Extracellular Vesicles in Lung Cancer: Implementation in Diagnosis and Therapeutic Perspectives
Abstract
Simple Summary
Abstract
1. Introduction
2. Extracellular Vesicles
3. EVs in Diagnosis
| CircRNA # | Function | Pathway | Reference |
|---|---|---|---|
| Circ_0012673 | Promote cell proliferation | Sponge miR-22; upregulate ErbB3 | [106] |
| Circ_0067934 | Promote cell proliferation, migration, and invasion | Modulate markers of epithelial-to-mesenchymal transition (EMT) | [107] |
| Circ_007288 | Promote cell proliferation | Sponge miR-377-5p/NOVA2 | [90] |
| Circ_0000376 | To induce resistance to cisplatin and promote tumorigenesis | Sponge miR-1298-5p/KPNA4 | [91] |
| Circ_PDZD8 | Promote cell proliferation | Sponge miR330-5p/LARP1 | [95] |
| Circ_0072309 | To promote tumor progression and invasion | Sponge miR607/FTO | [108] |
| Circ_ATAD1 | Enhance cancer progression | Sponge miR-191-5p | [109] |
| Circ_0092887 | Induce resistance to taxane | Sponge miR490-5p/UBE2T | [110] |
| Circ_0007385 | Promote cell proliferation, migration, tumourigenesis, and invasion | Sponge miR-181 | [111] |
| Circ_0013958 | Promote cell proliferation and invasion and prevent apoptosis | Sponge miR-134/cyclin D1 | [112] |
| Circ_0020123 | Inhibit proliferation and invasion | Sponge miR1299/HMGB3 | [97] |
| Circ_008305 | Inhibit tumor metastasis | Sponge miR-429/miR-200b-3p/PTK2 | [113] |
| Circ_CRIM1 | Inhibit tumor metastasis and invasion | Sponge miR-93 and miR-182; | [114] |
| Circ_RNF13 | Inhibit tumor proliferation and metastasis | Sponge miR-93-5p | [115] |
| CircSH3PXD2A | Inhibit tumor chemoresistance | miR-375-3p/YAP1 | [116] |
4. EVs in Lung Cancer Therapy
| Target/Study Models | Subject | Description | Reference |
|---|---|---|---|
| (Advanced) NSCLC | Vaccination trial with tumor antigen-loaded dendritic cell-derived exosomes | Maintenance immunotherapy in 47 patients with dexosomes to improve their PFS. | NCT01159288 |
| Solid tumors: primary liver cancer, SCLC, lymphoma, melanoma, multiple myeloma, renal cell carcinoma, NSCLC | Multicenter phase I study of MRX34, microRNA miR-RX34 liposomal injection | Phase I, open-label, multicenter, dose escalation study to investigate the safety, pharmacokinetics, and pharmacodynamics of the micro ribonucleic acid (microRNA) MRX34 in patients with unresectable primary liver cancer or advanced or metastatic cancer with or without liver involvement or hematologic malignancies. | NCT01829971 [147] |
| (Advanced) NSCLC | Phase I study of dexosome immunotherapy | Phase I study to evaluate safety and efficacy of autologous dexosomes loaded with tumor antigens (MAGE-A3, -A4, -A10, and MAGE-3DPO4), administered in 4 doses. Measurement of the immunologic responses and monitoring the clinical outcomes in 13 patients at different stages. | [167] |
| H1299 and A549 (NSCLC) | Nanosomes carrying doxorubicin anticancer activity against human lung cancer cells | In vitro analysis of gold nanoparticles (GNPs) loaded with doxorubicin to evaluate the release kinetics and the cytotoxic activity. | [173] |
| Mice injected with B16F10 cells to produce lung metastasis | EVs melanoma gold conjugated nanoparticle targeting lung tumors | The study provided an application system where exosomes isolated from cancer cells incorporated gold nanoparticles were tested in a mouse model to improve targeting system in metastatic foci. | [174] |
| In vitro: murine carcinoma cell line (3LL-M27); in vivo: mouse model with pulmonary metastases | Paclitaxel-loaded EVs against cancer cells | In vitro and in vivo study aims to introduce a new formulation for Paclitaxel distribution through exosomes (PTX-exo, fom RAW 264.7 cell line), providing high stability in tumor environment and a better effectiveness in vivo murine model. | [171] |
| In vitro: A549 and H1299 (NSCLC); In vivo: mouse model with lung cancer xenograft | Celastrol EVs formulation against lung cancer | Study focused on the effect of the natural compound celastrol loaded into exosomes, a new delivery system improved efficacy and reduced dose toxicity. | [175] |
| In vitro: A549 and H1299 (NSCLC); In vivo: nude mice with xenograft | Anthocyanidins EVs against multiple cancer types | The study aimed to obtain a nano-formulation of the natural derived compound, anthos, with exosomes. Exosomes enhanced the anti-proliferative and anti-inflammatory activity of anthos (vs the free compound) and the therapeutic affect toward lung cancer. | [176] |
| Nude mice with lung tumor xenografts | Milk-derived exosomes for oral delivery of paclitaxel | A study on chemotherapeutic paclitaxel delivery through exosomes in a formulation for oral administration, which exhibited greater therapeutic efficacy and lower systemic toxicity. | [177] |
5. Conclusions and Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef]
- Miceli, V.; Zito, G.; Bulati, M.; Gallo, A.; Busa, R.; Iannolo, G.; Conaldi, P.G. Different priming strategies improve distinct therapeutic capabilities of mesenchymal stromal/stem cells: Potential implications for their clinical use. World J. Stem Cells 2023, 15, 400–420. [Google Scholar] [CrossRef]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011, 71, 5346–5356. [Google Scholar] [CrossRef]
- Zhang, C.; Qin, C.; Dewanjee, S.; Bhattacharya, H.; Chakraborty, P.; Jha, N.K.; Gangopadhyay, M.; Jha, S.K.; Liu, Q. Tumor-derived small extracellular vesicles in cancer invasion and metastasis: Molecular mechanisms, and clinical significance. Mol. Cancer 2024, 23, 18. [Google Scholar] [CrossRef]
- Jothimani, G.; Pathak, S.; Dutta, S.; Duttaroy, A.K.; Banerjee, A. A Comprehensive Cancer-Associated MicroRNA Expression Profiling and Proteomic Analysis of Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosomes. Tissue Eng. Regen. Med. 2022, 19, 1013–1031. [Google Scholar] [CrossRef]
- Xiao, H.; Lasser, C.; Shelke, G.V.; Wang, J.; Radinger, M.; Lunavat, T.R.; Malmhall, C.; Lin, L.H.; Li, J.; Li, L.; et al. Mast cell exosomes promote lung adenocarcinoma cell proliferation—Role of KIT-stem cell factor signaling. Cell Commun. Signal. CCS 2014, 12, 64. [Google Scholar] [CrossRef]
- Bhatta, B.; Luz, I.; Krueger, C.; Teo, F.X.; Lane, D.P.; Sabapathy, K.; Cooks, T. Cancer Cells Shuttle Extracellular Vesicles Containing Oncogenic Mutant p53 Proteins to the Tumor Microenvironment. Cancers 2021, 13, 2985. [Google Scholar] [CrossRef]
- Di Giuseppe, F.; Carluccio, M.; Zuccarini, M.; Giuliani, P.; Ricci-Vitiani, L.; Pallini, R.; De Sanctis, P.; Di Pietro, R.; Ciccarelli, R.; Angelucci, S. Proteomic Characterization of Two Extracellular Vesicle Subtypes Isolated from Human Glioblastoma Stem Cell Secretome by Sequential Centrifugal Ultrafiltration. Biomedicines 2021, 9, 146. [Google Scholar] [CrossRef]
- Cammarata, G.; de Miguel-Perez, D.; Russo, A.; Peleg, A.; Dolo, V.; Rolfo, C.; Taverna, S. Emerging noncoding RNAs contained in extracellular vesicles: Rising stars as biomarkers in lung cancer liquid biopsy. Ther. Adv. Med. Oncol. 2022, 14, 17588359221131229. [Google Scholar] [CrossRef]
- Kato, T.; Vykoukal, J.V.; Fahrmann, J.F.; Hanash, S. Extracellular Vesicles in Lung Cancer: Prospects for Diagnostic and Therapeutic Applications. Cancers 2021, 13, 4604. [Google Scholar] [CrossRef]
- Liu, Y.; Xia, Y.; Smollar, J.; Mao, W.; Wan, Y. The roles of small extracellular vesicles in lung cancer: Molecular pathology, mechanisms, diagnostics, and therapeutics. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188539. [Google Scholar] [CrossRef]
- Tine, M.; Biondini, D.; Damin, M.; Semenzato, U.; Bazzan, E.; Turato, G. Extracellular Vesicles in Lung Cancer: Bystanders or Main Characters? Biology 2023, 12, 246. [Google Scholar] [CrossRef]
- Ginini, L.; Billan, S.; Fridman, E.; Gil, Z. Insight into Extracellular Vesicle-Cell Communication: From Cell Recognition to Intracellular Fate. Cells 2022, 11, 1375. [Google Scholar] [CrossRef]
- Alberti, G.; Russo, E.; Corrao, S.; Anzalone, R.; Kruzliak, P.; Miceli, V.; Conaldi, P.G.; Di Gaudio, F.; La Rocca, G. Current Perspectives on Adult Mesenchymal Stromal Cell-Derived Extracellular Vesicles: Biological Features and Clinical Indications. Biomedicines 2022, 10, 2822. [Google Scholar] [CrossRef]
- Russo, E.; Alberti, G.; Corrao, S.; Borlongan, C.V.; Miceli, V.; Conaldi, P.G.; Di Gaudio, F.; La Rocca, G. The Truth Is Out There: Biological Features and Clinical Indications of Extracellular Vesicles from Human Perinatal Stem Cells. Cells 2023, 12, 2347. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Di Bella, M.A. Overview and Update on Extracellular Vesicles: Considerations on Exosomes and Their Application in Modern Medicine. Biology 2022, 11, 804. [Google Scholar] [CrossRef]
- Sailliet, N.; Ullah, M.; Dupuy, A.; Silva, A.K.A.; Gazeau, F.; Le Mai, H.; Brouard, S. Extracellular Vesicles in Transplantation. Front. Immunol. 2022, 13, 800018. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. CCS 2021, 19, 47. [Google Scholar] [CrossRef]
- Guo, H.; Chitiprolu, M.; Roncevic, L.; Javalet, C.; Hemming, F.J.; Trung, M.T.; Meng, L.; Latreille, E.; Tanese de Souza, C.; McCulloch, D.; et al. Atg5 Disassociates the V(1)V(0)-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev. Cell 2017, 43, 716–730.e717. [Google Scholar] [CrossRef]
- Zubkova, E.; Kalinin, A.; Bolotskaya, A.; Beloglazova, I.; Menshikov, M. Autophagy-Dependent Secretion: Crosstalk between Autophagy and Exosome Biogenesis. Curr. Issues Mol. Biol. 2024, 46, 2209–2235. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021, 17, 163–177. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, aau6977. [Google Scholar] [CrossRef]
- Raposo, G.; Stahl, P.D. Extracellular vesicles—On the cusp of a new language in the biological sciences. Extracell. Vesicles Circ. Nucl. Acids 2023, 4, 240–254. [Google Scholar] [CrossRef]
- Cable, J.; Witwer, K.W.; Coffey, R.J.; Milosavljevic, A.; von Lersner, A.K.; Jimenez, L.; Pucci, F.; Barr, M.M.; Dekker, N.; Barman, B.; et al. Exosomes, microvesicles, and other extracellular vesicles-a Keystone Symposia report. Ann. N. Y. Acad. Sci. 2023, 1523, 24–37. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef]
- Clancy, J.W.; Sheehan, C.S.; Boomgarden, A.C.; D’Souza-Schorey, C. Recruitment of DNA to tumor-derived microvesicles. Cell Rep. 2022, 38, 110443. [Google Scholar] [CrossRef] [PubMed]
- Schmidtmann, M.; D’Souza-Schorey, C. Extracellular Vesicles: Biological Packages That Modulate Tumor Cell Invasion. Cancers 2023, 15, 5617. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Ferrari, P.; Biava, P.M. Exosomes and Cell Communication: From Tumour-Derived Exosomes and Their Role in Tumour Progression to the Use of Exosomal Cargo for Cancer Treatment. Cancers 2021, 13, 822. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lasser, C.; Szabo, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzas, E.I.; Lotvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2, 20677. [Google Scholar] [CrossRef]
- Liu, D.; Kou, X.; Chen, C.; Liu, S.; Liu, Y.; Yu, W.; Yu, T.; Yang, R.; Wang, R.; Zhou, Y.; et al. Circulating apoptotic bodies maintain mesenchymal stem cell homeostasis and ameliorate osteopenia via transferring multiple cellular factors. Cell Res. 2018, 28, 918–933. [Google Scholar] [CrossRef]
- Tang, H.; Luo, H.; Zhang, Z.; Yang, D. Mesenchymal Stem Cell-Derived Apoptotic Bodies: Biological Functions and Therapeutic Potential. Cells 2022, 11, 3879. [Google Scholar] [CrossRef]
- Li, A.; Zhang, T.; Zheng, M.; Liu, Y.; Chen, Z. Exosomal proteins as potential markers of tumor diagnosis. J. Hematol. Oncol. 2017, 10, 175. [Google Scholar] [CrossRef]
- Ghanam, J.; Chetty, V.K.; Barthel, L.; Reinhardt, D.; Hoyer, P.F.; Thakur, B.K. DNA in extracellular vesicles: From evolution to its current application in health and disease. Cell Biosci. 2022, 12, 37. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef]
- Soekmadji, C.; Li, B.; Huang, Y.; Wang, H.; An, T.; Liu, C.; Pan, W.; Chen, J.; Cheung, L.; Falcon-Perez, J.M.; et al. The future of Extracellular Vesicles as Theranostics—An ISEV meeting report. J. Extracell. Vesicles 2020, 9, 1809766. [Google Scholar] [CrossRef]
- Berckmans, R.J.; Nieuwland, R.; Boing, A.N.; Romijn, F.P.; Hack, C.E.; Sturk, A. Cell-derived microparticles circulate in healthy humans and support low grade thrombin generation. Thromb. Haemost. 2001, 85, 639–646. [Google Scholar]
- Zhang, J.; Li, H.; Fan, B.; Xu, W.; Zhang, X. Extracellular vesicles in normal pregnancy and pregnancy-related diseases. J. Cell. Mol. Med. 2020, 24, 4377–4388. [Google Scholar] [CrossRef]
- Akbar, N.; Azzimato, V.; Choudhury, R.P.; Aouadi, M. Extracellular vesicles in metabolic disease. Diabetologia 2019, 62, 2179–2187. [Google Scholar] [CrossRef]
- Bewicke-Copley, F.; Mulcahy, L.A.; Jacobs, L.A.; Samuel, P.; Akbar, N.; Pink, R.C.; Carter, D.R.F. Extracellular vesicles released following heat stress induce bystander effect in unstressed populations. J. Extracell. Vesicles 2017, 6, 1340746. [Google Scholar] [CrossRef]
- Chiaradia, E.; Tancini, B.; Emiliani, C.; Delo, F.; Pellegrino, R.M.; Tognoloni, A.; Urbanelli, L.; Buratta, S. Extracellular Vesicles under Oxidative Stress Conditions: Biological Properties and Physiological Roles. Cells 2021, 10, 1763. [Google Scholar] [CrossRef]
- Sproviero, D.; Gagliardi, S.; Zucca, S.; Arigoni, M.; Giannini, M.; Garofalo, M.; Fantini, V.; Pansarasa, O.; Avenali, M.; Ramusino, M.C.; et al. Extracellular Vesicles Derived from Plasma of Patients with Neurodegenerative Disease Have Common Transcriptomic Profiling. Front. Aging Neurosci. 2022, 14, 785741. [Google Scholar] [CrossRef]
- Thompson, A.G.; Gray, E.; Heman-Ackah, S.M.; Mager, I.; Talbot, K.; Andaloussi, S.E.; Wood, M.J.; Turner, M.R. Extracellular vesicles in neurodegenerative disease—Pathogenesis to biomarkers. Nat. Rev. Neurol. 2016, 12, 346–357. [Google Scholar] [CrossRef]
- Martucci, G.; Arcadipane, A.; Tuzzolino, F.; Occhipinti, G.; Panarello, G.; Carcione, C.; Bonicolini, E.; Vitiello, C.; Lorusso, R.; Conaldi, P.G.; et al. Identification of a Circulating miRNA Signature to Stratify Acute Respiratory Distress Syndrome Patients. J. Pers. Med. 2020, 11, 15. [Google Scholar] [CrossRef]
- Charla, E.; Mercer, J.; Maffia, P.; Nicklin, S.A. Extracellular vesicle signalling in atherosclerosis. Cell Signal. 2020, 75, 109751. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Moreno, L.; Mleczko, J.; Palomo, L.; Gonzalez, E.; Cabrera, D.; Cogolludo, A.; Vizcaino, F.P.; van-Liempd, S.; Falcon-Perez, J.M. Hepatocyte-secreted extracellular vesicles modify blood metabolome and endothelial function by an arginase-dependent mechanism. Sci. Rep. 2017, 7, 42798. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Hou, J.; Yang, C.; Wang, H.; Wu, S.; Wu, Y.; Zhao, X.; Lu, C. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery. J. Exp. Clin. Cancer Res. CR 2019, 38, 62. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Chen, H.; He, K.; Wang, J. Human bone marrow mesenchymal stem cells-derived exosomes attenuated prostate cancer progression via the miR-99b-5p/IGF1R axis. Bioengineered 2022, 13, 2004–2016. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yan, G.; Yue, M.; Wang, L. Extracellular vesicles-derived microRNA-222 promotes immune escape via interacting with ATF3 to regulate AKT1 transcription in colorectal cancer. BMC Cancer 2021, 21, 349. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, A.; Alini, M.; Baghaban Eslaminejad, M.; Hosseini, S. Engineering strategies for customizing extracellular vesicle uptake in a therapeutic context. Stem Cell Res. Ther. 2022, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Yue, S.; Stadel, D.; Zoller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Pazos, R.; Royo, F.; Gonzalez, E.; Roura-Ferrer, M.; Martinez, A.; Gamiz, J.; Reichardt, N.C.; Falcon-Perez, J.M. Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci. Rep. 2019, 9, 11920. [Google Scholar] [CrossRef] [PubMed]
- Limoni, S.K.; Moghadam, M.F.; Moazzeni, S.M.; Gomari, H.; Salimi, F. Engineered Exosomes for Targeted Transfer of siRNA to HER2 Positive Breast Cancer Cells. Appl. Biochem. Biotechnol. 2019, 187, 352–364. [Google Scholar] [CrossRef]
- Cheng, Q.; Shi, X.; Han, M.; Smbatyan, G.; Lenz, H.J.; Zhang, Y. Reprogramming Exosomes as Nanoscale Controllers of Cellular Immunity. J. Am. Chem. Soc. 2018, 140, 16413–16417. [Google Scholar] [CrossRef]
- Hong, Y.; Kim, Y.K.; Kim, G.B.; Nam, G.H.; Kim, S.A.; Park, Y.; Yang, Y.; Kim, I.S. Degradation of tumour stromal hyaluronan by small extracellular vesicle-PH20 stimulates CD103(+) dendritic cells and in combination with PD-L1 blockade boosts anti-tumour immunity. J. Extracell. Vesicles 2019, 8, 1670893. [Google Scholar] [CrossRef]
- Lee, T.H.; Chennakrishnaiah, S.; Audemard, E.; Montermini, L.; Meehan, B.; Rak, J. Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem. Biophys. Res. Commun. 2014, 451, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Miceli, V.; Bulati, M.; Iannolo, G.; Zito, G.; Gallo, A.; Conaldi, P.G. Therapeutic Properties of Mesenchymal Stromal/Stem Cells: The Need of Cell Priming for Cell-Free Therapies in Regenerative Medicine. Int. J. Mol. Sci. 2021, 22, 763. [Google Scholar] [CrossRef] [PubMed]
- Miceli, V.; Bertani, A. Mesenchymal Stromal/Stem Cells and Their Products as a Therapeutic Tool to Advance Lung Transplantation. Cells 2022, 11, 826. [Google Scholar] [CrossRef]
- Bulati, M.; Gallo, A.; Zito, G.; Busa, R.; Iannolo, G.; Cuscino, N.; Castelbuono, S.; Carcione, C.; Centi, C.; Martucci, G.; et al. 3D Culture and Interferon-gamma Priming Modulates Characteristics of Mesenchymal Stromal/Stem Cells by Modifying the Expression of Both Intracellular and Exosomal microRNAs. Biology 2023, 12, 1063. [Google Scholar] [CrossRef]
- Bulati, M.; Miceli, V.; Gallo, A.; Amico, G.; Carcione, C.; Pampalone, M.; Conaldi, P.G. The Immunomodulatory Properties of the Human Amnion-Derived Mesenchymal Stromal/Stem Cells Are Induced by INF-gamma Produced by Activated Lymphomonocytes and Are Mediated by Cell-To-Cell Contact and Soluble Factors. Front. Immunol. 2020, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Miceli, V.; Pampalone, M.; Vella, S.; Carreca, A.P.; Amico, G.; Conaldi, P.G. Comparison of Immunosuppressive and Angiogenic Properties of Human Amnion-Derived Mesenchymal Stem Cells between 2D and 3D Culture Systems. Stem Cells Int. 2019, 2019, 7486279. [Google Scholar] [CrossRef]
- Miceli, V.; Chinnici, C.M.; Bulati, M.; Pampalone, M.; Amico, G.; Schmelzer, E.; Gerlach, J.C.; Conaldi, P.G. Comparative study of the production of soluble factors in human placenta-derived mesenchymal stromal/stem cells grown in adherent conditions or as aggregates in a catheter-like device. Biochem. Biophys. Res. Commun. 2020, 522, 171–176. [Google Scholar] [CrossRef]
- Liang, B.; Liang, J.M.; Ding, J.N.; Xu, J.; Xu, J.G.; Chai, Y.M. Dimethyloxaloylglycine-stimulated human bone marrow mesenchymal stem cell-derived exosomes enhance bone regeneration through angiogenesis by targeting the AKT/mTOR pathway. Stem Cell Res. Ther. 2019, 10, 335. [Google Scholar] [CrossRef]
- Lu, Z.; Chen, Y.; Dunstan, C.; Roohani-Esfahani, S.; Zreiqat, H. Priming Adipose Stem Cells with Tumor Necrosis Factor-Alpha Preconditioning Potentiates Their Exosome Efficacy for Bone Regeneration. Tissue Eng. Part A 2017, 23, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Casagrande, G.M.S.; Silva, M.O.; Reis, R.M.; Leal, L.F. Liquid Biopsy for Lung Cancer: Up-to-Date and Perspectives for Screening Programs. Int. J. Mol. Sci. 2023, 24, 2505. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Hill, A.F. Therapeutically harnessing extracellular vesicles. Nat. Rev. Drug Discov. 2022, 21, 379–399. [Google Scholar] [CrossRef] [PubMed]
- Albanese, M.; Chen, Y.A.; Huls, C.; Gartner, K.; Tagawa, T.; Mejias-Perez, E.; Keppler, O.T.; Gobel, C.; Zeidler, R.; Shein, M.; et al. MicroRNAs are minor constituents of extracellular vesicles that are rarely delivered to target cells. PLoS Genet. 2021, 17, e1009951. [Google Scholar] [CrossRef] [PubMed]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.A.; Andahur, E.I.; Valenzuela, R.; Castellon, E.A.; Fulla, J.A.; Ramos, C.G.; Trivino, J.C. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget 2016, 7, 3993–4008. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.A.; Hur, J.Y.; Kim, H.J.; Kim, W.S.; Lee, K.Y. Extracellular Vesicle-Based Bronchoalveolar Lavage Fluid Liquid Biopsy for EGFR Mutation Testing in Advanced Non-Squamous NSCLC. Cancers 2022, 14, 2744. [Google Scholar] [CrossRef]
- Girard, N. Optimizing outcomes in EGFR mutation-positive NSCLC: Which tyrosine kinase inhibitor and when? Future Oncol. 2018, 14, 1117–1132. [Google Scholar] [CrossRef]
- Kim, I.A.; Hur, J.Y.; Kim, H.J.; Kim, W.S.; Lee, K.Y. A prospective phase 2 study of expeditious EGFR genotyping and immediate therapeutic initiation through extracellular vesicles (EV)-based bronchoalveolar lavage fluid (BALF) liquid biopsy in advanced NSCLC patients. Transl. Lung Cancer Res. 2023, 12, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Zhu, Y.; Zhang, J.; Zhang, W.; Wang, H.; Chen, H.; Wu, C.; Ni, J.; Xu, X.; Nian, B.; et al. Identification and evaluation of circulating small extracellular vesicle microRNAs as diagnostic biomarkers for patients with indeterminate pulmonary nodules. J. Nanobiotechnol. 2022, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guo, W.; Liu, T.; Liang, N.; Ma, Q.; Gao, Y.; Tan, F.; Xue, Q.; He, J. Plasma extracellular vesicle microRNA profiling and the identification of a diagnostic signature for stage I lung adenocarcinoma. Cancer Sci. 2022, 113, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Eom, J.S.; Kim, W.Y.; Jo, E.J.; Mok, J.; Lee, K.; Kim, K.U.; Park, H.K.; Lee, M.K.; Kim, M.H. Diagnostic value of microRNAs derived from exosomes in bronchoalveolar lavage fluid of early-stage lung adenocarcinoma: A pilot study. Thorac. Cancer 2018, 9, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS ONE 2016, 11, e0148407. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, 1413. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Yao, J.; Wang, Y.; Ni, B. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 2022, 29, 1257–1271. [Google Scholar] [CrossRef]
- Tan, Z.; Cao, F.; Jia, B.; Xia, L. Circ_0072088 promotes the development of non-small cell lung cancer via the miR-377-5p/NOVA2 axis. Thorac. Cancer 2020, 11, 2224–2236. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, Q.; Sun, J.; Xue, J.; Wang, C. Circular RNA circ_0000376 promotes paclitaxel resistance and tumorigenesis of non-small cell lung cancer via positively modulating KPNA4 by sponging miR-1298-5p. Thorac. Cancer 2023, 14, 2116–2126. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, L.; Wang, Y. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by sponging miR-590-5p to regulate THBS2. Cancer Cell Int. 2020, 20, 387. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Zeng, J.; Rong, F.; Xu, Y.; Wei, R.; Zou, C. Circ_0020123 enhances the cisplatin resistance in non-small cell lung cancer cells partly by sponging miR-140-3p to regulate homeobox B5 (HOXB5). Bioengineered 2022, 13, 5126–5140. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wang, C.; Wang, L.; Zhang, J. Circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8. Open Med. 2022, 17, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Du, T.; Chen, X.; Hu, P. Circ-PDZD8 promotes cell growth and glutamine metabolism in non-small cell lung cancer by enriching LARP1 via sequestering miR-330-5p. Thorac. Cancer 2023, 14, 2187–2197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, T.; Yuan, S.; Long, Y.; Tan, S.; Niu, G.; Zhang, P.; Yang, M. Circ_0020123 plays an oncogenic role in non-small cell lung cancer depending on the regulation of miR-512-3p/CORO1C. Thorac. Cancer 2022, 13, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Yang, X.; Song, W.; Yu, N.; Lin, Q. Tanshinone IIA (TSIIA) represses the progression of non-small cell lung cancer by the circ_0020123/miR-1299/HMGB3 pathway. Mol. Cell. Biochem. 2023, 478, 1973–1986. [Google Scholar] [CrossRef] [PubMed]
- de Fraipont, F.; Gazzeri, S.; Cho, W.C.; Eymin, B. Circular RNAs and RNA Splice Variants as Biomarkers for Prognosis and Therapeutic Response in the Liquid Biopsies of Lung Cancer Patients. Front. Genet. 2019, 10, 390. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Pedraz-Valdunciel, C.; Giannoukakos, S.; Gimenez-Capitan, A.; Fortunato, D.; Filipska, M.; Bertran-Alamillo, J.; Bracht, J.W.P.; Drozdowskyj, A.; Valarezo, J.; Zarovni, N.; et al. Multiplex Analysis of CircRNAs from Plasma Extracellular Vesicle-Enriched Samples for the Detection of Early-Stage Non-Small Cell Lung Cancer. Pharmaceutics 2022, 14, 2034. [Google Scholar] [CrossRef]
- Pedraz-Valdunciel, C.; Giannoukakos, S.; Potie, N.; Gimenez-Capitan, A.; Huang, C.Y.; Hackenberg, M.; Fernandez-Hilario, A.; Bracht, J.; Filipska, M.; Aldeguer, E.; et al. Digital multiplexed analysis of circular RNAs in FFPE and fresh non-small cell lung cancer specimens. Mol. Oncol. 2022, 16, 2367–2383. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shen, L.; Xia, Q.; Tao, H.; Liu, Z.; Wang, M.; Zhang, X.; Zhang, J.; Lv, J. Extracellular vesicle-derived circHIPK3: Novel diagnostic biomarker for lung cancer. Adv. Med. Sci. 2023, 68, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Jiang, J.; Qian, H.; Yan, Y.; Xu, W. Exosomal circRNA: Emerging insights into cancer progression and clinical application potential. J. Hematol. Oncol. 2023, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Cammarata, G.; Barraco, N.; Giusti, I.; Gristina, V.; Dolo, V.; Taverna, S. Extracellular Vesicles-ceRNAs as Ovarian Cancer Biomarkers: Looking into circRNA-miRNA-mRNA Code. Cancers 2022, 14, 3404. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Wang, J.; Liu, L.; Zou, C.; Zhao, Y.; Xue, Z.; Sun, X.; Jiang, T.; Song, J. Presence and prospects of exosomal circRNAs in cancer (Review). Int. J. Oncol. 2023, 62, 5495. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, X.; Zhang, H.; Wei, S.; Chen, Y.; Chen, Y.; Wang, F.; Fan, X.; Han, S.; Wu, G. Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem. Biophys. Res. Commun. 2018, 496, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3053–3060. [Google Scholar] [CrossRef]
- Mo, W.L.; Deng, L.J.; Cheng, Y.; Yu, W.J.; Yang, Y.H.; Gu, W.D. Circular RNA hsa_circ_0072309 promotes tumorigenesis and invasion by regulating the miR-607/FTO axis in non-small cell lung carcinoma. Aging 2021, 13, 11629–11645. [Google Scholar] [CrossRef]
- Wan, Z.; Jia, S.; Lu, J.; Ge, X.; Chen, Q. circ-ATAD1 as Competing Endogenous RNA for miR-191-5p Forces Non-small Cell Lung Cancer Progression. Appl. Biochem. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Z.; Tian, H. Hsa_circ_0092887 targeting miR-490-5p/UBE2T promotes paclitaxel resistance in non-small cell lung cancer. J. Clin. Lab. Anal. 2023, 37, e24781. [Google Scholar] [CrossRef]
- Jiang, M.M.; Mai, Z.T.; Wan, S.Z.; Chi, Y.M.; Zhang, X.; Sun, B.H.; Di, Q.G. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J. Cancer Res. Clin. Oncol. 2018, 144, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, X.; Wei, S.; Chen, Y.; Chen, Y.; Fan, X.; Han, S.; Wu, G. hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017, 284, 2170–2182. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tong, X.; Zhou, Z.; Wang, S.; Lei, Z.; Zhang, T.; Liu, Z.; Zeng, Y.; Li, C.; Zhao, J.; et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol. Cancer 2018, 17, 140. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, Y.; Mao, Q.; Xia, W.; Chen, B.; Shen, H.; Xu, L.; Jiang, F.; Dong, G. Circular RNA circCRIM1 inhibits invasion and metastasis in lung adenocarcinoma through the microRNA (miR)-182/miR-93-leukemia inhibitory factor receptor pathway. Cancer Sci. 2019, 110, 2960–2972. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, S.; Mao, Y.; Xu, J.; Yang, S.; Shen, H.; Xu, W.; Fan, W.; Wang, J. CircRNF13 regulates the invasion and metastasis in lung adenocarcinoma by targeting miR-93-5p. Gene 2018, 671, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Chao, F.; Zhang, Y.; Lv, L.; Wei, Y.; Dou, X.; Chang, N.; Yi, Q.; Li, M. Extracellular Vesicles Derived circSH3PXD2A Inhibits Chemoresistance of Small Cell Lung Cancer by miR-375-3p/YAP1. Int. J. Nanomed. 2023, 18, 2989–3006. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e1018. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, K.R.; Paulsen, B.S.; Baek, R.; Varming, K.; Sorensen, B.S.; Jorgensen, M.M. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J. Extracell. Vesicles 2015, 4, 26659. [Google Scholar] [CrossRef] [PubMed]
- Malla, R.R.; Pandrangi, S.; Kumari, S.; Gavara, M.M.; Badana, A.K. Exosomal tetraspanins as regulators of cancer progression and metastasis and novel diagnostic markers. Asia-Pac. J. Clin. Oncol. 2018, 14, 383–391. [Google Scholar] [CrossRef]
- Taverna, S.; Pucci, M.; Giallombardo, M.; Di Bella, M.A.; Santarpia, M.; Reclusa, P.; Gil-Bazo, I.; Rolfo, C.; Alessandro, R. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci. Rep. 2017, 7, 3170. [Google Scholar] [CrossRef]
- Thuya, W.L.; Kong, L.R.; Syn, N.L.; Ding, L.W.; Cheow, E.S.H.; Wong, R.T.X.; Wang, T.; Goh, R.M.W.; Song, H.; Jayasinghe, M.K.; et al. FAM3C in circulating tumor-derived extracellular vesicles promotes non-small cell lung cancer growth in secondary sites. Theranostics 2023, 13, 621–638. [Google Scholar] [CrossRef]
- Dou, X.; Hua, Y.; Chen, Z.; Chao, F.; Li, M. Extracellular vesicles containing PD-L1 contribute to CD8+ T-cell immune suppression and predict poor outcomes in small cell lung cancer. Clin. Exp. Immunol. 2022, 207, 307–317. [Google Scholar] [CrossRef]
- Xu, J.; Wang, L.; Yin, N.; Chen, A.; Yi, J.; Tang, J.; Xiang, J. Proteomic profiling of extracellular vesicles and particles reveals the cellular response to cisplatin in NSCLC. Thorac. Cancer 2021, 12, 2601–2610. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Kang, B.; Kim, O.Y.; Choi, D.S.; Lee, J.; Kim, S.R.; Go, G.; Yoon, Y.J.; Kim, J.H.; Jang, S.C.; et al. EVpedia: An integrated database of high-throughput data for systemic analyses of extracellular vesicles. J. Extracell. Vesicles 2013, 2, 20384. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.; Li, Y.; Zhang, H.; Hu, J.; Liao, J.; Su, Y.; Li, Q.; Chen, B.; Li, C.; Wang, Z.; et al. exoRBase 2.0: An atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 2022, 50, D118–D128. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Carvalho, A.S.; Moraes, M.C.S.; Hyun Na, C.; Fierro-Monti, I.; Henriques, A.; Zahedi, S.; Bodo, C.; Tranfield, E.M.; Sousa, A.L.; Farinho, A.; et al. Is the Proteome of Bronchoalveolar Lavage Extracellular Vesicles a Marker of Advanced Lung Cancer? Cancers 2020, 12, 3450. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Park, H.Y.; Hur, J.Y.; Kim, H.J.; Kim, I.A.; Kim, W.S.; Lee, K.Y. Genomic profiling of extracellular vesicle-derived DNA from bronchoalveolar lavage fluid of patients with lung adenocarcinoma. Transl. Lung Cancer Res. 2021, 10, 104–116. [Google Scholar] [CrossRef]
- Hur, J.Y.; Kim, H.J.; Lee, J.S.; Choi, C.M.; Lee, J.C.; Jung, M.K.; Pack, C.G.; Lee, K.Y. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol. Cancer 2018, 17, 15. [Google Scholar] [CrossRef]
- Park, J.; Lee, C.; Eom, J.S.; Kim, M.H.; Cho, Y.K. Detection of EGFR Mutations Using Bronchial Washing-Derived Extracellular Vesicles in Patients with Non-Small-Cell Lung Carcinoma. Cancers 2020, 12, 2822. [Google Scholar] [CrossRef] [PubMed]
- Javadi, J.; Gorgens, A.; Vanky, H.; Gupta, D.; Hjerpe, A.; El-Andaloussi, S.; Hagey, D.; Dobra, K. Diagnostic and Prognostic Utility of the Extracellular Vesicles Subpopulations Present in Pleural Effusion. Biomolecules 2021, 11, 1606. [Google Scholar] [CrossRef] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.F.; Faivre-Finn, C.; Mok, T.S.; Reck, M.; Van Schil, P.E.; Hellmann, M.D.; et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, iv192–iv237. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Wu, Y.L.; Ding, P.N.; Lord, S.J.; Inoue, A.; Zhou, C.; Mitsudomi, T.; Rosell, R.; Pavlakis, N.; Links, M.; et al. Impact of Specific Epidermal Growth Factor Receptor (EGFR) Mutations and Clinical Characteristics on Outcomes After Treatment with EGFR Tyrosine Kinase Inhibitors Versus Chemotherapy in EGFR-Mutant Lung Cancer: A Meta-Analysis. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 1958–1965. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, F.; Hirsch, F.R.; Pirker, R.; Felip, E.; Valencia, C.; Smit, E.F. The role of anti-EGFR therapies in EGFR-TKI-resistant advanced non-small cell lung cancer. Cancer Treat. Rev. 2024, 122, 102664. [Google Scholar] [CrossRef]
- Lei, Y.; Lei, Y.; Shi, X.; Wang, J. EML4-ALK fusion gene in non-small cell lung cancer. Oncol. Lett. 2022, 24, 277. [Google Scholar] [CrossRef]
- Baglivo, S.; Ricciuti, B.; Ludovini, V.; Metro, G.; Siggillino, A.; De Giglio, A.; Chiari, R. Dramatic Response to Lorlatinib in a Heavily Pretreated Lung Adenocarcinoma Patient Harboring G1202R Mutation and a Synchronous Novel R1192P ALK Point Mutation. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2018, 13, e145–e147. [Google Scholar] [CrossRef]
- Shaw, A.T.; Ou, S.H.; Bang, Y.J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.B.; et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef]
- Drilon, A.; Somwar, R.; Wagner, J.P.; Vellore, N.A.; Eide, C.A.; Zabriskie, M.S.; Arcila, M.E.; Hechtman, J.F.; Wang, L.; Smith, R.S.; et al. A Novel Crizotinib-Resistant Solvent-Front Mutation Responsive to Cabozantinib Therapy in a Patient with ROS1-Rearranged Lung Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 2351–2358. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Brambilla, E.; Faivre-Finn, C.; Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Primers 2021, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Su, J.; Li, F.L.; Chen, T.; Mayner, J.; Engler, A.; Ma, S.; Li, Q.; Guan, K.L. YAP silencing by RB1 mutation is essential for small-cell lung cancer metastasis. Nat. Commun. 2023, 14, 5916. [Google Scholar] [CrossRef] [PubMed]
- Wildey, G.; Shay, A.M.; McColl, K.S.; Yoon, S.; Shatat, M.A.; Perwez, A.; Spainhower, K.B.; Kresak, A.M.; Lipka, M.; Yang, M.; et al. Retinoblastoma Expression and Targeting by CDK4/6 Inhibitors in Small Cell Lung Cancer. Mol. Cancer Ther. 2023, 22, 264–273. [Google Scholar] [CrossRef]
- Paramanantham, A.; Asfiya, R.; Das, S.; McCully, G.; Srivastava, A. Extracellular Vesicle (EVs) Associated Non-Coding RNAs in Lung Cancer and Therapeutics. Int. J. Mol. Sci. 2022, 23, 13637. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Badami, E.; Carcione, C.; Chinnici, C.M.; Tinnirello, R.; Conaldi, P.G.; Iannolo, G. HCV Interplay with Mir34a: Implications in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 803278. [Google Scholar] [CrossRef]
- Vakhshiteh, F.; Rahmani, S.; Ostad, S.N.; Madjd, Z.; Dinarvand, R.; Atyabi, F. Exosomes derived from miR-34a-overexpressing mesenchymal stem cells inhibit in vitro tumor growth: A new approach for drug delivery. Life Sci. 2021, 266, 118871. [Google Scholar] [CrossRef]
- Shan, C.; Liang, Y.; Wang, K.; Li, P. Mesenchymal Stem Cell-Derived Extracellular Vesicles in Cancer Therapy Resistance: From Biology to Clinical Opportunity. Int. J. Biol. Sci. 2024, 20, 347–366. [Google Scholar] [CrossRef]
- Sohrabi, B.; Dayeri, B.; Zahedi, E.; Khoshbakht, S.; Nezamabadi Pour, N.; Ranjbar, H.; Davari Nejad, A.; Noureddini, M.; Alani, B. Mesenchymal stem cell (MSC)-derived exosomes as novel vehicles for delivery of miRNAs in cancer therapy. Cancer Gene Ther. 2022, 29, 1105–1116. [Google Scholar] [CrossRef]
- Wu, H.; Mu, X.; Liu, L.; Wu, H.; Hu, X.; Chen, L.; Liu, J.; Mu, Y.; Yuan, F.; Liu, W.; et al. Bone marrow mesenchymal stem cells-derived exosomal microRNA-193a reduces cisplatin resistance of non-small cell lung cancer cells via targeting LRRC1. Cell Death Dis. 2020, 11, 801. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ding, C.; Yang, X.; Zhao, J. BMSCs-Derived Exosomal MiR-126-3p Inhibits the Viability of NSCLC Cells by Targeting PTPN9. JBUON Off. J. Balk. Union Oncol. 2021, 26, 1832–1841. [Google Scholar]
- Li, X.; Wu, F. Mesenchymal stem cell-derived extracellular vesicles transfer miR-598 to inhibit the growth and metastasis of non-small-cell lung cancer by targeting THBS2. Cell Death Discov. 2023, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Yang, X.; Yang, H.; Lv, M.; Sun, X.; Zhou, B. Exosomal miR-338-3p suppresses non-small-cell lung cancer cells metastasis by inhibiting CHL1 through the MAPK signaling pathway. Cell Death Dis. 2021, 12, 1030. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Xu, M.; Wang, Z.; Yang, M. Engineered exosomes loaded with miR-449a selectively inhibit the growth of homologous non-small cell lung cancer. Cancer Cell Int. 2021, 21, 485. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Xie, X.; Zhang, D.; Zhou, Y.; Li, B.; Li, F.; Li, F.; Cheng, Y.; Mei, H.; Meng, H.; et al. Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale 2020, 12, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Anthiya, S.; Ozturk, S.C.; Yanik, H.; Tavukcuoglu, E.; Sahin, A.; Datta, D.; Charisse, K.; Alvarez, D.M.; Loza, M.I.; Calvo, A.; et al. Targeted siRNA lipid nanoparticles for the treatment of KRAS-mutant tumors. J. Control. Release Off. J. Control. Release Soc. 2023, 357, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.H.; Li, Y.; Zhang, J.; Rong, J.; Ye, S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Investig. 2013, 31, 330–335. [Google Scholar] [CrossRef] [PubMed]
- de Miguel-Perez, D.; Russo, A.; Arrieta, O.; Ak, M.; Barron, F.; Gunasekaran, M.; Mamindla, P.; Lara-Mejia, L.; Peterson, C.B.; Er, M.E.; et al. Extracellular vesicle PD-L1 dynamics predict durable response to immune-checkpoint inhibitors and survival in patients with non-small cell lung cancer. J. Exp. Clin. Cancer Res. CR 2022, 41, 186. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Q.; Liu, J.T.; Fan, L.L.; Liu, Y.; Cheng, L.; Wang, F.; Yu, H.Q.; Gao, J.; Wei, W.; Wang, H.; et al. Exosomes derived from gefitinib-treated EGFR-mutant lung cancer cells alter cisplatin sensitivity via up-regulating autophagy. Oncotarget 2016, 7, 24585–24595. [Google Scholar] [CrossRef]
- Li, W.; Hu, Y.; Jiang, T.; Han, Y.; Han, G.; Chen, J.; Li, X. Rab27A regulates exosome secretion from lung adenocarcinoma cells A549: Involvement of EPI64. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2014, 122, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guo, J.; Yang, M.; Zhu, X.; Cao, X. Chemokine-containing exosomes are released from heat-stressed tumor cells via lipid raft-dependent pathway and act as efficient tumor vaccine. J. Immunol. 2011, 186, 2219–2228. [Google Scholar] [CrossRef] [PubMed]
- Hirschowitz, E.A.; Foody, T.; Kryscio, R.; Dickson, L.; Sturgill, J.; Yannelli, J. Autologous dendritic cell vaccines for non-small-cell lung cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2004, 22, 2808–2815. [Google Scholar] [CrossRef] [PubMed]
- Markov, O.; Oshchepkova, A.; Mironova, N. Immunotherapy Based on Dendritic Cell-Targeted/-Derived Extracellular Vesicles-A Novel Strategy for Enhancement of the Anti-tumor Immune Response. Front. Pharmacol. 2019, 10, 1152. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.M.; Andre, F.; Amigorena, S.; Soria, J.C.; Eggermont, A.; Kroemer, G.; Zitvogel, L. Dendritic cell-derived exosomes for cancer therapy. J. Clin. Investig. 2016, 126, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.A.; Delcayre, A.; et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Escudier, B.; Angevin, E.; Tursz, T.; Zitvogel, L. Exosomes for cancer immunotherapy. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2004, 15 (Suppl. S4), iv141–iv144. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Yuan, D.; Deygen, I.; Klyachko, N.L.; Kabanov, A.V.; Batrakova, E.V. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: In vitro and in vivo evaluations. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 195–204. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, N.; Wang, J. M1 macrophage-derived exosome-encapsulated cisplatin can enhance its anti-lung cancer effect. Minerva Medica 2023, 114, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Amreddy, N.; Babu, A.; Panneerselvam, J.; Mehta, M.; Muralidharan, R.; Chen, A.; Zhao, Y.D.; Razaq, M.; Riedinger, N.; et al. Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells. Sci. Rep. 2016, 6, 38541. [Google Scholar] [CrossRef] [PubMed]
- Lara, P.; Palma-Florez, S.; Salas-Huenuleo, E.; Polakovicova, I.; Guerrero, S.; Lobos-Gonzalez, L.; Campos, A.; Munoz, L.; Jorquera-Cordero, C.; Varas-Godoy, M.; et al. Gold nanoparticle based double-labeling of melanoma extracellular vesicles to determine the specificity of uptake by cells and preferential accumulation in small metastatic lung tumors. J. Nanobiotechnol. 2020, 18, 20. [Google Scholar] [CrossRef]
- Aqil, F.; Kausar, H.; Agrawal, A.K.; Jeyabalan, J.; Kyakulaga, A.H.; Munagala, R.; Gupta, R. Exosomal formulation enhances therapeutic response of celastrol against lung cancer. Exp. Mol. Pathol. 2016, 101, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Agrawal, A.K.; Mudd, A.M.; Kyakulaga, A.H.; Singh, I.P.; Vadhanam, M.V.; Gupta, R.C. Exosomal formulation of anthocyanidins against multiple cancer types. Cancer Lett. 2017, 393, 94–102. [Google Scholar] [CrossRef]
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S.; et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Bari, E.; Ferrarotti, I.; Torre, M.L.; Corsico, A.G.; Perteghella, S. Mesenchymal stem/stromal cell secretome for lung regeneration: The long way through “pharmaceuticalization” for the best formulation. J. Control. Release Off. J. Control. Release Soc. 2019, 309, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Chinnici, C.M.; Russelli, G.; Bulati, M.; Miceli, V.; Gallo, A.; Busa, R.; Tinnirello, R.; Conaldi, P.G.; Iannolo, G. Mesenchymal stromal cell secretome in liver failure: Perspectives on COVID-19 infection treatment. World J. Gastroenterol. 2021, 27, 1905–1919. [Google Scholar] [CrossRef]
- Yao, X.; Liao, B.; Chen, F.; Liu, L.; Wu, K.; Hao, Y.; Li, Y.; Wang, Y.; Fan, R.; Yin, J.; et al. Comparison of proteomic landscape of extracellular vesicles in pleural effusions isolated by three strategies. Front. Bioeng. Biotechnol. 2023, 11, 1108952. [Google Scholar] [CrossRef]
- French, K.C.; Antonyak, M.A.; Cerione, R.A. Extracellular vesicle docking at the cellular port: Extracellular vesicle binding and uptake. Semin. Cell Dev. Biol. 2017, 67, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
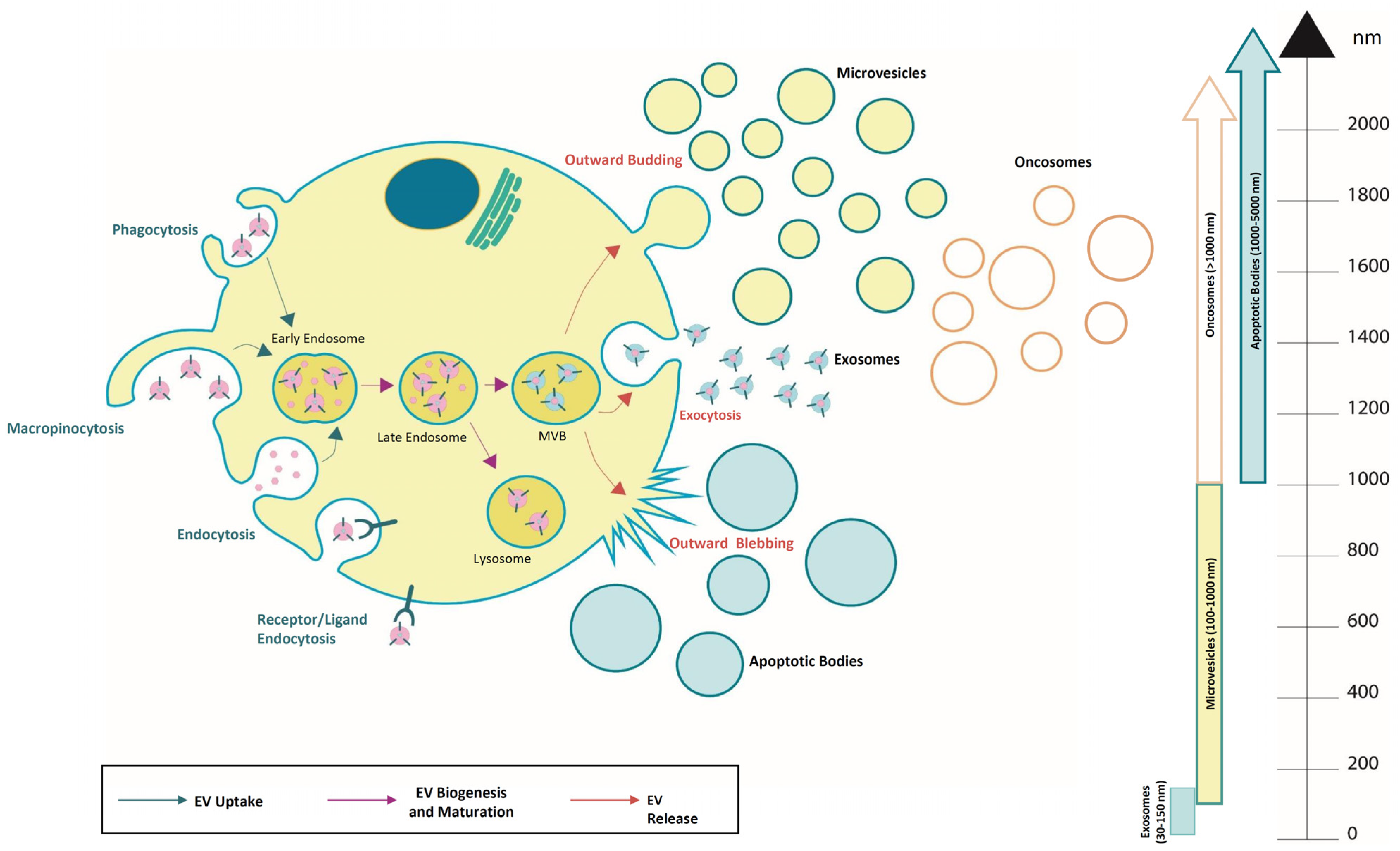

| Characteristics of Extracellular Vesicles (EVs) Subtypes | ||||
|---|---|---|---|---|
| EV Subtypes | Origin | Markers | Cargo | Reference |
| Exosomes | MVBs fuse with plasma membrane | CD63, CD81, CD9, HSP60, HSP70, Alix, TSG101 | Genomic DNA, mRNA, miRNA, circRNA, lncRNA, MHC class I and II | [23,24,25] |
| Microvesicles | Outward budding of plasma membrane | Anneximìn A1, Integrins, CD62, CD40 ligand | mRNA, miRNA, circRNA, lncRNA, Lipids, Adesion proteins | [26,27,28] |
| Oncosomes | Exclusively shed by cancer cells;Outward budding of plasma membrane | CAV-1, Keratin 18, ARF6, GAPDH | Genomic DNA, mRNA, miRNA, circRNA, lncRNA, MHC calss I and II | [29,30,31,32] |
| Apoptotic bodies | Outward blebbing from cells in apoptosis | Caspase 3, Annexin V, CD63, CD81 | miRNA, mRNA, Fragmented DNA, Histones | [33,34,35] |
| Disease | Body Fluid Samples Source | Description | Reference |
|---|---|---|---|
| Lung Cancer | BALF | LC-MS analysis of proteome profile. DNMT3B protein complex as potential therapeutic target. | [128] |
| Early-Stage Lung Adenocarcinoma | BALF | Quantitative analysis of miRNAs with diagnostic value. miR-126 and Let-7a possible diagnostic biomarkers: higher levels in lung adenocarcinoma than in control subjects. | [84] |
| Early-Stage Lung Adenocarcinoma/Invasive Stage Lung Adenocarcinoma | Plasma | A signature drawn up with four miRNAs (hsa-miR-106b-3p, hsa-miR-125a-5p, hsa-miR-3615, and hsa-miR-450b-5p) for early diagnosis. | [83] |
| Advanced-Stage Lung Adenocarcinoma | BALF | Next-Generation Sequencing (NGS) of EV DNA content to identify genetic alterations, suitable for a clinical approach. | [129] |
| (Advanced) NSCLC | BALF | EGFR mutation analysis on BALF EVs as method more accurate, specific and rapid than cfDNA evaluation. | [79] |
| (Advanced) NSCLC | Plasma and BALF | BALF EV DNA analysis as alternative diagnostic method in accordance with tissue biopsy and greater efficiency for detecting the p.T790 M mutation in the patients resistant to EGFR-TKIs. | [130] |
| (Advanced) NSCLC | BALF | A phase 2 study on BALF EV as platform for EGFR genotyping and rapid therapeutic intervention. | [81] |
| Adenocarcinoma, Squamous Cell Carcinoma, NSCLC | Bronchial Washing | Detection of EGFR mutation and evaluation of its prognostic value. | [131] |
| Early-Stage Malignant Pleural Mesothelioma (MPM) vs Benign Conditions and Metastatic Adenocarcinomas | Pleural Effusions | Characterization of surface marker or proteins differentially expressed as diagnostic markers. | [132] |
| Indeterminate Pulmonary Nodules (IPNs) | Plasma | CircEV-miR profile as a molecular model to distinguish the benign and malignant IPNs. miR-30c-5p, miR-30e-5p, miR-500a-3p, miR-125a-5p, and miR-99a-5p: five miRNAs differentially expressed and associated to an overall survival. | [82] Chinese Clinical Trials: ChiCTR1800019877 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carreca, A.P.; Tinnirello, R.; Miceli, V.; Galvano, A.; Gristina, V.; Incorvaia, L.; Pampalone, M.; Taverna, S.; Iannolo, G. Extracellular Vesicles in Lung Cancer: Implementation in Diagnosis and Therapeutic Perspectives. Cancers 2024, 16, 1967. https://doi.org/10.3390/cancers16111967
Carreca AP, Tinnirello R, Miceli V, Galvano A, Gristina V, Incorvaia L, Pampalone M, Taverna S, Iannolo G. Extracellular Vesicles in Lung Cancer: Implementation in Diagnosis and Therapeutic Perspectives. Cancers. 2024; 16(11):1967. https://doi.org/10.3390/cancers16111967
Chicago/Turabian StyleCarreca, Anna Paola, Rosaria Tinnirello, Vitale Miceli, Antonio Galvano, Valerio Gristina, Lorena Incorvaia, Mariangela Pampalone, Simona Taverna, and Gioacchin Iannolo. 2024. "Extracellular Vesicles in Lung Cancer: Implementation in Diagnosis and Therapeutic Perspectives" Cancers 16, no. 11: 1967. https://doi.org/10.3390/cancers16111967
APA StyleCarreca, A. P., Tinnirello, R., Miceli, V., Galvano, A., Gristina, V., Incorvaia, L., Pampalone, M., Taverna, S., & Iannolo, G. (2024). Extracellular Vesicles in Lung Cancer: Implementation in Diagnosis and Therapeutic Perspectives. Cancers, 16(11), 1967. https://doi.org/10.3390/cancers16111967








