Simple Summary
Endometrial cancer is the most common gynecological cancer. However, advanced and recurrent cancers are less sensitive to chemotherapy and have poor prognoses. Therefore, new treatment strategies are being explored. Recently, immune checkpoint inhibitors (ICIs) have been used in treating various cancers. Although AT-rich interaction domain 1A (ARID1A) negativity has been proposed as a new biomarker for immune checkpoint inhibitors, there have been no reports on ARID1A biomarkers in endometrial cancer. Therefore, we investigated whether ARID1A negativity predicts the efficacy of ICIs in treating endometrial cancer. We assessed ARID1A expression and tumor-infiltrating lymphocytes (CD8+) and immune checkpoint molecules (PD-L1/PD-1) using immunostaining and MSI analysis. Throughout our experiment, CD8 and PD-1 expression did not differ significantly between the ARID1A-negative and ARID1A-positive groups. Our findings suggest that ARID1A negativity may not be a suitable biomarker for ICI efficacy in endometrial cancer.
Abstract
Background: AT-rich interaction domain 1A (ARID1A) has been proposed as a new biomarker for predicting response to immune checkpoint inhibitors (ICIs). The predictive value of ARID1A for predicting ICI effectiveness has not been reported for endometrial cancer. Therefore, we investigated whether ARID1A negativity predicts ICI effectiveness for endometrial cancer treatment. Methods: We evaluated ARID1A expression, tumor-infiltrating lymphocytes (CD8+), and immune checkpoint molecules (PD-L1/PD-1) by immunostaining endometrial samples from patients with endometrial cancer. Samples in which any of the four mismatch repair proteins (MLH1, MSH2, MSH6, and PMS2) were determined to be negative via immunostaining were excluded. In the ARID1A-negative group, microsatellite instability (MSI) status was confirmed via MSI analysis. Results: Of the 102 samples investigated, 25 (24.5%) were ARID1A-negative. CD8 and PD-1 expression did not differ significantly between the ARID1A-negative group and the ARID1A-positive group; however, the ARID1A-negative group showed significantly lower PD-L1 expression. Only three samples (14.2%) in the ARID1A-negative group showed high MSI. Sanger sequencing detected three cases of pathological mutation in the MSH2-binding regions. We also established an ARID1A-knockout human ovarian endometriotic epithelial cell line (HMOsisEC7 ARID1A KO), which remained microsatellite-stable after passage. Conclusion: ARID1A negativity is not suitable as a biomarker for ICI effectiveness in treating endometrial cancer.
1. Introduction
With the advent of next-generation sequencing (NGS), the rapid genetic analysis of tumor tissues has become possible. As a result, treatments personalized according to genetic mutation are gradually being implemented in clinical practice. Regarding lung cancer, genetic tests (epidermal growth factor receptor [EGFR] mutation, anaplastic lymphoma kinase [ALK] fusion gene, ROS1 fusion gene, and BRAF V600E mutation) and their respective treatments are covered by insurance, and precision medicine is employed according to the specific genetic mutations observed [1,2,3,4,5,6,7]. However, treatment according to genetic mutations has not yet been established in endometrial cancer treatment. Endometrial cancer is one of the most common gynecologic cancers, and its incidence is gradually increasing [8]. The primary treatment for endometrial cancer is surgery with postoperative adjuvant chemotherapy. However, recurrent and advanced cancers are extremely resistant to chemotherapy and have poor prognoses [9]. Therefore, new therapeutic strategies are required.
The anti-programmed cell death-1 (PD-1) antibody pembrolizumab, which is an immune checkpoint inhibitor (ICI), is used to treat microsatellite instability (MSI)-high solid tumors [10]. Based on the results of the comprehensive genetic analysis of endometrial cancer, The Cancer Genome Atlas Research Network classified endometrial cancer into four molecular phenotypes (polymerase epsilon [POLE]-ultramutated, MSI-hypermutated, copy number high, and copy number low) [11]. The prevalence of MSI-high in endometrial cancer is approximately 30%. Because the proportion of MSI-high tumors is higher in endometrial carcinoma than in other carcinomas, ICIs can be expected to be effective for treating endometrial cancer [12,13].
Recently, it was reported that a deficiency of AT-rich interaction domain 1A (ARID1A) is potentially useful as a biomarker for predicting response to ICIs [14]. ARID1A is a tumor suppressor gene that encodes a subunit of the chromatin remodeling complex SWI/SNF [15]. ARID1A mutations lead to loss of function, and there are currently no effective therapeutic drugs targeting ARID1A. However, ARID1A is mutated in many cancers, including ovarian cancer, endometrial cancer, gastric cancer, and bladder cancer [16,17,18,19]. ARID1A mutations have been reported to be a poor prognostic factor in various cancers, including ovarian cancer, breast cancer, and gastric cancer [20,21,22]. Therefore, therapeutic drugs targeting ARID1A are needed. Shen et al. [14] reported that ARID1A interacts with one of the mismatch repair (MMR) proteins, namely, MSH2. If ARID1A is deficient, MSH2 fails to function and exhibits MSI-high. ARID1A mutations are associated with high expression of programmed cell death-ligand 1 (PD-L1) and MSI-high, and ICIs are generally effective in treating ARID1A-mutated cancers [23,24]. The relationship between ARID1A deficiency and the efficacy of ICIs in endometrial cancer has not yet been analyzed. Therefore, we investigated the suitability of ARID1A deficiency as a biomarker for predicting response to ICIs in endometrial cancer.
2. Materials and Methods
2.1. Ethics Statement
This study was conducted in accordance with the ethical standards of national and international guidelines and the Declaration of Helsinki. It was also approved by the Institutional Review Board of Shimane University Hospital (approval number 2004-0381). After approval by the Institutional Review Board, patients’ written consent was obtained to collect tumor specimens.
2.2. Tissue Samples
We collected tissue samples from 102 patients with endometrial carcinoma (62 with Grade 1, 32 with Grade 2, and 8 with Grade 3) treated between January 2006 and January 2017 in the Department of Obstetrics and Gynecology of Shimane University Hospital in Japan. The patients were initially treated as follows: total hysterectomy for 39 patients, modified radical hysterectomy for 51, and radical hysterectomy for 9. Three patients underwent chemotherapy and/or radiotherapy without surgery due to complications. Retroperitoneal lymph node dissection was performed in 86 patients. Radiotherapy (whole pelvic irradiation) and/or chemotherapy (using 175 mg/m2 of paclitaxel and with a carboplatin area under the curve = 5 mg/mL*min) was performed postoperatively for patients with high recurrence risk (deep myometrial invasion, Grade 2 or 3; lymph node metastasis; or lymphovascular space invasion). The samples were formalin-fixed and converted to paraffin-embedded tissue blocks. The samples were assessed by pathologists after being stained with hematoxylin and eosin. To evaluate the interaction between ARID1A and MSH2, we excluded samples in which one or more of the four MMR proteins (MLH1, MSH2, MSH6, PMS2) were negative. We confirmed MMR protein expression in 102 samples via immunohistochemistry (Supplementary Figure S1).
We used the International Federation of Gynecology and Obstetrics (FIGO) 2014 guidelines for endometrial cancer to stage the tumors [25]. The histological diagnosis of endometrial carcinomas was performed according to the 2014 World Health Organization criteria. The clinical data were collected by retrospective review.
2.3. Immunohistochemistry
We evaluated the expression of ARID1A, immune checkpoint molecules (PD-L1 and PD-1), CD8 tumor infiltration, and MMR proteins (MLH1, MSH2, MSH6, and PMS2) using immunohistochemistry.
Formalin-fixed and paraffin-embedded sections (4 μm thick) were immunostained as previously described [26]. We used antibodies against ARID1A (anti-ARID1A antibody sc-32761, mouse monoclonal antibody; Santa Cruz Biotechnology, Dallas, TX, USA), PD-L1 (SP263, Rabbit Monoclonal Primary Antibody; Roche, Basel, Switzerland), PD-1 (NAT105 Mouse Monoclonal Antibody; Roche), CD8 (SP57 Rabbit Monoclonal Primary Antibody; Roche), MutL Protein Homolog 1 (1:50; Dako, Santa Clara, CA, USA), MutS Protein Homolog 2 (1:50; Dako), MutS Protein Homolog 6 (1:50; Dako), and Postmeiotic Segregation Increased 2 (PMS2; 1:40; Dako).
Samples in which no ARID1A expression was found in the nuclei of the tumor cells were evaluated as negative (Supplementary Figure S2). ARID1A’s C-terminal region (1600–1800 amino acids) is essential for its interaction with MSH2 [14].
An anti-ARID1A antibody that binds to 600–1018 amino acids of ARID1A was used for immunohistochemistry. Therefore, it can be inferred that the interaction with MSH2 was also lost in the samples that were determined to be ARID1A negative via immunohistochemistry (Supplementary Figure S3).
The immunostaining of PD-L1, PD-1, and CD8 was performed as previously described [26]. Samples were evaluated as PD-L1-positive if 5% or more of the tumor cells (membrane and cytoplasmic) were stained. Samples were evaluated as PD-1-positive if 5% or more of the tumor-infiltrating lymphocytes were stained. Tumor-infiltrating lymphocyte expression was evaluated in 4 levels (0, undetectable; 1+, low density; 2+, moderate density; and 3+, high density). Samples that stained 2+ or 3+ were evaluated as CD8-positive. Since PD-L1 can be expressed not only in tumor cells but also in immune cells, it can be suspected as being positive in cases with a mixture of tumor cells and immune-related cells. We compared HE staining with PD-1 and PD-L1 immunostaining and carefully observed the cell morphology to clearly distinguish tumor cells from immune cells.
2.4. Microsatellite Instability Analysis
MSI analysis was performed to investigate whether samples that were determined ARID1A-negative via immunostaining were MSI-high. DNA was extracted from paraffin-embedded sections of normal tissues and tumor tissues, respectively. MSI analysis was performed using the MSI test (CDx) (BML, Tokyo, Japan) [27], in which a polymerase chain reaction (PCR) with five microsatellite markers (BAT25, BAT26, NR21, MONO27, and NR24) is performed. Samples were assessed as being MSI-high if two or more markers showed length differences between normal tissue and tumor tissue. The remaining samples were assessed as microsatellite-stable (MSS).
2.5. Sanger Sequencing
ARID1A mutations that are located on the C-terminal side of amino acids 600–1018 are considered ARID1A-positive in immunostaining (The anti-ARID1A antibody used in immunostaining binds to amino acids 600–1018 of ARID1A). Therefore, we performed Sanger sequencing of ARID1A at the regions that bind to MSH2 in samples that were determined ARID1A-positive via immunohistochemistry.
DNA was extracted from endometrial carcinomas and amplified with PCR using primers for exons 18-c, 18-d, 19, and 20a of ARID1A. The primers are shown in Supplementary Figure S4. Sequencing was performed using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). All mutations identified in tumors were evaluated for pathogenic variant by reference to the Catalogue of Somatic Mutations in Cancer (COSMIC).
2.6. ARID1A Knockout Human Ovarian Endometriotic Epithelial Cell Line
Epithelial cells derived from ovarian endometriotic cysts were extracted via primary culturing; then, mutant CDK4 (CDK4R24C, an inhibitor-resistant form of CDK4), Cyclin D1, and hTERT were introduced using lentiviral vectors, and the immortalized cell line HMOsisEC7 was successfully established [28]. Subsequently, we aimed to establish HMOsisEC7 (ARID1A knockout) by using CRISPR-Cas9. HMOsisEC7 was transfected with piggyback vectors, PB-TAC-ERN-3xFlag-hCas9, PB-TKbsd-U6/H1R-ARID1A-gRNA401-394, and PB-TKbsd-U6/H1R-ARID1A-gRNA416-423, and pCAG-PBase-M282V was introduced via electroporation using an NEPA21 instrument (Nepagene, Chiba, Japan). Detailed information on plasmids is provided in the Supplementary Materials [29,30,31]. HMOsisEC7 cells mixed with 2 μg of PB-TAC-ERN-3xFlag-hCas9, 2 μg of PB-TKbsd-U6/H1R-ARID1A-gRNA401-394, 2 μg of PB-TKbsd-U6/H1R-ARID1A-gRNA416-423, 4 μg of pCAG-PBase-M282V, and 0.1 μg of pCMV-EGFP in 100 μL of OptiMEM medium were pulsed using NEPA21. The parameters were as follows: voltage, 175 V; pulse length, 5 ms; pulse interval, 50 ms; number of pulses, 2; decay rate, 10%; polarity + as poring pulse and voltage, 20 V; pulse length, 50 ms; pulse interval, 50 ms; number of pulses, 5; decay rate, 40%; and polarity +/− as the transfer pulse. Subsequently, the mixtures were rapidly transferred to three wells in a six-well plate with the complete culture medium. The cells were cultivated in the presence of 8 μg/mL of Blasticidin S and 100 μg/mL of G418 for 7 days and then treated with 1 μg/mL of doxycycline for 2 weeks. The cells were seeded into three 90 mm dishes at a density of 100 cells/dish, and 24-well-isolated colonies were trypsinized with cloning cylinders and transferred into a 24-well plate. After propagation in a 6-well plate, cellular protein and DNA were extracted from each clone and subjected to Western blot and Sanger sequencing with genomic PCR to confirm the ARID1A status. The genomic DNAs were amplified using PCR with forward primer 5′-GATCAGATGGGCAAGATGAGAC-3′ and reverse primer 5′-GTACCTGTGACCAGGGAGTAAGTAGT-3′. Clones #1, 4, 13, 15, 17, and 19 were confirmed to be ARID1A KO clones.
Clone #19, which had homologous 86 bp deletion between NT 1185 and 1270 of the ARID1A coding sequence, was further propagated, and the cells were transfected with 10 μg of pCAG-hyperPBase-i7EX via electroporation with NEPA21, followed by selection in the presence of 8 μg/mL of ganciclovir. Ganciclovir-resistant clones #19–30 were propagated and used for further experiments (Figure 1, Supplementary Figure S5). We performed MSI analysis of the ARID1A-knockout human ovarian endometriotic epithelial cell line (HMOsisEC7 ARID1A KO).
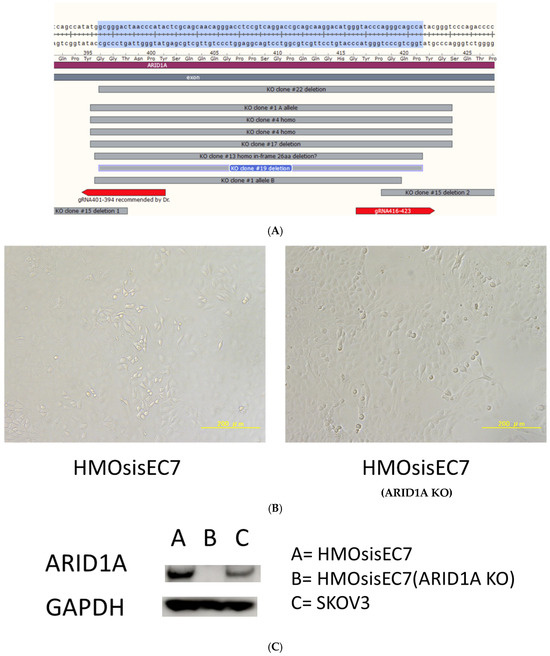
Figure 1.
(A) Genomic analysis of endometrial cancer samples. Clones #1, 4, 13, 15, 17, and 19 are ARID1A KO clones. We designated clone #19 as HMOsisEC7 (ARID1A KO) and used it for experiments. (B) Micrographs of HMOsisEC7 and HMOsisEC7 (ARID1A KO). (C) Western blot showing lack of ARID1A expression in HMOsisEC7 (ARID1A KO). The uncropped blots are shown in Figure S5.
2.7. Statistical Analyses
We performed univariable analysis for progression-free survival (PFS) and overall survival (OS). PFS and OS were calculated between the date of diagnosis and the date of first relapse and last follow-up, respectively. The data are shown as Kaplan–Meier curves. The log-rank test was used to test for the statistical significance of differences in survival between groups. The chi-squared test was used to assess the significance of the association between ARID1A and the expression of CD8, PD-1, and PD-L1. p-values < 0.05 were considered statistically significant. The statistical calculations were performed using Statistical Package for the Social Sciences 23.0 (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Relationship between ARID1A and Clinicopathological Factors
Immunostaining for ARID1A showed the loss of ARID1A expression in 25 of 102 samples (24.5%). We compared the clinicopathological characteristics of the ARID1A-negative and ARID1A-positive groups. There were no significant differences between the two groups in any of the factors compared. The clinicopathological characteristics of the study participants and their tumors are shown in Table 1.

Table 1.
Relationship between ARID1A and clinicopathological factors.
3.2. Relationship between ARID1A and Expression of PD-L1, PD-1 and CD8
The statistical significance of the association between ARID1A expression and immune checkpoint molecules/intratumoral CD8 infiltration was evaluated using the chi-squared test. The ARID1A-negative group had significantly lower PD-L1 expression than the ARID1A-positive group. There was no significant difference in the expression of PD-1 and CD8 between the two groups (Table 2). In the univariable analysis, there was no significant difference in PFS or OS between the two groups (Figure 2).

Table 2.
Relationship between ARID1A and expression of PD-L1, PD-1, and CD8.
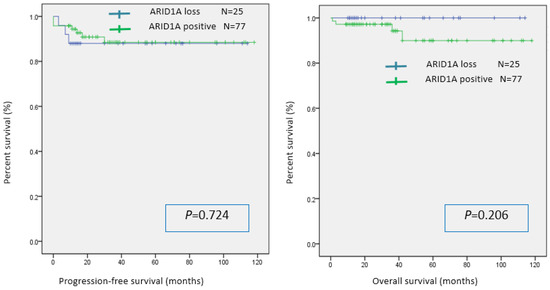
Figure 2.
Kaplan–Meier analysis of progression-free survival and overall survival in the ARID1A-positive and ARID1A-negative groups.
3.3. Microsatellite Instability
MSI analysis was performed for the ARID1A-negative group. In total, 3 of the 21 samples (14.2%) were determined to be MSI-high (Figure 3).
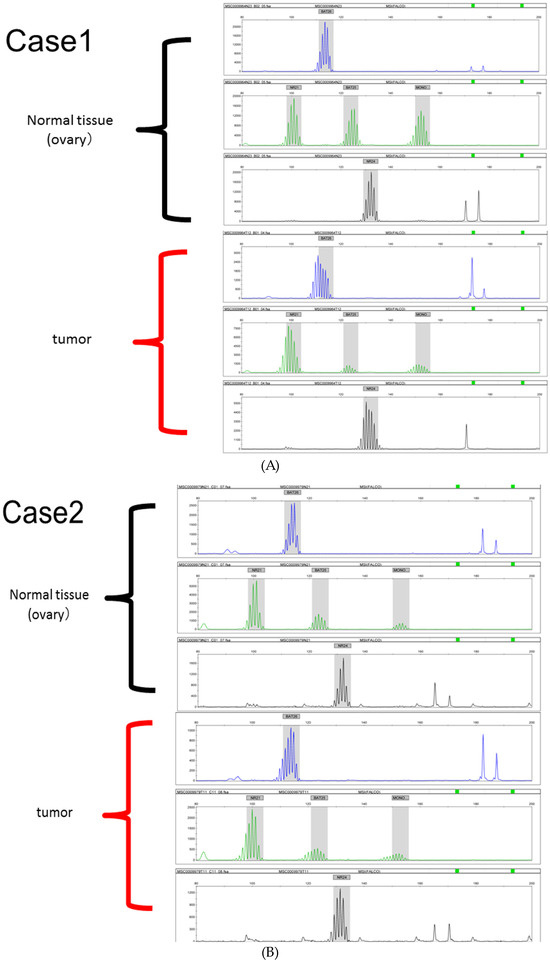
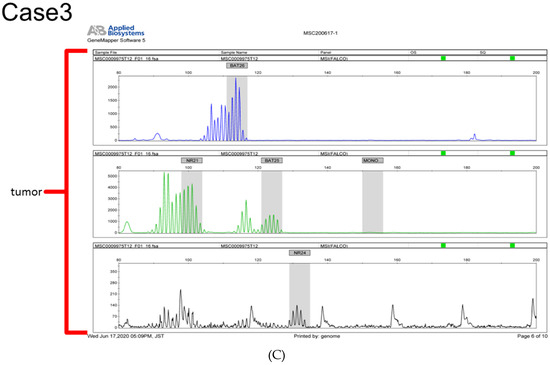
Figure 3.
(A–C) Microsatellite instability (MSI) analysis of three endometrial carcinoma samples determined to be MSI-high.
3.4. Sanger Sequencing of ARID1A
The anti-ARID1A antibody used for immunostaining recognizes amino acids 600–1018 of ARID1A. As amino acids 1600–1800 of ARID1A interact with MSH2, the ARID1A-negative group identified through immunostaining cannot interact with MSH2. However, tumors with ARID1A mutations in the C-terminal side of the site recognized by the anti-ARID1A antibody show ARID1A expression through immunostaining but cannot interact with MSH2. Therefore, we performed the Sanger sequencing of ARID1A at the regions that bind to MSH2 (the region from amino acids 1600–1800 of ARID1A) in samples that were ARID1A-positive using immunohistochemistry. Of the 77 ARID1A-positive samples determined in this manner, 3 had pathogenic mutations (Figure 4A–C). However, the MSI analysis revealed that all three samples were MSS (Figure 4D–F). In addition, all three cases had low CD8 lymphocyte infiltration within the tumors.
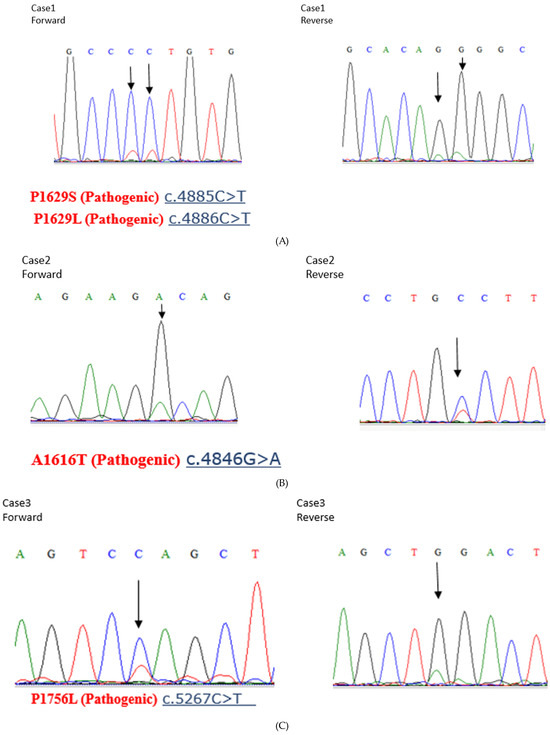
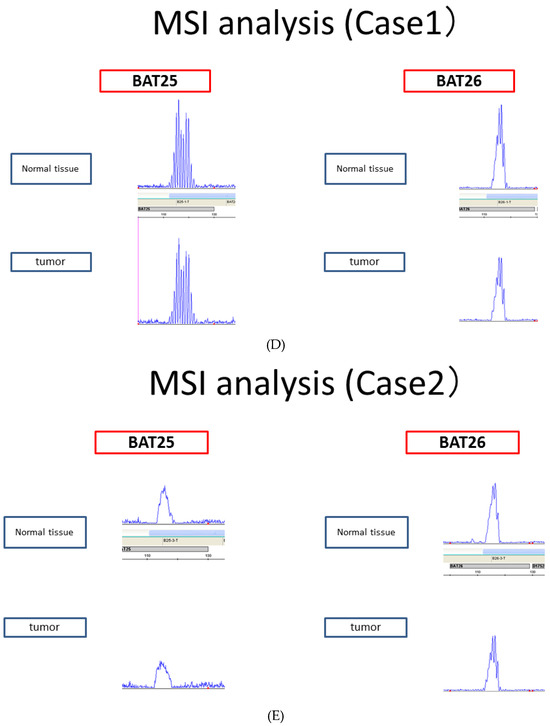
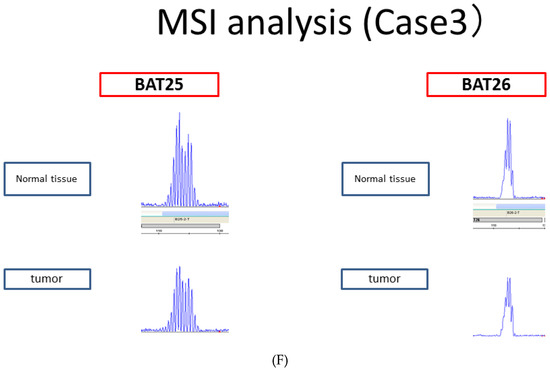
Figure 4.
(A–C) Sanger sequencing of three endometrial carcinoma samples with pathogenic ARID1A mutations. (D–F). Microsatellite instability (MSI) analysis of the three endometrial carcinoma samples with pathogenic ARID1A mutations, showing negativity for all microsatellite markers.
3.5. MSI Analysis of HMOsis EC7
HMOsisEC7 ARID1A KO was negative for all microsatellite markers at both population doublings (PDs) 3 and 255 (Supplementary Figure S6).
4. Discussion
ARID1A mutations have been reported in various carcinomas. They are commonly found in gynecologic cancers, including ovarian cancer and endometrial cancer, and are present in 30–57% of tumors [19]. In this study, 24.5% of endometrial cancer samples were determined to be ARID1A-negative according to immunostaining, a result consistent with previous reports. Correlations between ARID1A expression and prognosis have been reported for various carcinomas. Several studies have found that ARID1A negativity is a poor prognostic factor [20,21,22,32,33,34], but other studies have found that it is a good prognostic factor [35,36]. In this study, there was no significant difference in PFS and OS between the ARID1A-negative and ARID1A-positive groups. Therefore, the relationship between ARID1A and prognosis is unclear. However, recurrent and advanced cancers are extremely resistant to chemotherapy and have poor prognoses. Therefore, it is thought that precision medicine employed according to genetic mutation is necessary. The discovery of new therapeutic strategies against ARID1A may improve the prognosis of endometrial cancer.
Shen et al. [14] reported that ARID1A interacts with MSH2 and that ICIs may be effective in tumors with ARID1A mutations. Since the publication of the report by Shen et al., ARID1A mutations have been reported to be biomarkers for ICIs in various cancers. However, most of the reports have not analyzed the expression of MMR proteins [24,37,38,39,40,41,42,43]. It has been reported that tumors with ARID1A mutations are significantly more likely than ARID1A wild-type tumors to have mutations of MMR genes [24]. A deficiency of MMR proteins may cause ARID1A mutations, thereby misleadingly indicating that ARID1A mutations are biomarkers for ICIs. There was no association between ARID1A mutations and OS in patients with MSS solid tumors in an ICI treatment cohort [42]. This result suggests that ARID1A alterations contribute to impaired MMR and mutator phenotypes in cancer. However, this contradicts the hypothesis that ARID1A interacts with MSH2. Therefore, it is unclear whether ARID1A is useful as a biomarker for predicting response to ICI therapy in patients with endometrial cancer.
To date, the association between ARID1A and immune-related molecules has been investigated only in relation to colorectal cancer, targeting MSS cases. Among the MSS colorectal cancers, tumors with ARID1A mutations showed higher expression of immune-related molecules (PD-1, cytotoxic T-lymphocyte antigen 4 (CTLA4)) and cytotoxic T cell infiltration compared to the ARID1A wild type [44]. Our study is the first report to investigate the relationship between ARID1A expression and immune-related molecules in MSS endometrial cancer. However, in this study, no correlation was found between ARID1A expression and CD8 lymphocyte expression in MSS endometrial cancer. It has been reported that high PD-L1 expression is a biomarker for ICIs [45,46,47]. However, the ARID1A-negative group had significantly lower PD-L1 expression than the ARID1A-positive group.
In addition, only three samples (14.2%) in the ARID1A-negative group were MSI-high. The Sanger sequencing of ARID1A in the region that binds to MSH2 detected pathogenic mutations in three samples. All three tumors were determined to be MSS according to MSI analysis. These results suggest that only a small proportion of patients with ARID1A mutations are likely to benefit from ICIs. Our results differ from those of previous studies on MSS colorectal cancers [44]. Therefore, whether ARID1A serves as a useful biomarker for predicting a patient’s response to ICI therapy may vary according to cancer type.
We subcultured HMOsisEC7 (ARID1A KO) and hypothesized that if ARID1A interacts with MSH2, the passage of HMOsisEC7 (ARID1A KO) would eventually result in an MSI-high status. However, contrary to expectations, HMosisEC7 (ARID1A KO) remained MSS even after passage to PDs255. Based on the results of this study, it is too early to consider ARID1A a biomarker for ICIs in endometrial cancer.
The main limitation of this study was its small sample size. Therefore, it is necessary to accumulate more cases and continue to examine whether ARID1A is unsuitable as a biomarker for ICIs. In addition, to evaluate immune cells, immunostaining was only performed for CD8 and PD-1. Recently, it has been reported that the production levels of cytokines such as interferon and interleukin-12 contribute to the efficacy of ICIs. Therefore, we believe that it is important to include these cytokine production levels in future studies [48,49,50].
5. Conclusions
In MSS endometrial carcinoma, no significant PD-L1 expression or immune cell infiltration were observed in the ARID1A-negative group. Thus, ARID1A is not suitable for use as a biomarker to predict response to ICI therapy among patients with endometrial cancer because there are few cases of MSI-high in patients with ARID1A-negative tumors. The results of this study are contrary to previous reports that ARID1A is useful as a biomarker for predicting the response to ICI therapy in other types of cancer. If ARID1A mutations are detected through genetic testing in patients with endometrial cancer in clinical practice, ICIs should be considered the recommended treatment, based on previous reports. However, as ARID1A was found to be unsuitable for use as a biomarker for deciding on ICI therapy for patients with endometrial carcinoma in this study, it is necessary to reconsider its use in genomic medicine.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cancers16111999/s1. Figure S1: Cases studied; Figure S2: Representative images of ARID1A-positive and -negative cases; Figure S3: Relationship between anti ARID1A antibody used in the experiment and MSH2 binding site; Figure S4: Primer for exons 18-c, 18-d, 19, and 20a of ARID1A; Figure S5A: Original Images for Blots: ARID1A expression; Figure S5B: Original Images for Blots:GAPDH expression; Figure S6A: HMOsisEC7 ARID1A KO was negative for all microsatellite markers at PD 3; Figure S6B: HMOsisEC7 ARID1A KO was negative for all microsatellite markers at PD 255; Supplementary information: Plasmids.
Author Contributions
H.Y. and K.N. drafted the manuscript. K.K., T.I., M.I., K.I. and S.R. carried out the immunohistochemical analysis and molecular genetic studies. T.K. helped to develop the immortalized cell line. K.N. participated in the design of the study. S.K. conceived the study, participated in its design and coordination, and helped in drafting the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant numbers 21K09472 and 22K09596).
Institutional Review Board Statement
This study was conducted in accordance with the ethical standards of national and international guidelines and the Declaration of Helsinki. It was also approved by the Institutional Review Board of Shimane University Hospital (approval number 2004-0381). After approval by the Institutional Review Board (Shimane University Hospital, approval number 2004-0381). Written informed consent for sample collection was obtained from all the patients.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patients to publish this paper.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mitsudomi, T.; Morita, S.; Yatabe, Y.; Negoro, S.; Okamoto, I.; Tsurutani, J.; Seto, T.; Satouchi, M.; Tada, H.; Hirashima, T.; et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WITOG3405): An open label, randomized phase 3 trial. Lancet Oncol. 2010, 11, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated, E.G.F.R. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wu, Y.L.; Chen, G.; Feng, J.; Liu, X.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomized, phase 3 study. Lancet Oncol. 2011, 12, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Zhou, C.; Hu, C.P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Shi, J.H.; Lee, K.Y.; et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomized phase 3 trial. Lancet Oncol. 2014, 15, 213–222. [Google Scholar] [CrossRef]
- Solomon, B.J.; Mok, T.; Kim, D.W.; Wu, Y.L.; Nakagawa, K.; Mekhail, T.; Felip, E.; Cappuzzo, F.; Paolini, J.; Usari, T.; et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 2014, 371, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Ou, S.H.; Bang, Y.J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.B.; et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Besse, B.; Groen, H.J.M.; Souquet, P.J.; Quoix, E.; Baik, C.S.; Barlesi, F.; Kim, T.M.; Mazieres, J.; Novello, S.; et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: An open-label, multicentre phase 2 trial. Lancet Oncol. 2016, 17, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality world wide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial Cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- FDA Approves First Cancer Treatment for Any Solid Tumor with a Specific Genetic Feature. Available online: https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm (accessed on 20 May 2024).
- Cancer Genome Atlas Research, N.; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jürgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial cancer combined Analysis of the PORTEc Cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ju, Z.; Zhao, W.; Wang, L.; Peng, Y.; Ge, Z.; Nagel, Z.D.; Zou, J.; Wang, C.; Kapoor, P.; et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018, 24, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Nagl, N.G.; Wilsker, D.; Scoy, M.V.; Pacchione, S.; Yaciuk, P.; Dallas, P.B.; Moran, E. Two relate ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem. J. 2004, 383, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Wang, T.L.; Shih, I.M.; Mao, T.L.; Nakayama, K.; Roden, R.; Glas, R.; Slamon, D.; Diaz, L.J.; Vogelstein, B.; et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010, 330, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Kan, J.; Yuen, S.T.; Shi, S.T.; Chu, K.M.; Law, S.; Chan, T.L.; Kan, Z.; Chan, A.S.; Kan, Z.; et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 2011, 43, 1219–1223. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Guo, G.; Huang, Y.; Hu, X.; Tang, A.; Gao, S.; Wu, R.; Chen, C.; Li, X.; Zhou, L.; et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011, 43, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Takeda, T.; Banno, K.; Okawa, R.; Yanokura, M.; Iijima, M.; Irie, K.H.; Nakamura, K.; Iida, M.; Adachi, M.; Umene, K.; et al. ARID1A gene mutation in ovarian and endometrial cancers (Review). Oncol. Rep. 2016, 35, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, A.; Nakayama, K.; Rahman, M.T.; Rahman, M.; Katagiri, H.; Nakayama, N.; Ishikawa, M.; Ishibashi, T.; Iida, K.; Kobayashi, H.; et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod. Pathol. 2012, 25, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.D.; Lee, J.E.; Jung, H.Y.; Oh, M.H.; Lee, J.H.; Jang, S.H.; Kim, K.J.; Han, S.W.; Kim, S.Y.; Kim, H.J.; et al. Loss of tumor suppressor ARID1A protein expression correlates with poor prognosis in patients with primary breast cancer. J. Breast Cancer 2015, 18, 339–346. [Google Scholar] [CrossRef]
- Zhu, Y.P.; Sheng, L.L.; Wu, J.; Yang, M.; Cheng, X.F.; Wu, N.N.; Ye, X.B.; Cai, J.; Wang, L.; Shen, Q.; et al. Loss of ARID1A expression is associated with poor prognosis in patients with gastric cancer. Hum. Pathol. 2018, 78, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Ahn, J.M.; Bae, W.; Sung, C.O.; Lee, D. Functional loss of ARID1A is tightly associated with high PD-L1 expression in gastric cancer. Int. J. Cancer 2019, 145, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, M.; Jiang, Z.; Wang, X. ARID1A mutations are associated with increased immune activity in gastrointestinal cancer. Cells 2019, 8, 678. [Google Scholar] [CrossRef] [PubMed]
- Prat, J. Staging classification for of the ovary, fallopian tube, and peritoneum. Int. J. Gynaecol. Obstet. 2014, 124, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Nakayama, K.; Ishikawa, M.; Nakamura, K.; Ishibashi, T.; Sanuki, K.; Ono, R.; Sasamori, H.; Minamoto, T.; Iida, K.; et al. Microsatellite instability is a biomarker for immune checkpoint inhibitors in endometrial cancer. Oncotarget 2017, 9, 5652–5664. [Google Scholar] [CrossRef] [PubMed]
- BML Co., Ltd. Available online: http://www.bml.co.jp/ (accessed on 20 May 2024).
- Mohammad, M.H.; Kentaro, N.; Kamrunnahar, S.; Sultana, R.; Masako, I.; Tomoka, I.; Hitomi, Y.; Seiya, S.; Kouji, I.; Kosuke, K.; et al. Establishment of a Novel In Vitro Model of Endometriosis with Oncogenic KRAS and PIK3CA Mutations for Understanding the Underlying Biology and Molecular Pathogenesis. Cancers 2021, 13, 3174. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Oceguera-Yanez, F.; Sakurai, C.; Nakagawa, M.; Yamanaka, S.; Woltjen, K. Inducible Transgene Expression in Human iPS Cells Using Versatile All-in-One piggyBac Transposons. Methods Mol. Biol. 2016, 1357, 111–131. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Burnight, E.R.; Cooney, A.L.; Malani, N.; Brady, T.; Sander, J.D.; Staber, J.; Wheelan, S.J.; Joung, J.K.; McCray, P.B., Jr.; et al. piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. USA 2013, 110, E2279–E2287. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, C.; Suh, J.H.; Chae, J.Y.; Kim, H.W.; Moon, K.C. Decreased ARID1A expression correlates with poor prognosis of clear cell renal cell carcinoma. Hum. Pathol. 2015, 46, 454–460. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Sy, K.; Kalloger, S.E.; Li-Chang, H.; Woods, R.; Kumar, A.; Streutker, C.J.; Hafezi-Bakhtiari, S.; Zhou, C.; Lim, H.J.; et al. ARID1A/BAF250a as a prognostic marker for gastric carcinoma: A study of 2 cohorts. Hum. Pathol. 2014, 45, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Xu, P.; Fu, Z.; Chang, H.L.; Woods, R.; Kumar, A.; Streutker, C.J.; Bakhtiari, S.H.; Zhou, C.; Jim, H.J.; et al. Prognostic and clinicopathological significance of ARID1A in endometrium-related gynecological cancers: A meta-analysis. J. Cell Biochem. 2017, 118, 4517–4525. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.L.; Wang, D.S.; Xi, S.Y.; Wu, W.J.; Chen, D.L.; Zeng, Z.L.; Wang, R.Y.; Huang, Y.X.; Jin, Y.; Wang, F.; et al. Clinicopathologic and prognostic relevance of ARID1A protein loss in colorectal cancer. World J. Gastroenterol. 2014, 20, 18404–18412. [Google Scholar] [CrossRef] [PubMed]
- Faraj, S.F.; Chaux, A.; Gonzalez-Roibon, N.; Munari, E.; Ellis, C.; Driscoll, T.; Schoenberg, M.P.; Bivalacqua, T.J.; Shih, I.M.; Netto, G.J. ARID1A immunohistochemistry improves outcome prediction in invasive urothelial carcinoma of urinary bladder. Hum. Pathol. 2014, 45, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Shi, R.; Li, Y.; Zhang, Z.; Xu, S.; Chen, C.; Cao, P.; Zhang, H.; Liu, M.; Pan, Z.; et al. ARID1A, ARID1B, and ARID2 mutations serve as potential biomarkers for immune checkpoint blockade in patients with non-small cell lung cancer. Front. Immunol. 2021, 12, 670040. [Google Scholar] [CrossRef] [PubMed]
- Yuka, K.; Tatsuyuki, C.; Miho, K.; Nakamura, K.; Aimono, E.; Yoshimura, T.; Takahashi, M.; Saotome, K.; Yoshihama, T.; Iwasa, N.; et al. ARID1A mutation/ARID1A loss is associated with a high immunogenic profile in clear cell ovarian cancer. Gynecol. Oncol. 2021, 162, 679–685. [Google Scholar]
- Okamura, R.; Kato, S.; Lee, S.; Jimenez, R.E.; Sicklick, J.K.; Kurzrock, R. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J. Immunother. Cancer. 2020, 8, e00438. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Tu, W.; Yang, L.; Peng, G.; Yang, L. ARID1A deficiency and immune checkpoint blockade therapy: From mechanisms to clinical application. Cancer Lett. 2020, 473, 148–155. [Google Scholar] [CrossRef]
- Kir, G.; Soylemez, T.; Olgun, Z.C.; Aydin, A.; McCluggage, W.G. Correlation of PD-L1 expression with immunohistochemically determined molecular profile in endometrial carcinomas. Virchows Arch. 2020, 477, 845–856. [Google Scholar] [CrossRef]
- Jiang, T.; Chen, X.; Su, C.; Ren, S.; Zhou, C. Pan-cancer analysis of ARID1A alterations as biomarkers for immunotherapy outcomes. J. Cancer. 2020, 11, 31949479. [Google Scholar] [CrossRef]
- Sun, D.; Tian, L.; Zhu, Y.; Wo, Y.; Liu, Q.; Liu, S.; Li, H.; Hou, H. Subunits of ARID1 serve as novel biomarkers for the sensitivity to immune checkpoint inhibitors and prognosis of advanced non-small cell lung cancer. Mol. Med. 2020, 26, 78. [Google Scholar] [CrossRef] [PubMed]
- Mehrvarz Sarshekeh, A.; Alshenaifi, J.; Roszik, J.; Manyam, G.C.; Advani, S.M.; Katkhuda, R.; Verma, A.; Lam, M.; Willis, J.; Shen, J.P.; et al. ARID1A mutation may define an immunologically active subgroup in patients with microsatellite stable colorectal cancer. Clin. Cancer Res. 2021, 27, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.K.; Wu Yl Kudaba, I.; Kowalski, D.M.; Cho, B.C.; Turna, H.Z.; Castro, G., Jr.; Srimuninnimit, V.; Laktionov, K.K.; Bondarenko, I.; Kubota, K.; et al. Pembrolizumab versus chemotherapy for preciously untreated, PD-L1-epressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): A randomized, open-label, controlled, phase3 trial. Lancet 2019, 393, 1819–1830. [Google Scholar] [CrossRef] [PubMed]
- Ready, N.; Hellmann, M.D.; Awad, M.M.; Otterson, G.A.; Gutierrez, M.; Gainor, J.F.; Borghaei, H.; Jolivet, J.; Horn, L.; Mates, M.; et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): Outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J. Clin. Oncol. 2019, 37, 992–1000. [Google Scholar] [CrossRef]
- Ott, P.A.; Bang, Y.J.; Piha-Paul, S.A.; Abdul Razak, A.R.; Bennouna, J.; Soria, J.C.; Rugo, H.S.; Cohen, R.B.; O’Neil, B.H.; Mehnert, J.M.; et al. T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J. Clin. Oncol. 2019, 37, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Tsoi, J.; Onyshchenko, M.; Abril-Rodriguez, G.; Ross-Macdonald, P.; Wind-Rotolo, M.; Champhekar, A.; Medina, E.; Torrejon, D.Y.; Shin, D.S.; et al. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2020, 38, 500–515. [Google Scholar] [CrossRef] [PubMed]
- Renren, Y.; Bo, Z.; Degao, C. Type I interferon-mediated tumor immunity and its role in immunotherapy. Cell Mol. Life Sci. 2022, 79, 191. [Google Scholar]
- Cirella, A.; Luri-Rey, C.; Di Trani, C.A.; Teijeira, A.; Olivera, I.; Bolaños, E.; Castañón, E.; Palencia, B.; Brocco, D.; Fernández-Sendin, M.; et al. Novel strategies exploiting interleukin-12 in cancer immunotherapy. Pharmacol. Ther. 2022, 239, 108189. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).