epigenetic-based blood test, HCCBloodTest (Epigenomics AG), has been developed, utilizing NGS bisulfite sequencing targeting the methylation status of the SEPT7 gene [85], achieving, in clinical study NCT03804593, a sensitivity and specificity of 76.7% and 64.1%, respectively.
Another test focusing specifically on hepatic malignancies is IvyGene Liver, with a sensitivity of 80% and a specificity of 86% [86].
It is worth mentioning that methylation-based noninvasive tests have also been developed for the detection of cervical cancer based on the methylation status of the ZNF582 gene, namely, Cervi-M® [87], as well as for head and neck cancer, namely, Oral-M®, with additional PAX1 gene methylation detection. [88,89]. Since both tests are swab-based, they do not qualify as liquid biopsy methods, and they are not further studied in this review, although the Cervi-M® test is discussed to possibly utilize tampon blood as a source of DNA, thus qualifying as a liquid biopsy [88,89].
3.2.2. Multi-Cancer Detection Tests
Lately, there has been a surge in research addressing the screening gap through the exploration of multi-cancer early detection (MCED) tests designed to generate cancer signal without offering a conclusive diagnosis [90]. Leveraging blood-based ctDNA for the simultaneous detection and localization of multiple cancer types may offer a solution to this significant unmet demand. Implementing such a multi-cancer detection strategy in large-scale population screening necessitates high specificity, clinically relevant sensitivity, and precise tissue of origin identification to manage the scale, expenses, and intricacies involved in evaluating asymptomatic patients [91,92].
Galleri® Test
The Galleri® test (GRAIL) is the first commercially available MCED able to detect more than 50 types of cancer [93]. The test utilizes a focused NGS bisulfite sequencing methylation assay, covering over 100,000 distinct regions and more than 1,000,000 cytosine–guanine dinucleotides coupled with machine learning techniques. The system can identify cancer signals across various cancer types and accurately predict the origin of these signals, known as Cancer Signal Origin (CSO) [94,95].
Its ability was investigated in the well-known PATHFINDER study (NCT04241796), a prospective clinical trial across seven US health networks that enrolled 6662 adults aged 50 or older without cancer symptoms for MCED testing from 12 December 2019 to 4 December 2020. Among the 6621 participants with analyzable results, 4204 (63.5%) were women and 2417 (36.5%) were men. A cancer signal was detected in 92 (1.4%) participants, with 35 (38%) receiving a cancer diagnosis (true positives) and 57 (62%) not diagnosed with cancer (false positives). Excluding two participants whose diagnostic assessments began before the MCED test results, the median time to diagnostic resolution was 79 days (IQR 37–219), with 57 days (33–143) in true-positive participants and 162 days (44–248) in false-positive participants. Most participants underwent both laboratory tests (26 [79%] of 33 true-positive participants and 50 [88%] of 57 false-positive participants) and imaging (30 [91%] of 33 true-positive participants and 53 [93%] of 57 false-positive participants) [96].
Another randomized controlled trial, called NHS-Galleri (ISRCTN91431511), is being conducted to evaluate the effectiveness of a blood test in detecting cancers early, potentially reducing the occurrence of late-stage cancers. Over 140,000 individuals from the general population of England in the age span of from 50 to 77 years who have neither been diagnosed with nor are under investigation for cancer have been enrolled in the NHS-Galleri trial. Blood samples are being collected up to three times: initially upon study enrollment and subsequently at 12- and 24-month intervals [97]. The results of the study are expected to be released in the spring or summer of 2024 [98]. To this day, only limited data from ongoing clinical trials are available. The revealed data project the positive predictive value (PPV) of this test to be below 10%, a level that may not adequately facilitate an effective screening initiative, along with modest sensitivities for early-stage cancer (17% for stage I, 40% for stage II, and 27.5% for stages I-II) [95]. A good overview of sensitivities and PPV in a setting of 99% specificity provides a reply to published data by Pons-Belda et al. [99].
PanSeer
The PanSeer test employs a comparable methodology to the Galleri test, encompassing 11,787 CpG sites spanning 595 genomic regions [94]. In the Taizhou Longitudinal Study (TZL), plasma samples from 123,115 initially healthy individuals were collected and stored for future analysis, with subsequent cancer monitoring. The preliminary findings from PanSeer, a noninvasive blood test utilizing ctDNA methylation, were evaluated using TZL plasma samples from 605 asymptomatic individuals, including 191 later diagnosed with various cancers within four years. PanSeer exhibited a detection rate of 88% (95% CI: 80–93%) in post-diagnosis patients with a specificity of 96% (95% CI: 93–98%) and 95% (95% CI: 89–98%) in asymptomatic individuals later diagnosed, warranting further longitudinal studies for confirmation [71,100].
IvyGene
IvyGene, developed by the Laboratory for Advanced Medicine, is a diagnostic test designed to assess the levels of four common cancers (breast, colon, liver, and lung) by analyzing the methylation status of cfDNA extracted from patients’ blood samples using a panel of 46 markers [101]. Positioned as a supplementary clinical tool, physicians can order this test to complement patient evaluations, and it is currently available in the USA as a direct-to-consumer service [102]. Currently, the test is offered as IvyGeneCORE, targeting four mentioned cancers, producing a sensitivity of 84% and specificity of 90% [86].
CancerRadar
A recently developed MCED test (not commercially available yet) is CancerRadar, which combines cfMethyl-Seq for genome-wide cfDNA methylation profiling, offering an over 12-fold enrichment compared to whole-genome bisulfite sequencing in CpG islands with a computational platform for extracting diverse methylation data and patient diagnosis [94,103]. In a study involving 408 colon, liver, lung, and stomach cancer patients and controls, a specificity of 97.9% and sensitivities of 85.9% for detecting all-stage cancer and 81.4% for early-stage (I and II) cancers have been achieved. Additionally, accuracies of 89.6% for identifying the tissue of origin of all-stage cancer and 84.4% for early-stage cancer were attained [94].

Table 1.
Overview of available methylation-based liquid biopsy tests analyzing cell-free DNA for cancer detection with their respective focus, specificity, sensitivity, and source of cfDNA.
Table 1.
Overview of available methylation-based liquid biopsy tests analyzing cell-free DNA for cancer detection with their respective focus, specificity, sensitivity, and source of cfDNA.
| Test | Cancer Type | Specificity | Sensitivity | Sample Type | Ref. |
|---|---|---|---|---|---|
| Epi proColon® 2.0 CE | Colorectal Cancer | 79.1% | 68.2% | Blood (plasma) | [65] |
| COLVERA | Colorectal Cancer | 89.3% | 73.1% | Blood (plasma) | [69] |
| Nu.Q™ Colorectal Cancer Test | Colorectal Cancer | 70% | 75% | Blood (serum) | [71,72] |
| EarlyTect® L | Lung Cancer | 92.3% | 77.8% | Blood (serum) | [73,74] |
| Epi ProLung | Lung Cancer | 96% | 78% | Blood (plasma) | [75] |
| Urodiag® | Bladder Cancer | 75.9% | 94.5% | Urine | [77] |
| AssureMDx | Bladder Cancer | 83% | 97% | Urine | [78] |
| Bladder Epicheck | Bladder Cancer | 83% | 90% | Urine | [79] |
| UroMark | Bladder Cancer | 97% | 98% | Urine | [81] |
| EarlyTect® B | Bladder Cancer | N/A | N/A | Urine | [70,82] |
| Urifind | Bladder Cancer | 77.3% | 91.7% | Urine | [83] |
| Bladmetrix | Bladder Cancer | 93.3% | 92.1% | Urine | [84] |
| HCCBloodTest | Liver Cancer | 64.1% | 76.7% | Blood | [85] |
| IvyGene Liver | Liver Cancer | 86% | 80% | Blood | [86] |
| Galleri® Test | Multi-cancer Early Detection | N/A * | N/A * | Blood | [96] |
| PanSeer | Multi-cancer Early Detection | 96% | 88% | Blood | [71,100] |
| IvyGeneCORE | Breast, Colon, Liver, and Lung Cancer | 90% | 84% | Blood | [86] |
| CancerRadar | Multi-cancer Early Detection | 97.9% | 85.9% | Blood | [94] |
* Specificity and sensitivity vary among cancer types and stages, as discussed in Section 3.2.
3.2.3. Methylation-Based Methods in Companion Diagnostics
The variability in pharmacotherapy can be substantial, often attributed to the heterogeneity of cancer, posing the necessity to develop predictive assays based on specific biomarkers to guide targeted therapy use, known as companion diagnostics [104]. Epigenomics holds significant potential in predicting treatment response and cancer susceptibility. An exemplary illustration of the practical application of DNA methylation in personalized medicine is demonstrated through the utilization of MGMT promoter hypermethylation. This method effectively categorizes the glioblastoma patients in clinical trials and aids in determining their response to alkylating agents. MGMT, a pivotal DNA repair enzyme, plays a crucial role in eliminating specific and abnormal DNA adducts [105]. As this method is not liquid-biopsy-based and invasive biopsy is necessary, its broad clinical application remains challenging [106].
For breast cancer methylation-based companion diagnostics, interesting research was conducted by Mastoraki et al., assessing the ESR1 gene methylation status as a predictive biomarker, as gene methylation was correlated with non-responsiveness to the everolimus/exemestane treatment. Further investigation into ESR1 methylation is warranted to assess its potential as a liquid-biopsy-based biomarker [107].
In brain tumors, the methylation status of MGMT-STP27 was shown as a promising predictive biomarker to assess the response to a PCV chemotherapy regimen (procarbazine, CCNU, and vincristine) in anaplastic oligodendrogliomas and oligoastrocytomas [106,108,109]. For the purpose of decision-making, PyroMark Therascreen MGMT and the PyroMark Q96 CpG MGMT kits were developed [110]. Great promise for liquid-biopsy-based diagnostics in brain tumors lies in the utilization of cerebrospinal fluid, as discussed in a review by Eibl and Schneemann [111].
4. Biological Factors Influencing DNA Methylation
Apart from the technology-related limitations mentioned in Section 3.1, there are also biological and physiological factors influencing the methylation of somatic cells, which must be counted on, especially when assessing global methylation status for cancer diagnostics. Inter- and intra-tumor heterogeneity exists due to variability in the morphology, genetics, epigenetics, and phenotype of cell populations. Comprehending the variations in phenotypes requires knowledge of variations in DNA methylation variability amongst cell groups [112,113,114]. Heterogeneity within profiled cell populations is often ignored in research because they only look at average DNA methylation levels of individual CpGs. Liquid biopsies are more useful in such a situation, as tissue biopsies can capture only part of this heterogeneity [115].
The potential for false-positive or false-negative results due to methylation changes arises with aging, inflammation, or other non-cancerous conditions. Other physiological characteristics that may arise in particular clinical circumstances, such as a cardiac infarct, may also modify the epigenetic and biological aspects of cfDNA.
Individual differences in DNA methylation patterns can result from both hereditary and environmental influences. Variations in DNA segments or genes are associated with approximately 20% of the interindividual variation in DNA methylation patterns [15,16]. Dietary components that affect DNA methylation include folate absorption [116]. Age is another interesting factor of methylation, with studies referring to changing patterns in the methylation of tissues over time as a methylation clock successfully determining age [117]. Gender is also significantly correlated with tissue-specific DNA methylation levels [118,119], which is particularly important in autoimmune disorders with a strong sex bias. For example, X-chromosome DNA methylation variations determined in the manner of the original parent may account for the higher frequency of autoimmune disorders in women than in men [120].
Metabolic disorders, such as diabetes mellitus [121] and obesity [121,122], are associated with differential methylation phenotypes compared to healthy individuals.
5. Conclusions
While the utilization of cfDNA methylation analysis in cancer diagnostics holds significant promise, it is currently hindered by various biological limitations and technical challenges. Circulating DNA originating from rare tumor subpopulations might only be detectable in ctDNA at extremely low concentrations, posing challenges along with inter- and intra-tumor heterogeneity and the stage of its development, along with the factors of age, gender, environmental influences, and biological activities in normal cells introducing variability in methylation patterns, leading to misdiagnosis. In clinical practice, currently, there is a limited market of epigenetic-based liquid biopsy tests screening for the global presence of cancer in the body, and more clinical studies and improved performance for early-stage cancers must be achieved. This review discussed the most common techniques for methylation detection in cfDNA, their limitations, and their suitability for specific clinical use. For diagnostic tests focusing on methylation detection on a single or limited number of sites on the genome related to a particular cancer type, PCR remains the most robust, economical, and sensitive method. On the other hand, MCED tests, which are set to target multiple cancers or general cancer signals, have to utilize genome-wide methylation sequencing or array-based methods, coming with challenges associated with reproducibility and higher costs. From a cancer-type perspective, the biggest number of tests were developed for bladder cancer, despite being only the 10th most common cancer worldwide. For this cancer, no screening tests for the population at average risk have been developed to this day. Following these are the tests for colorectal cancer, where methylation-based liquid biopsy has to compete with already established and relatively inexpensive fecal occult blood tests (FOBTs). Following these are tests for lung cancer, a disease with the highest mortality and missing efficient and noninvasive screening tests. Cancer-specific liquid biopsy test limitations remain in terms of poor performance, a lack of clinical and post-market data, and education of clinicians in the possibility of their adoption, especially for patients refusing standard diagnostic methods. The use of MCED tests is usually limited to a certain number of cancers. Alternatively, they produce a tissue-specific or non-specific cancer signal, which can navigate healthcare providers for further testing. Their use for screening on the general public remains controversial, raising questions about the benefits, harms, and high costs involved, as seen from already ongoing trials. The standardization of pre-analytical techniques, improved assay methodologies, and an enhanced understanding of the biological mechanisms underlying methylation patterns are essential steps toward overcoming these challenges and realizing the full clinical potential of ctDNA methylation analysis in oncology, encouraging further basic as well as clinical research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers16112001/s1, Table S1: Overview of methylation detection methods in cfDNA listed with their characteristic, major advantages and disadvantages in practical use.
Author Contributions
Conceptualization, T.R. and T.D.; methodology, O.P.; investigation, R.S., T.D. and T.R.; writing—original draft preparation, T.R.; writing—review and editing, O.P. and J.B.; visualization, R.S.; supervision, T.S.; project administration, V.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by project 101160008 (FORGENOM II) funded by the Horizon Europe program, and support was also provided by the Slovak Research and Development Agency grant APVV-21-0296 (INCAM).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the Supplementary Material. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
Authors O.P., J.B., and T.S. are employees of Geneton Ltd. and are involved in numerous research and development efforts for adapting new technologies to better understand genomic data and facilitate their implementation in patient care. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Dagenais, G.R.; Leong, D.P.; Rangarajan, S.; Lanas, F.; Lopez-Jaramillo, P.; Gupta, R.; Diaz, R.; Avezum, A.; Oliveira, G.B.F.; Wielgosz, A.; et al. Variations in Common Diseases, Hospital Admissions, and Deaths in Middle-Aged Adults in 21 Countries from Five Continents (PURE): A Prospective Cohort Study. Lancet 2020, 395, 785–794. [Google Scholar] [CrossRef]
- Parkinson, D.R.; McCormack, R.T.; Keating, S.M.; Gutman, S.I.; Hamilton, S.R.; Mansfield, E.A.; Piper, M.A.; DeVerka, P.; Frueh, F.W.; Jessup, J.M.; et al. Evidence of Clinical Utility: An Unmet Need in Molecular Diagnostics for Patients with Cancer. Clin. Cancer Res. 2014, 20, 1428–1444. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, R.C.; Antoniou, A.C.; Fruk, L.; Rosenfeld, N. The Future of Early Cancer Detection. Nat. Med. 2022, 28, 666–677. [Google Scholar] [CrossRef]
- Clarke, C.A.; Hubbell, E.; Kurian, A.W.; Colditz, G.A.; Hartman, A.-R.; Gomez, S.L. Projected Reductions in Absolute Cancer-Related Deaths from Diagnosing Cancers Before Metastasis, 2006–2015. Cancer Epidemiol. Biomark. Prev. 2020, 29, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Wang, X.; Jin, P. Developing DNA Methylation-Based Diagnostic Biomarkers. J. Genet. Genom. 2018, 45, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in Cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Vogelstein, B. Hypomethylation Distinguishes Genes of Some Human Cancers from Their Normal Counterparts. Nature 1983, 301, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Reik, W.; Surani, A. Genomic Imprinting; IRL Press: Oxford, UK, 1997; ISBN 9780199636259. [Google Scholar]
- Xie, W.; Barr, C.L.; Kim, A.; Yue, F.; Lee, A.Y.; Eubanks, J.; Dempster, E.L.; Ren, B. Base-Resolution Analyses of Sequence and Parent-of-Origin Dependent DNA Methylation in the Mouse Genome. Cell 2012, 148, 816–831. [Google Scholar] [CrossRef]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A Genome-Wide Analysis of CpG Dinucleotides in the Human Genome Distinguishes Two Distinct Classes of Promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef]
- DNA Methylation and Cancer. Advances in Genetics; Academic Press: Cambridge, MA, USA, 2010; Volume 70, pp. 27–56. [Google Scholar]
- Insua, Y.V.; De la Cámara, J.; Vázquez, E.B.; Fernández, A.; Rivera, F.V.; Silva, M.J.V.; Barbazán, J.; Muinelo-Romay, L.; Folgar, S.C.; Abalo, A.; et al. Predicting Outcome and Therapy Response in mCRC Patients Using an Indirect Method for CTCs Detection by a Multigene Expression Panel: A Multicentric Prospective Validation Study. Int. J. Mol. Sci. 2017, 18, 1265. [Google Scholar] [CrossRef] [PubMed]
- García-Saenz, J.A.; Ayllón, P.; Laig, M.; Acosta-Eyzaguirre, D.; García-Esquinas, M.; Montes, M.; Sanz, J.; Barquín, M.; Moreno, F.; Garcia-Barberan, V.; et al. Tumor Burden Monitoring Using Cell-Free Tumor DNA Could Be Limited by Tumor Heterogeneity in Advanced Breast Cancer and Should Be Evaluated Together with Radiographic Imaging. BMC Cancer 2017, 17, 210. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.V.; Turner, S.T.; Smith, J.A.; Hammond, P.I.; Lazarus, A.; Van De Rostyne, J.L.; Cunningham, J.M.; Kardia, S.L.R. Comparison of the DNA Methylation Profiles of Human Peripheral Blood Cells and Transformed B-Lymphocytes. Hum. Genet. 2010, 127, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Reinius, L.E.; Acevedo, N.; Joerink, M.; Pershagen, G.; Dahlén, S.-E.; Greco, D.; Söderhäll, C.; Scheynius, A.; Kere, J. Differential DNA Methylation in Purified Human Blood Cells: Implications for Cell Lineage and Studies on Disease Susceptibility. PLoS ONE 2012, 7, e41361. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Epigenetics in Cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.T.; Brandes, J.C.; Vertino, P.M. Cancer DNA Methylation: Molecular Mechanisms and Clinical Implications. Clin. Cancer Res. 2009, 15, 3927–3937. [Google Scholar] [CrossRef] [PubMed]
- Opavsky, R.; Wang, S.-H.; Trikha, P.; Raval, A.; Huang, Y.; Wu, Y.-Z.; Rodriguez, B.; Keller, B.; Liyanarachchi, S.; Wei, G.; et al. CpG Island Methylation in a Mouse Model of Lymphoma Is Driven by the Genetic Configuration of Tumor Cells. PLoS Genet. 2007, 3, 1757–1769. [Google Scholar] [CrossRef] [PubMed]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Kin Sung, K.W.; Rigoutsos, I.; Loring, J.; et al. Dynamic Changes in the Human Methylome during Differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef]
- Zane, L.; Sharma, V.; Misteli, T. Common Features of Chromatin in Aging and Cancer: Cause or Coincidence? Trends Cell Biol. 2014, 24, 686–694. [Google Scholar] [CrossRef]
- Miranda, T.B.; Jones, P.A. DNA Methylation: The Nuts and Bolts of Repression. J. Cell. Physiol. 2007, 213, 384–390. [Google Scholar] [CrossRef]
- Costello, J.F.; Frühwald, M.C.; Smiraglia, D.J.; Rush, L.J.; Robertson, G.P.; Gao, X.; Wright, F.A.; Feramisco, J.D.; Peltomäki, P.; Lang, J.C.; et al. Aberrant CpG-Island Methylation Has Non-Random and Tumour-Type-Specific Patterns. Nat. Genet. 2000, 24, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-C.; Giannopoulou, E.G.; Park, K.; Mosquera, J.M.; Sboner, A.; Tewari, A.K.; Garraway, L.A.; Beltran, H.; Rubin, M.A.; Elemento, O. Epigenomic Alterations in Localized and Advanced Prostate Cancer. Neoplasia 2013, 15, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Jerónimo, C.; Bastian, P.J.; Bjartell, A.; Carbone, G.M.; Catto, J.W.F.; Clark, S.J.; Henrique, R.; Nelson, W.G.; Shariat, S.F. Epigenetics in Prostate Cancer: Biologic and Clinical Relevance. Eur. Urol. 2011, 60, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Beggs, A.D.; Jones, A.; El-Bahrawy, M.; Abulafi, M.; Hodgson, S.V.; Tomlinson, I.P.M. Whole-Genome Methylation Analysis of Benign and Malignant Colorectal Tumours. J. Pathol. 2013, 229, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Salta, S.; Nunes, S.P.; Fontes-Sousa, M.; Lopes, P.; Freitas, M.; Caldas, M.; Antunes, L.; Castro, F.; Antunes, P.; Palma de Sousa, S.; et al. A DNA Methylation-Based Test for Breast Cancer Detection in Circulating Cell-Free DNA. J. Clin. Med. Res. 2018, 7, 420. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.; Hauge, T.; Jeanmougin, M.; Pharo, H.D.; Kresse, S.H.; Honne, H.; Winge, S.B.; Five, M.-B.; Kumar, T.; Mala, T.; et al. Targeted Genetic and Epigenetic Profiling of Esophageal Adenocarcinomas and Non-Dysplastic Barrett’s Esophagus. Clin. Epigenetics 2022, 14, 77. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Dang, S.; Hou, P. Gene Methylation in Gastric Cancer. Clin. Chim. Acta 2013, 424, 53–65. [Google Scholar] [CrossRef]
- Hoang, P.H.; Landi, M.T. DNA Methylation in Lung Cancer: Mechanisms and Associations with Histological Subtypes, Molecular Alterations, and Major Epidemiological Factors. Cancers 2022, 14, 961. [Google Scholar] [CrossRef]
- Rademakers, G.; Massen, M.; Koch, A.; Draht, M.X.; Buekers, N.; Wouters, K.A.D.; Vaes, N.; De Meyer, T.; Carvalho, B.; Meijer, G.A.; et al. Identification of DNA Methylation Markers for Early Detection of CRC Indicates a Role for Nervous System-Related Genes in CRC. Clin. Epigenetics 2021, 13, 80. [Google Scholar] [CrossRef]
- Kisiel, J.B.; Dukek, B.A.; Kanipakam, R.V.S.R.; Ghoz, H.M.; Yab, T.C.; Berger, C.K.; Taylor, W.R.; Foote, P.H.; Giama, N.H.; Onyirioha, K.; et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology 2019, 69, 1180–1192. [Google Scholar] [CrossRef]
- Stewart, C.M.; Tsui, D.W.Y. Circulating Cell-Free DNA for Non-Invasive Cancer Management. Cancer Genet. 2018, 228–229, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Delgado, P.O.; Alves, B.C.A.; Gehrke, F.d.S.; Kuniyoshi, R.K.; Wroclavski, M.L.; Del Giglio, A.; Fonseca, F.L.A. Characterization of Cell-Free Circulating DNA in Plasma in Patients with Prostate Cancer. Tumour Biol. 2013, 34, 983–986. [Google Scholar] [CrossRef] [PubMed]
- Frattini, M.; Gallino, G.; Signoroni, S.; Balestra, D.; Battaglia, L.; Sozzi, G.; Leo, E.; Pilotti, S.; Pierotti, M.A. Quantitative Analysis of Plasma DNA in Colorectal Cancer Patients: A Novel Prognostic Tool. Ann. N. Y. Acad. Sci. 2006, 1075, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Marsavela, G.; McEvoy, A.C.; Pereira, M.R.; Reid, A.L.; Al-Ogaili, Z.; Warburton, L.; Khattak, M.A.; Abed, A.; Meniawy, T.M.; Millward, M.; et al. Detection of Clinical Progression through Plasma ctDNA in Metastatic Melanoma Patients: A Comparison to Radiological Progression. Br. J. Cancer 2022, 126, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Roadmap Epigenomics Consortium; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative Analysis of 111 Reference Human Epigenomes. Nature 2015, 518, 317–330. [Google Scholar]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid Biopsy: A Step Closer to Transform Diagnosis, Prognosis and Future of Cancer Treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, A.; Bartlett, J.; Cheng, Y.; Pasic, M.D.; Yousef, G.M. Liquid Biopsy: A Step Forward towards Precision Medicine in Urologic Malignancies. Mol. Cancer 2017, 16, 80. [Google Scholar] [CrossRef] [PubMed]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef] [PubMed]
- Shames, D.S.; Girard, L.; Gao, B.; Sato, M.; Lewis, C.M.; Shivapurkar, N.; Jiang, A.; Perou, C.M.; Kim, Y.H.; Pollack, J.R.; et al. A Genome-Wide Screen for Promoter Methylation in Lung Cancer Identifies Novel Methylation Markers for Multiple Malignancies. PLoS Med. 2006, 3, e486. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Jensen, T.; Oshiro, M.M.; Watts, G.S.; Kim, C.J.; Futscher, B.W. Agglomerative Epigenetic Aberrations Are a Common Event in Human Breast Cancer. Cancer Res. 2008, 68, 8616–8625. [Google Scholar] [CrossRef]
- Heeke, S.; Gay, C.M.; Estecio, M.R.; Tran, H.; Morris, B.B.; Zhang, B.; Tang, X.; Raso, M.G.; Rocha, P.; Lai, S.; et al. Tumor- and Circulating-Free DNA Methylation Identifies Clinically Relevant Small Cell Lung Cancer Subtypes. Cancer Cell 2024, 42, 225–237.e5. [Google Scholar] [CrossRef]
- Pham, T.M.Q.; Phan, T.H.; Jasmine, T.X.; Tran, T.T.T.; Huynh, L.A.K.; Vo, T.L.; Nai, T.H.T.; Tran, T.T.; Truong, M.H.; Tran, N.C.; et al. Multimodal Analysis of Genome-Wide Methylation, Copy Number Aberrations, and End Motif Signatures Enhances Detection of Early-Stage Breast Cancer. Front. Oncol. 2023, 13, 1127086. [Google Scholar] [CrossRef]
- Brenne, S.S.; Madsen, P.H.; Pedersen, I.S.; Hveem, K.; Skorpen, F.; Krarup, H.B.; Giskeødegård, G.F.; Laugsand, E.A. Colorectal Cancer Detected by Liquid Biopsy 2 Years prior to Clinical Diagnosis in the HUNT Study. Br. J. Cancer 2023, 129, 861–868. [Google Scholar] [CrossRef]
- Dillinger, T.; Sheibani-Tezerji, R.; Pulverer, W.; Stelzer, I.; Hassler, M.R.; Scheibelreiter, J.; Pérez Malla, C.U.; Kuroll, M.; Domazet, S.; Redl, E.; et al. Identification of Tumor Tissue-Derived DNA Methylation Biomarkers for the Detection and Therapy Response Evaluation of Metastatic Castration Resistant Prostate Cancer in Liquid Biopsies. Mol. Cancer 2022, 21, 7. [Google Scholar] [CrossRef]
- Cheng, F.; Su, L.; Qian, C. Circulating Tumor DNA: A Promising Biomarker in the Liquid Biopsy of Cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef]
- Pajares, M.J.; Palanca-Ballester, C.; Urtasun, R.; Alemany-Cosme, E.; Lahoz, A.; Sandoval, J. Methods for Analysis of Specific DNA Methylation Status. Methods 2021, 187, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Martisova, A.; Holcakova, J.; Izadi, N.; Sebuyoya, R.; Hrstka, R.; Bartosik, M. DNA Methylation in Solid Tumors: Functions and Methods of Detection. Int. J. Mol. Sci. 2021, 22, 4247. [Google Scholar] [CrossRef]
- Perry, N.; Wasko, K.; Cheng, J.; Tabbaa, D.; Marco, E.; Giannoukos, G.; Albright, C.F.; Borges, C.M. Methylation-Sensitive Restriction Enzyme Quantitative Polymerase Chain Reaction Enables Rapid, Accurate, and Precise Detection of Methylation Status of the Regulatory T Cell (Treg)-Specific Demethylation Region in Primary Human Tregs. J. Immunol. 2021, 206, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A Genomic Sequencing Protocol That Yields a Positive Display of 5-Methylcytosine Residues in Individual DNA Strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Mikeska, T.; Krypuy, M.; Dobrovic, A. Sensitive Melting Analysis after Real Time- Methylation Specific PCR (SMART-MSP): High-Throughput and Probe-Free Quantitative DNA Methylation Detection. Nucleic Acids Res. 2008, 36, e42. [Google Scholar] [CrossRef] [PubMed]
- Sigalotti, L.; Covre, A.; Colizzi, F.; Fratta, E. Quantitative Methylation-Specific PCR: A Simple Method for Studying Epigenetic Modifications of Cell-Free DNA. Methods Mol. Biol. 2019, 1909, 137–162. [Google Scholar]
- Esteller, M. The Necessity of a Human Epigenome Project. Carcinogenesis 2006, 27, 1121–1125. [Google Scholar] [CrossRef]
- Boscolo Bielo, L.; Trapani, D.; Repetto, M.; Crimini, E.; Valenza, C.; Belli, C.; Criscitiello, C.; Marra, A.; Subbiah, V.; Curigliano, G. Variant Allele Frequency: A Decision-Making Tool in Precision Oncology? Trends Cancer Res. 2023, 9, 1058–1068. [Google Scholar] [CrossRef]
- Leitão, E.; Beygo, J.; Zeschnigk, M.; Klein-Hitpass, L.; Bargull, M.; Rahmann, S.; Horsthemke, B. Locus-Specific DNA Methylation Analysis by Targeted Deep Bisulfite Sequencing. Methods Mol. Biol. 2018, 1767, 351–366. [Google Scholar] [PubMed]
- Söderhäll, C.; Reinius, L.E.; Salmenperä, P.; Gentile, M.; Acevedo, N.; Konradsen, J.R.; Nordlund, B.; Hedlin, G.; Scheynius, A.; Myllykangas, S.; et al. High-Resolution Targeted Bisulfite Sequencing Reveals Blood Cell Type-Specific DNA Methylation Patterns in IL13 and ORMDL3. Clin. Epigenetics 2021, 13, 106. [Google Scholar] [CrossRef]
- Schenk, D.; Song, G.; Ke, Y.; Wang, Z. Amplification of Overlapping DNA Amplicons in a Single-Tube Multiplex PCR for Targeted next-Generation Sequencing of BRCA1 and BRCA2. PLoS ONE 2017, 12, e0181062. [Google Scholar] [CrossRef]
- Zhu, G.; Guo, Y.A.; Ho, D.; Poon, P.; Poh, Z.W.; Wong, P.M.; Gan, A.; Chang, M.M.; Kleftogiannis, D.; Lau, Y.T.; et al. Tissue-Specific Cell-Free DNA Degradation Quantifies Circulating Tumor DNA Burden. Nat. Commun. 2021, 12, 2229. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, C.M.; Borsu, L.; Cankovic, M.; Earle, J.S.L.; Gocke, C.D.; Hameed, M.; Jordan, D.; Lopategui, J.R.; Pullambhatla, M.; Reuther, J.; et al. Recommendations for Cell-Free DNA Assay Validations: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2023, 25, 876–897. [Google Scholar] [CrossRef]
- Chen, Z.; Li, C.; Zhou, Y.; Yao, Y.; Liu, J.; Wu, M.; Su, J. Liquid Biopsies for Cancer: From Bench to Clinic. MedComm (2020) 2023, 4, e329. [Google Scholar] [CrossRef]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 Methylated DNA Is a Sensitive and Specific Blood Test for Colorectal Cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef]
- Onieva-García, M.Á.; Llanos-Méndez, A.; Baños-Álvarez, E.; Isabel-Gómez, R. A Systematic Review of the Clinical Validity of the CologuardTM Genetic Test for Screening Colorectal Cancer. Rev. Clin. Esp. 2015, 215, 527–536.
- Ned, R.M.; Melillo, S.; Marrone, M. Fecal DNA Testing for Colorectal Cancer Screening: The ColoSureTM Test. PLoS Curr. 2011, 3, RRN1220. [Google Scholar] [CrossRef] [PubMed]
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a Real-Time PCR-Based Qualitative Assay for the Detection of Methylated SEPT9 DNA in Human Plasma. Clin. Chem. 2014, 60, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Lamb, Y.N.; Dhillon, S. Epi proColon® 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol. Diagn. Ther. 2017, 21, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Kang, Q.; Wang, X.; Yang, L.; Yu, Y.; Li, N.; He, Y.-Q.; Han, X.; Hang, J.; Zhang, J.; et al. Performance of a Second-Generation Methylated SEPT9 Test in Detecting Colorectal Neoplasm. J. Gastroenterol. Hepatol. 2015, 30, 830–833. [Google Scholar] [CrossRef]
- Vucetic, Z.; Loayza, N.; Pedersen, S.K.; Tuck, M.; LaPointe, L.C. Clinical Performance of Methylation-Based Liquid Biopsy Test COLVERA after Optimization of Test Interpretation Rules. J. Clin. Oncol. 2021, 39, 3546. [Google Scholar] [CrossRef]
- Murray, D.H.; Baker, R.T.; Gaur, S.; Young, G.P.; Pedersen, S.K. Validation of a Circulating Tumor-Derived DNA Blood Test for Detection of Methylated BCAT1 and IKZF1 DNA. J. Appl. Lab. Med. 2017, 2, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Ladd-Acosta, C.; Wen, B.; Wu, Z.; Montano, C.; Onyango, P.; Cui, H.; Gabo, K.; Rongione, M.; Webster, M.; et al. The Human Colon Cancer Methylome Shows Similar Hypo- and Hypermethylation at Conserved Tissue-Specific CpG Island Shores. Nat. Genet. 2009, 41, 178–186. [Google Scholar] [CrossRef]
- Holdenrieder, S.; Dharuman, Y.; Standop, J.; Trimpop, N.; Herzog, M.; Hettwer, K.; Simon, K.; Uhlig, S.; Micallef, J. Novel Serum Nucleosomics Biomarkers for the Detection of Colorectal Cancer. Anticancer. Res. 2014, 34, 2357–2362. [Google Scholar]
- Herzog, M.; Eccleston, M.; Micallef, J.; Pamart, D.; Cuvelier, B.; Josseaux, E.; Terrell, J.; Christensen, I.; Nielsen, H. Validation of Nu.QTM Colorectal Cancer Screening Triage Test to Identify FIT Positive Individuals at Low Risk of Screen Relevant Neoplasia. Ann. Oncol. 2017, 28 (Suppl. S3), iii146. [Google Scholar] [CrossRef]
- Jeong, I.B.; Yoon, Y.S.; Park, S.Y.; Cha, E.J.; Na, M.J.; Kwon, S.J.; Kim, J.H.; Oh, T.J.; An, S.; Park, C.R.; et al. Methylation Biomarker in Bronchial Washing Specimens as an Adjunctive Diagnostic Tool to Bronchoscopy in Lung Cancer. Oncol. Lett. 2018, 16, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- EarlyTect® Lung Cancer. Available online: https://genomictree.com/eng/products/earlytect-lung-cancer/ (accessed on 25 March 2024).
- Weiss, G.; Schlegel, A.; Kottwitz, D.; König, T.; Tetzner, R. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J. Thorac. Oncol. 2017, 12, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Roperch, J.-P.; Hennion, C. A Novel Ultra-Sensitive Method for the Detection of FGFR3 Mutations in Urine of Bladder Cancer Patients—Design of the Urodiag® PCR Kit for Surveillance of Patients with Non-Muscle-Invasive Bladder Cancer (NMIBC). BMC Med. Genet. 2020, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Roperch, J.P.; Grandchamp, B.; Desgrandchamps, F. Promoter hypermethylation of HS3ST2, SEPTIN9 and SLIT2 combined with FGFR3 mutations as a sensitive/specific urinary assay for diagnosis and surveillance in patients with low or high-risk non-muscle-invasive bladder cancer. BMC Cancer 2016, 16, 704. [Google Scholar] [CrossRef]
- van Kessel, K.E.M.; Van Neste, L.; Lurkin, I.; Zwarthoff, E.C.; Van Criekinge, W. Evaluation of an Epigenetic Profile for the Detection of Bladder Cancer in Patients with Hematuria. J. Urol. 2016, 195, 601–607. [Google Scholar] [CrossRef]
- Wasserstrom, A.; Frumkin, D.; Dotan, Z.; Bukin, E.; Gadish, T.; Hanuka, S.; Knirsh, R.; Darawsha, A.E.; Leibovitch, I. Mp13-15 Molecular Urine Cytology—Bladder Epicheck Is a Novel Molecular Diagnostic Tool for Monitoring of Bladder Cancer Patients. J. Urol. 2016, 195, e140. [Google Scholar] [CrossRef][Green Version]
- Witjes, J.A.; Morote, J.; Cornel, E.B.; Gakis, G.; van Valenberg, F.J.P.; Lozano, F.; Sternberg, I.A.; Willemsen, E.; Hegemann, M.L.; Paitan, Y.; et al. Performance of the Bladder EpiCheckTM Methylation Test for Patients Under Surveillance for Non–muscle-Invasive Bladder Cancer: Results of a Multicenter, Prospective, Blinded Clinical Trial. Eur. Urol. Oncol. 2018, 1, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Feber, A.; Dhami, P.; Dong, L.; de Winter, P.; Tan, W.S.; Martínez-Fernández, M.; Paul, D.S.; Hynes-Allen, A.; Rezaee, S.; Gurung, P.; et al. UroMark-a Urinary Biomarker Assay for the Detection of Bladder Cancer. Clin. Epigenetics 2017, 9, 8. [Google Scholar] [CrossRef]
- EarlyTect® Bladder Cancer. Available online: https://genomictree.com/eng/products/earlytect-bladder-cancer/ (accessed on 26 March 2024).
- Chen, X.; Zhang, J.; Ruan, W.; Huang, M.; Wang, C.; Wang, H.; Jiang, Z.; Wang, S.; Liu, Z.; Liu, C.; et al. Urine DNA Methylation Assay Enables Early Detection and Recurrence Monitoring for Bladder Cancer. J. Clin. Invest. 2020, 130, 6278–6289. [Google Scholar] [CrossRef]
- Pharo, H.D.; Jeanmougin, M.; Ager-Wick, E.; Vedeld, H.M.; Sørbø, A.K.; Dahl, C.; Larsen, L.K.; Honne, H.; Brandt-Winge, S.; Five, M.-B.; et al. BladMetrix: A Novel Urine DNA Methylation Test with High Accuracy for Detection of Bladder Cancer in Hematuria Patients. Clin. Epigenetics 2022, 14, 115. [Google Scholar] [CrossRef]
- Lewin, J.; Kottwitz, D.; Aoyama, J.; deVos, T.; Garces, J.; Hasinger, O.; Kasielke, S.; Knaust, F.; Rathi, P.; Rausch, S.; et al. Plasma Cell Free DNA Methylation Markers for Hepatocellular Carcinoma Surveillance in Patients with Cirrhosis: A Case Control Study. BMC Gastroenterol. 2021, 21, 136. [Google Scholar] [CrossRef] [PubMed]
- The IvyGene Liver Test—IvyGene, Non-Invasive Early Cancer Detection Test. Available online: https://www.ivygenelabs.co.za/the-ivygene-liver-test/ (accessed on 11 March 2024).
- Lin, H.; Chen, T.-C.; Chang, T.-C.; Cheng, Y.-M.; Chen, C.-H.; Chu, T.-Y.; Hsu, S.-T.; Liu, C.-B.; Yeh, L.-S.; Wen, K.-C.; et al. Methylated ZNF582 Gene as a Marker for Triage of Women with Pap Smear Reporting Low-Grade Squamous Intraepithelial Lesions—A Taiwanese Gynecologic Oncology Group (TGOG) Study. Gynecol. Oncol. 2014, 135, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Bakkum-Gamez, J.N.; Wentzensen, N.; Maurer, M.J.; Hawthorne, K.M.; Voss, J.S.; Kroneman, T.N.; Famuyide, A.O.; Clayton, A.C.; Halling, K.C.; Kerr, S.E.; et al. Detection of Endometrial Cancer via Molecular Analysis of DNA Collected with Vaginal Tampons. Gynecol. Oncol. 2015, 137, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-García, J.; Osca-Verdegal, R.; Mena-Mollá, S.; García-Giménez, J.L. Epigenetic IVD Tests for Personalized Precision Medicine in Cancer. Front. Genet. 2019, 10, 621. [Google Scholar] [CrossRef] [PubMed]
- Loomans-Kropp, H.A.; Umar, A.; Minasian, L.M.; Pinsky, P.F. Multi-Cancer Early Detection Tests: Current Progress and Future Perspectives. Cancer Epidemiol. Biomark. Prev. 2022, 31, 512–514. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; CCGA Consortium. Sensitive and Specific Multi-Cancer Detection and Localization Using Methylation Signatures in Cell-Free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Oxnard, G.R.; Klein, E.A.; Seiden, M.V.; Hubbell, E.; Venn, O.; Jamshidi, A.; Zhang, N.; Beausang, J.F.; Gross, S.; Kurtzman, K.N.; et al. Simultaneous Multi-Cancer Detection and Tissue of Origin (TOO) Localization Using Targeted Bisulfite Sequencing of Plasma Cell-Free DNA (cfDNA). Ann. Oncol. 2019, 30, v912. [Google Scholar] [CrossRef]
- Blood Test for Cancer Screening. Available online: https://www.galleri.com/ (accessed on 25 March 2024).
- Hackshaw, A.; Clarke, C.A.; Hartman, A.-R. New Genomic Technologies for Multi-Cancer Early Detection: Rethinking the Scope of Cancer Screening. Cancer Cell 2022, 40, 109–113. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical Validation of a Targeted Methylation-Based Multi-Cancer Early Detection Test Using an Independent Validation Set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Schrag, D.; Beer, T.M.; McDonnell, C.H., 3rd; Nadauld, L.; Dilaveri, C.A.; Reid, R.; Marinac, C.R.; Chung, K.C.; Lopatin, M.; Fung, E.T.; et al. Blood-Based Tests for Multicancer Early Detection (PATHFINDER): A Prospective Cohort Study. Lancet 2023, 402, 1251–1260. [Google Scholar] [CrossRef]
- Neal, R.D.; Johnson, P.; Clarke, C.A.; Hamilton, S.A.; Zhang, N.; Kumar, H.; Swanton, C.; Sasieni, P. Cell-Free DNA-Based Multi-Cancer Early Detection Test in an Asymptomatic Screening Population (NHS-Galleri): Design of a Pragmatic, Prospective Randomised Controlled Trial. Cancers 2022, 14, 4818. [Google Scholar] [CrossRef]
- Al-Obeidee, M.; Al-Obeidee, E. Exploring the Potential of Multi-Cancer Early Detection Tests as Triage Tools in Urgent Referrals: Insights from Recent Clinical Trial. Postgrad. Med. J. 2024, 140, qgae033. [Google Scholar] [CrossRef] [PubMed]
- Pons-Belda, O.D.; Fernandez-Uriarte, A.; Diamandis, E.P. Multi Cancer Early Detection by Using Circulating Tumor DNA-The Galleri Test. Reply to Klein et Al. The Promise of Multicancer Early Detection. Comment on “Pons-Belda et al. Can Circulating Tumor DNA Support a Successful Screening Test for Early Cancer Detection? The Grail Paradigm. 2021, 11, 2171”. Diagnostics 2022, 12, 1244. [Google Scholar] [CrossRef]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-Invasive Early Detection of Cancer Four Years before Conventional Diagnosis Using a Blood Test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef] [PubMed]
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.C.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Luo, H.; Krawczyk, M.; Wei, W.; Wang, W.; Wang, J.; Flagg, K.; Hou, J.; Zhang, H.; Yi, S.; et al. DNA Methylation Markers for Diagnosis and Prognosis of Common Cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 7414–7419. [Google Scholar] [CrossRef] [PubMed]
- CancerRadar. Available online: https://earlydx.com/product/cancer-radar/ (accessed on 25 March 2024).
- Jørgensen, J.T.; Hersom, M. Companion Diagnostics—A Tool to Improve Pharmacotherapy. Ann. Transl. Med. 2016, 4, 482. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-Repair Gene MGMT and the Clinical Response of Gliomas to Alkylating Agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT Promoter Methylation Status Testing to Guide Therapy for Glioblastoma: Refining the Approach Based on Emerging Evidence and Current Challenges. Neuro. Oncol. 2018, 21, 167–178. [Google Scholar] [CrossRef]
- Mastoraki, S.; Strati, A.; Tzanikou, E.; Chimonidou, M.; Politaki, E.; Voutsina, A.; Psyrri, A.; Georgoulias, V.; Lianidou, E. Methylation: A Liquid Biopsy-Based Epigenetic Assay for the Follow-up of Patients with Metastatic Breast Cancer Receiving Endocrine Treatment. Clin. Cancer Res. 2018, 24, 1500–1510. [Google Scholar] [CrossRef]
- Hafazalla, K.; Sahgal, A.; Jaja, B.; Perry, J.R.; Das, S. Procarbazine, CCNU and Vincristine (PCV) versus Temozolomide Chemotherapy for Patients with Low-Grade Glioma: A Systematic Review. Oncotarget 2018, 9, 33623–33633. [Google Scholar] [CrossRef] [PubMed]
- van den Bent, M.J.; Erdem-Eraslan, L.; Idbaih, A.; de Rooi, J.; Eilers, P.H.C.; Spliet, W.G.M.; den Dunnen, W.F.A.; Tijssen, C.; Wesseling, P.; Sillevis Smitt, P.A.E.; et al. MGMT-STP27 Methylation Status as Predictive Marker for Response to PCV in Anaplastic Oligodendrogliomas and Oligoastrocytomas—A Report from EORTC Study 26951. Clin. Cancer Res. 2013, 19, 5513–5522. [Google Scholar] [CrossRef] [PubMed]
- Gusyatiner, O.; Hegi, M.E. Glioma Epigenetics: From Subclassification to Novel Treatment Options. Semin. Cancer Biol. 2018, 51, 50–58. [Google Scholar] [CrossRef]
- Eibl, R.H.; Schneemann, M. Liquid Biopsy and Primary Brain Tumors. Cancers 2021, 13, 5429. [Google Scholar] [CrossRef] [PubMed]
- Sheffield, N.C.; Pierron, G.; Klughammer, J.; Datlinger, P.; Schönegger, A.; Schuster, M.; Hadler, J.; Surdez, D.; Guillemot, D.; Lapouble, E.; et al. DNA Methylation Heterogeneity Defines a Disease Spectrum in Ewing Sarcoma. Nat. Med. 2017, 23, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Garrett-Bakelman, F.E.; Chung, S.S.; Sanders, M.A.; Hricik, T.; Rapaport, F.; Patel, J.; Dillon, R.; Vijay, P.; Brown, A.L.; et al. Distinct Evolution and Dynamics of Epigenetic and Genetic Heterogeneity in Acute Myeloid Leukemia. Nat. Med. 2016, 22, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Mazor, T.; Pankov, A.; Song, J.S.; Costello, J.F. Intratumoral Heterogeneity of the Epigenome. Cancer Cell 2016, 29, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Snow, A.; Chen, D.; Lang, J.E. The Current Status of the Clinical Utility of Liquid Biopsies in Cancer. Expert. Rev. Mol. Diagn. 2019, 19, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Mierziak, J.; Kostyn, K.; Boba, A.; Czemplik, M.; Kulma, A.; Wojtasik, W. Influence of the Bioactive Diet Components on the Gene Expression Regulation. Nutrients 2021, 13, 3673. [Google Scholar] [CrossRef]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Kaushik, A.; Chaudhary, V.; Longkumer, I.; Saraswathy, K.N.; Jain, S. Sex-Specific Variations in Global DNA Methylation Levels with Age: A Population-Based Exploratory Study from North India. Front. Genet. 2023, 14, 1038529. [Google Scholar] [CrossRef] [PubMed]
- Yusipov, I.; Bacalini, M.G.; Kalyakulina, A.; Krivonosov, M.; Pirazzini, C.; Gensous, N.; Ravaioli, F.; Milazzo, M.; Giuliani, C.; Vedunova, M.; et al. Age-Related DNA Methylation Changes Are Sex-Specific: A Comprehensive Assessment. Aging 2020, 12, 24057–24080. [Google Scholar] [CrossRef] [PubMed]
- Golden, L.C.; Itoh, Y.; Itoh, N.; Iyengar, S.; Coit, P.; Salama, Y.; Arnold, A.P.; Sawalha, A.H.; Voskuhl, R.R. Parent-of-Origin Differences in DNA Methylation of X Chromosome Genes in T Lymphocytes. Proc. Natl. Acad. Sci. USA 2019, 116, 26779–26787. [Google Scholar] [CrossRef] [PubMed]
- Dayeh, T.; Volkov, P.; Salö, S.; Hall, E.; Nilsson, E.; Olsson, A.H.; Kirkpatrick, C.L.; Wollheim, C.B.; Eliasson, L.; Rönn, T.; et al. Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion. PLoS Genet. 2014, 10, e1004160. [Google Scholar] [CrossRef]
- Dick, K.J.; Nelson, C.P.; Tsaprouni, L.; Sandling, J.K.; Aïssi, D.; Wahl, S.; Meduri, E.; Morange, P.-E.; Gagnon, F.; Grallert, H.; et al. DNA Methylation and Body-Mass Index: A Genome-Wide Analysis. Lancet 2014, 383, 1990–1998. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).