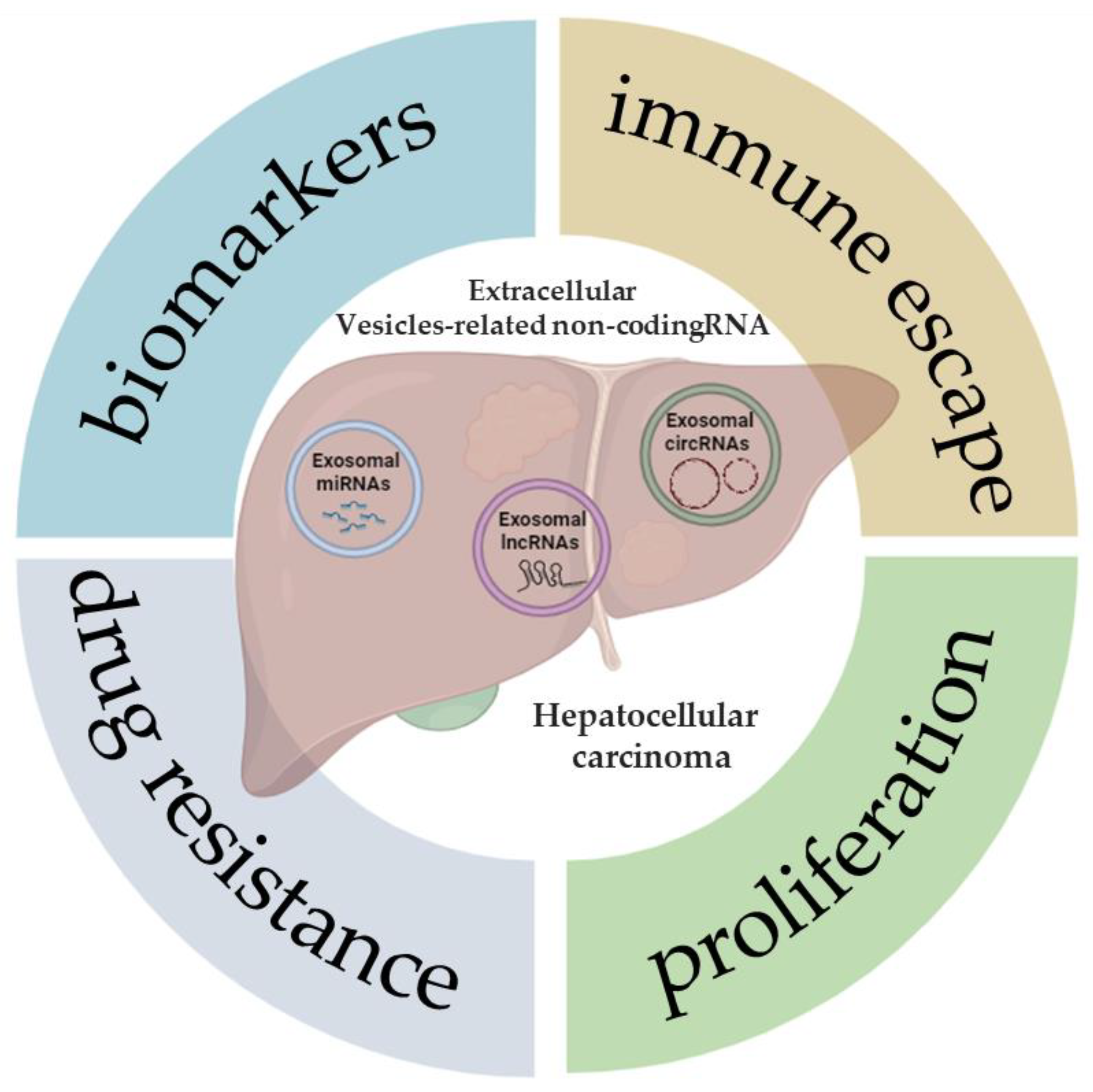
Extracellular Vesicle-Related Non-Coding RNAs in Hepatocellular Carcinoma: An Overview
Abstract
Simple Summary
Abstract
1. Introduction
2. miRNA-Extracellular Vesicles
2.1. miRNA-Extracellular Vesicles and Hepatocellular Carcinoma Progression
2.2. miRNA-Extracellular Vesicles and Immune Escape in Hepatocellular Carcinoma
2.3. miRNA-Extracellular Vesicles and Hepatocellular Carcinoma Drug Resistance
2.4. miRNA-Extracellular Vesicles as Biomarkers in Hepatocellular Carcinoma
| mRNA | Gene Target | Function | Type of Extraction | Reference |
|---|---|---|---|---|
| miR-9-3p | HBGF-5 | cell proliferation | ultracentrifugation | [62] |
| miR-27° | PPAR-γ | cell proliferation | ultracentrifugation | [61] |
| miR-429 | RBBP4 | cell formation/progression | ultracentrifugation | [63] |
| miR-221 | p27/Kip1 | cell proliferation/development | ultracentrifugation | [65] |
| miR-665 | MAPK/ERK | cell proliferation/biomarker | precipitation | [66] |
| miR-21 | PTEN | cell proliferation/progression | ultracentrifugation | [67] |
| miR-93 | TIMP2/TP53INP1/ CDKN1A | cell proliferation/invasion | precipitation | [68] |
| miR-3129 | TXNIP | cell proliferation/ EMT/biomarker | ultracentrifugation | [69] |
| miR-23a-3p | PTEN | immune escape | - | [72] |
| miR-146a-5p | - | immune escape | ultracentrifugation | [73] |
| miR-452-5p | TIMP3 | immune escape | gradient centrifugation | [74] |
| miR-21-5p | RhoB | immune escape/biomarker | ultrafiltration/ ultracentrifugation/ precipitation | [75,88] |
| miRNA let-7b | IL-6 | immune escape | ultracentrifugation | [76] |
| miR-92b | CD69 | immune escape/angiogenesis/metastasis | precipitation | [77] |
| miR-25 | p53 | drug resistance | ultracentrifugation | [81] |
| miR-744 | PAX2 | cell proliferation/drug resistance | ultracentrifugation | [82] |
| miR-1247-3p | B4GALT3 | cell proliferation/EMT/drug resistance | ultracentrifugation | [83] |
| miR-224 | GNMT | cell proliferation/biomarker | precipitation | [87] |
| miR-92°-3p | PTEN | cell proliferation/EMT/metastasis/ biomarker | precipitation | [88] |
| miR-4661-5p | IL10 | biomarker | precipitation | [89] |
| miR-10b-5p | - | biomarker | ultracentrifugation | [90] |
| miR-19-3-p | - | biomarker | precipitation | [91] |
| miR-23a | - | cell proliferation/biomarker | ultracentrifugation | [92] |
3. lncRNA-Extracellular Vesicles
3.1. lncRNA-Extracellular Vesicles and Hepatocellular Carcinoma Tumor Progression
3.2. lncRNA-Extracellular Vesicles as Biomarkers in Hepatocellular Carcinoma
3.3. lncRNA-Extracellular Vesicles and Immune Escape
3.4. lncRNA-Extracellular Vesicles and Drug Resistance in Hepatocellular Carcinoma
| lncRNA | Gene Target | Function | Type of Extraction | Reference |
|---|---|---|---|---|
| lncRNA FAL1 | miR-1236 | cell proliferation/migration | precipitation | [99] |
| lncRNA FAM138B | miR-765 | cell proliferation/migration/invasion | gradient centrifugation | [100] |
| lncRNA RP11-85G21.1 (lnc85) | miR-324-5p | cell proliferation/migration | precipitation | [101] |
| LINC00161 | miR-590-3p | tumorigenesis/metastasis | precipitation | [102] |
| lncRNA ASMTL-AS1 | miR-342-3p | cell proliferation/migration/invasion EMT | precipitation | [103] |
| lncH19 | miR-520a-3p | tumor development | precipitation | [104] |
| lncRNA MKLN1-AS | miR-22-3p | cell growth/angiogenesis/ migration/ | precipitation | [105,120] |
| Linc-ROR | HIF | regulate hypoxia condition/ drug resistance | ultracentrifugation | [106] |
| lncRNA TUG1 | PTEN | migration/invasion/glycolysis | ultracentrifugation | [107] |
| lnc HEIH | cell proliferation/metastasis/biomarker | ultracentrifugation and density gradient separation | [108,109,120] | |
| lncTuc339 | cell proliferation/metastasis Immune escape | ultracentrifugation and density gradient separation | [108,109] | |
| Lnc HOTAIR | cell proliferation/metastasis | ultracentrifugation and density gradient separation | [108,110] | |
| TMCC1-AS1, AL031985.3, LINC01138, AC099850.3 | biomarkers | precipitation | [111] | |
| DLEU2, HOTTIP, MALAT1, SNHG1 | biomarkers | precipitation | [112] | |
| lncSENP3-EIF4A1 | miR-9-5p | immune escape | precipitation | [114] |
| lncRNA CRNDE | cell proliferation/migration/invasion biomarker | precipitation | [115] | |
| THEMIS2-211 | miR-940, modulation of proteoglycan SPOK1 | proliferation/migration/invasion/EMT/biomarker | ultracentrifugation | [117] |
| PRKACA-202 | biomarker | ultracentrifugation | [117] | |
| CTD-2116N20.1 | biomarker cell proliferation/tumor metastasis | [118] | ||
| RP11-136I14.5 | biomarker | [118] | ||
| RP11-583F2.2 | miR-1298 | biomarker | precipitation | [119] |
| ENSG00000248932.1, ENST00000440688.1, ENST00000457302.2 | biomarkers | precipitation | [121] | |
| ENSG00000258332.1 (LINC02394), LINC00635 | biomarkers | precipitation | [122] | |
| lncRNA-ATB | biomarkers | precipitation | [123] | |
| lnc-FAM72D-3, lnc-GPR89B-15, lncZEB2-19 | Has-miR-5787 | biomarkers/oncogene | ultracentrifugation | [124] |
| lnc-EPC1-4 | SLCO1B1v and miR-29b-1-5p | biomarker/tumor suppressor | ultracentrifugation | [124] |
| lncRNA-VLDLR | drug resistance | ultracentrifugation | [129] |
4. circRNA-Extracellular Vesicles
4.1. circRNA-Extracellular Vesicles and Hepatocellular Carcinoma Tumor Progression
4.2. circRNA-Extracellular Vesicles and Immune Escape
4.3. circRNA-Extracellular Vesicles and Drug Resistance in Hepatocellular Carcinoma
4.4. circRNA-Extracellular Vesicles as Biomarkers in Hepatocellular Carcinoma
| circRNA | Gene Target | Function | Type of Extraction | Reference |
|---|---|---|---|---|
| circRNA_100284 | miR-217 | cell proliferation | precipitation | [138] |
| circRNA Cdr1 | miR-1270 | cell proliferation/invasion | precipitation | [139] |
| circ_0061395 | miR-877 | cell proliferation | precipitation | [140] |
| circTTLL5 | miR-136-5p | cell proliferation | precipitation | [141] |
| circ_0003028 | miR-498 | cell proliferation/EMT | precipitation | [144] |
| circWDR25 | miR-4474-3p | cell proliferation/EMT | ultracentrifugation | [145] |
| circ-0004277 | ZO-1 | cell proliferation/EMT | precipitation | [146] |
| circ_002136 | miR-19a-3p | cell proliferation | precipitation | [147] |
| circWHSC1 | miR-142-3p | cell proliferation | precipitation | [148] |
| circ_0074854 | HuR | cell migration/immune escape | ultracentrifugation | [149] |
| circCCAR1 | miR-127-5p | immune escape | precipitation | [150] |
| circUHRF1 | miR-449c-5p | immune escape | precipitation | [151] |
| circASAP1 | miR-326/ miR-532-5p | cell proliferation/ immune escape | precipitation | [152] |
| circGSE1 | miR-324-5p | cell proliferation/immune scape | ultracentrifugation | [153] |
| circTMEM181 | miR-488-3p | immune escape | ultracentrifugation | [155,156] |
| circRNA-SORE | miR-660-3p | drug resistance | - | [158] |
| circPAK1 | 14–3-3 ζ | cell proliferation/ drug resistance | ultracentrifugation | [159] |
| circ_0004001 circ_0004123 circ_0075792 | - | biomarkers | - | [161] |
| circ_0072088 | - | biomarkers | gradient centrifugation | [162] |
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef]
- Llovet, J.M.; Pinyol, R.; Kelley, R.K.; El-Khoueiry, A.; Reeves, H.L.; Wang, X.W.; Gores, G.J.; Villanueva, A. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat. Cancer 2022, 3, 386–401. [Google Scholar] [CrossRef]
- Tampaki, M.; Papatheodoridis, G.V.; Cholongitas, E. Intrahepatic recurrence of hepatocellular carcinoma after resection: An update. Clin. J. Gastroenterol. 2021, 14, 699–713. [Google Scholar] [CrossRef]
- Tsuchiya, N.; Sawada, Y.; Endo, I.; Saito, K.; Uemura, Y.; Nakatsura, T. Biomarkers for the Early Diagnosis of Hepatocellular Carcinoma. World J. Gastroenterol. 2015, 21, 10573–10583. [Google Scholar] [CrossRef]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and Surveillance for Hepatocellular Carcinoma: New Trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef]
- Cersosimo, R.J. Systemic targeted and immunotherapy for advanced hepatocellular carcinoma. Am. J. Health Syst. Pharm. 2021, 78, 187–202. [Google Scholar] [CrossRef]
- Van de Wakker, S.I.; Meijers, F.M.; Sluijter, J.P.G.; Vader, P. Extracellular Vesicle Heterogeneity and Its Impact for Regenerative Medicine Applications. Pharmacol. Rev. 2023, 75, 1043–1061. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, S.; Yang, D.; Xu, W.; Qian, H. Extracellular vesicles: Emerging roles, biomarkers and therapeutic strategies in fibrotic diseases. J. Nanobiotechnol. 2023, 21, 164. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef]
- Rezaee, M.; Mohammadi, F.; Keshavarzmotamed, A.; Yahyazadeh, S.; Vakili, O.; Milasi, Y.E.; Veisi, V.; Dehmordi, R.M.; Asadi, S.; Ghorbanhosseini, S.S.; et al. The landscape of exosomal non-coding RNAs in breast cancer drug resistance, focusing on underlying molecular mechanisms. Front. Pharmacol. 2023, 14, 1152672. [Google Scholar] [CrossRef] [PubMed]
- Eitan, E.; Suire, C.; Zhang, S.; Mattson, M.P. Impact of lysosome status on extracellular vesicle content and release. Ageing Res. Rev. 2016, 32, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Ji, J.; Jin, D.; Wu, Y.; Wu, T.; Lin, R.; Zhu, S.; Jiang, F.; Ji, Y.; Bao, B.; et al. The biogenesis and secretion of exosomes and multivesicular bodies (MVBs): Intercellular shuttles and implications in human diseases. Genes Dis. 2022, 10, 1894–1907. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Poupardin, R.W.; Ebner-Peking, P.; Andrade, A.C.; Blöchl, C.; Obermayer, A.; Gomes, F.G.; Vari, B.; Maeding, N.; Eminger, E.; et al. A functional corona around extracellular vesicles enhances angiogenesis, skin regeneration and immunomodulation. J. Extracell. Vesicles 2022, 11, 12207. [Google Scholar] [CrossRef] [PubMed]
- Sohal, I.S.; Kasinski, A.L. Emerging diversity in extracellular vesicles and their roles in cancer. Front. Oncol. 2023, 13, 1167717. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Xing, J.; Xu, K.; Liu, D.; Zhuo, Y. Exosomes in the tumor microenvironment: Promoting cancer progression. Front. Immunol. 2022, 13, 1025218. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M. Exosomes: Revisiting their role as “garbage bags”. Traffic 2019, 20, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; de Wildt, B.W.M.; Vis, M.A.M.; de Korte, C.E.; Ito, K.; Hofmann, S.; Yuana, Y. Matrix Vesicles: Role in Bone Mineralization and Potential Use as Therapeutics. Pharmaceuticals 2021, 14, 289. [Google Scholar] [CrossRef]
- Jing, L.; Li, L.; Sun, Z.; Bao, Z.; Shao, C.; Yan, J.; Pang, Q.; Geng, Y.; Zhang, L.; Wang, X.; et al. Role of Matrix Vesicles in Bone-Vascular Cross-Talk. J. Cardiovasc. Pharmacol. 2019, 74, 372–378. [Google Scholar] [CrossRef]
- Rackov, G.; Garcia-Romero, N.; Esteban-Rubio, S.; Carrión-Navarro, J.; Belda-Iniesta, C.; Ayuso-Sacido, A. Vesicle-Mediated Control of Cell Function: The Role of Extracellular Matrix and Microenvironment. Front. Physiol. 2018, 9, 651. [Google Scholar] [CrossRef]
- Berumen Sánchez, G.; Bunn, K.E.; Pua, H.H.; Rafat, M. Extracellular vesicles: Mediators of intercellular communication in tissue injury and disease. Cell Commun. Signal. 2021, 19, 104. [Google Scholar] [CrossRef] [PubMed]
- Cavallari, C.; Camussi, G.; Brizzi, M.F. Extracellular Vesicles in the Tumour Microenvironment: Eclectic Supervisors. Int. J. Mol. Sci. 2020, 21, 6768. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, S.H.; Andrews, A.M.; Paul, D.; Pachter, J.S. Extracellular vesicles: Mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS 2018, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in Exosome Isolation and Analysis in Health and Disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [PubMed]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef] [PubMed]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Zhang, X.; Borg EG, F.; Liaci, A.M.; Vos, H.R.; Stoorvogel, W. A novel three step protocol to isolate extracellular vesicles from plasma or cell culture medium with both high yield and purity. J. Extracell. Vesicles 2020, 9, 1791450. [Google Scholar] [CrossRef] [PubMed]
- Mol, E.A.; Goumans, M.J.; Doevendans, P.A.; Sluijter, J.P.; Vader, P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 2017, 13, 2061–2065. [Google Scholar] [CrossRef]
- Linares, R.; Tan, S.; Gounou, C.; Arraud, N.; Brisson, A.R. High-speed centrifugation induces aggregation of extracellular vesicles. J. Extracell. Vesicles 2015, 4, 29509. [Google Scholar] [CrossRef]
- Grant, R.; Ansa-Addo, E.; Stratton, D.; Antwi-Baffour, S.; Jorfi, S.; Kholia, S.; Krige, L.; Lange, S.; Inal, J. A filtration-based protocol to isolate human plasma membrane-derived vesicles and exosomes from blood plasma. J. Immunol. Methods 2011, 371, 143–151. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen Wen, S.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.L.; Zhu, J.; Liu, J.X.; Jiang, F.; Ni, W.K.; Qu, L.S.; Ni, R.Z.; Lu, C.H.; Xiao, M.B. A comparison of traditional and novel methods for the separation of exosomes from human samples. BioMed Res. Int. 2018, 2018, 3634563. [Google Scholar] [CrossRef] [PubMed]
- Inal, J.M.; Kosgodage, U.; Azam, S.; Stratton, D.; Antwi-Baffour, S.; Lange, S. Blood/plasma secretome and microvesicles. Biochim. Biophys. Acta 2013, 1834, 2317–2325. [Google Scholar] [CrossRef]
- Benedikter, B.J.; Bouwman, F.G.; Vajen, T.; Heinzmann, A.C.; Grauls, G.; Mariman, E.C.; Wouters EF, M.; Savelkoul, P.H.; Lopez-Iglesias, C.; Koenen, R.R.; et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci. Rep. 2017, 7, 15297. [Google Scholar] [CrossRef] [PubMed]
- Foers, A.D.; Chatfield, S.; Dagley, L.F.; Scicluna, B.J.; Webb, A.I.; Cheng, L.; Hill, A.F.; Wicks, I.P.; Pang, K.C. Enrichment of extracellular vesicles from human synovial fluid using size exclusion chromatography. J. Extrecell. Vesicles 2018, 7, 1490145. [Google Scholar] [CrossRef]
- Gamez-Valero, A.; Mongui_o-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.; Beyer, K.; Borràs, F.E. Size-exclusion chromatography-based isolation minimally alters extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef]
- Hirschberg, Y.; Schildermans, K.; van Dam, A.; Sterck, K.; Boonen, K.; Nelissen, I.; Vermeiren, Y.; Mertens, I. Characterizing extracellular vesicles from cerebrospinal fluid by a novel size exclusion chromatography method. Alzheimer’s Dement. 2021, 17, e051264. [Google Scholar] [CrossRef]
- García-Romero, N.; Madurga, R.; Rackov, G.; Palacín-Aliana, I.; Núñez-Torres, R.; Asensi-Puig, A.; Carri_on-Navarro, J.; Esteban-Rubio, S.; Peinado, H.; Gonz_alez-Neira, A.; et al. Polyethylene glycol improves current methods for circulating extracellular vesicle-derived DNA isolation. J. Transl. Med. 2019, 17, 75. [Google Scholar] [CrossRef]
- Weng, Y.; Sui, Z.; Shan, Y.; Hu, Y.; Chen, Y.; Zhang, L.; Zhang, Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016, 141, 4640–4646. [Google Scholar] [CrossRef]
- Ludwig, A.K.; de Miroschedji, K.; Doeppner, T.R.; Börger, V.; Ruesing, J.; Rebmann, V.; Durst, S.; Jansen, S.; Bremer, M.; Behrmann, E.; et al. Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. J. Extracell. Vesicles 2018, 7, 1528109. [Google Scholar] [CrossRef] [PubMed]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef]
- Brambilla, D.; Sola, L.; Ferretti, A.M.; Chiodi, E.; Zarovni, N.; Fortunato, D.; Criscuoli, M.; Dolo, V.; Giusti, I.; Murdica, V.; et al. EV separation: Release of intact extracellular vesicles immunocaptured on magnetic particles. Anal. Chem. 2021, 93, 5476–5483. [Google Scholar] [CrossRef]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Gandham, S.; Su, X.; Wood, J.; Nocera, A.L.; Alli, S.C.; Milane, L.; Zimmerman, A.; Amiji, M.; Ivanov, A.R. Technologies and standardization in research on extracellular vesicles. Trends Biotechnol. 2020, 38, 1066–1098. [Google Scholar] [CrossRef] [PubMed]
- Paolini, L.; Zendrini, A.; Noto, G.D.; Busatto, S.; Lottini, E.; Radeghieri, A.; Dossi, A.; Caneschi, A.; Ricotta, D.; Bergese, P. Residual matrix from different separation techniques impacts exosome biological activity. Sci. Rep. 2016, 6, 23550. [Google Scholar] [CrossRef]
- Balaj, L.; Atai, N.A.; Chen, W.; Mu, D.; Tannous, B.A.; Breakefield, X.O.; Skog, J.; Maguire, C.A. Heparin affinity purification of extracellular vesicles. Sci. Rep. 2015, 5, 10266. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, Y.; Yamaguchi, H.; Hung, M.C.; Kameoka, J. Isolation of cancer-derived extracellular vesicle subpopulations by a size-selective microfluidic platform. Biomicrofluidics 2020, 14, 034113. [Google Scholar] [CrossRef] [PubMed]
- Iliescu, F.S.; Vrtačnik, D.; Neuzil, P.; Iliescu, C. Microfluidic technology for clinical applications of exosomes. Micromachines 2019, 10, 392. [Google Scholar] [CrossRef]
- Chiang, C.Y.; Chen, C. Toward characterizing extracellular vesicles at a single-particle level. J. Biomed. Sci. 2019, 26, 9. [Google Scholar] [CrossRef]
- Bonner, S.E.; Willms, E. Intercellular communication through extracellular vesicles in cancer and evolutionary biology. Prog. Biophys. Mol. Biol. 2021, 165, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ji, X.; Liu, J.; Fan, D.; Zhou, Q.; Chen, C.; Wang, W.; Wang, G.; Wang, H.; Yuan, W.; et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol. Cancer 2019, 18, 39. [Google Scholar] [CrossRef]
- Bamankar, S.; Londhe, V.Y. The Rise of Extracellular Vesicles as New Age Biomarkers in Cancer Diagnosis: Promises and Pitfalls. Technol. Cancer Res. Treat. 2023, 22, 15330338221149266. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Dang, W.; Zhang, S.; Yue, W.; Yang, L.; Zhai, X.; Yan, Q.; Lu, J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer 2019, 18, 37. [Google Scholar] [CrossRef]
- Wang, W.; Hao, L.P.; Song, H.; Chu, X.Y.; Wang, R. The Potential Roles of Exosomal Non-Coding RNAs in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 790916. [Google Scholar] [CrossRef] [PubMed]
- Wong, Q.W.; Lung, R.W.; Law, P.T.; Lai, P.B.; Chan, K.Y.; To, K.F.; Wong, N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology 2008, 135, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Cabiati, M.; Salvadori, C.; Basta, G.; Del Turco, S.; Aretini, P.; Cecchettini, A.; Del Ry, S. miRNA and long non-coding RNA transcriptional expression in hepatocellular carcinoma cell line-secreted extracellular vesicles. Clin. Exp. Med. 2022, 22, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Yip, K.W.; Spence, T.; Liu, F.F. MicroRNAs in extracellular vesicles: Potential cancer biomarkers. J. Hum. Genet. 2017, 62, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Lapitz, A.; Arbelaiz, A.; Olaizola, P.; Aranburu, A.; Bujanda, L.; Perugorria, M.J.; Banales, J.M. Extracellular Vesicles in Hepatobiliary Malignancies. Front. Immunol. 2018, 9, 2270. [Google Scholar] [CrossRef]
- Lee, Y.T.; Tran, B.V.; Wang, J.J.; Liang, I.Y.; You, S.; Zhu, Y.; Agopian, V.G.; Tseng, H.R.; Yang, J.D. The Role of Extracellular Vesicles in Disease Progression and Detection of Hepatocellular Carcinoma. Cancers 2021, 13, 3076. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Fei, B.Y.; Shao, D.; Pan, Y.; Mo, Z.H.; Sun, B.Z.; Zhang, D.; Zheng, X.; Zhang, M.; et al. MiR-27a promotes hepatocellular carcinoma cell proliferation through suppression of its target gene peroxisome proliferator-activated receptor γ. Chin. Med. J. 2015, 128, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Li, Y.; Liu, K.; Zhu, Q.; Yang, W.H.; Xiong, L.K.; Guo, D.L. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. 2018, 109, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Li, S.; Li, G.; Zhang, S.; Tang, X.; Ni, S.; Jian, X.; Xu, C.; Zhu, J.; Lu, M. The role of extracellular vesicles in mediating progression, metastasis and potential treatment of hepatocellular carcinoma. Oncotarget 2017, 8, 3683–3695. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tang, J.; Zhang, B.; Yang, W.; Liu Gao, M.; Wang, R.; Tan, Y.; Fan, J.; Chang, Y.; Fu, J.; et al. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut 2015, 64, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Sun, L.P.; Chen, X.M.; Li, H.Y.; Huang, S.A.; Jie, S.H. Comparison of microRNA expression profiles in HCC-derived microvesicles and the parental cells and evaluation of their roles in HCC. J. Huazhong Univ. Sci. Technol. Med. Sci. 2013, 33, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Wu, J.; Wu, J.; Ji, A.; Qiang, G.; Jiang, Y.; Jiang, C.; Ding, Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017, 8, 80666–80678. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ren, H.; Dai, B.; Li, J.; Shang, L.; Huang, J.; Shi, X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. 2018, 37, 324. [Google Scholar] [CrossRef]
- Xue, X.; Wang, X.; Zhao, Y.; Hu, R.; Qin, L. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem. Biophys. Res. Commun. 2018, 502, 515–521. [Google Scholar] [CrossRef]
- Yang, Y.; Mao, F.; Guo, L.; Shi, J.; Wu, M.; Cheng, S.; Guo, W. Tumor cells derived-extracellular vesicles transfer miR-3129 to promote hepatocellular carcinoma metastasis by targeting TXNIP. Dig. Liver Dis. 2021, 53, 474–485. [Google Scholar] [CrossRef]
- Chen, S.; Saeed, A.F.U.H.; Liu, Q.; Jiang, Q.; Xu, H.; Xiao, G.G.; Rao, L.; Duo, Y. Macrophages in immunoregulation and therapeutics. Signal Transduct. Target. Ther. 2023, 8, 207. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Liu, J.; Fan, L.; Yu, H.; Zhang, J.; He, Y.; Feng, D.; Wang, F.; Li, X.; Liu, Q.; Li, Y.; et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology 2019, 70, 241–258. [Google Scholar] [CrossRef]
- Yin, C.; Han, Q.; Xu, D.; Zheng, B.; Zhao, X.; Zhang, J. SALL4-mediated upregulation of exosomal miR-146a-5p drives T-cell exhaustion by M2 tumor-associated macrophages in HCC. Oncoimmunology 2019, 8, 1601479. [Google Scholar] [CrossRef]
- Zongqiang, H.; Jiapeng, C.; Yingpeng, Z.; Chuntao, Y.; Yiting, W.; Jiashun, Z.; Li, L. Exosomal miR-452-5p Induce M2 Macrophage Polarization to Accelerate Hepatocellular Carcinoma Progression by Targeting TIMP3. J. Immunol. Res. 2022, 2022, 1032106. [Google Scholar] [CrossRef]
- Yu, H.; Pan, J.; Zheng, S.; Cai, D.; Luo, A.; Xia, Z.; Huang, J. Hepatocellular Carcinoma Cell-Derived Exosomal miR-21-5p Induces Macrophage M2 Polarization by Targeting RhoB. Int. J. Mol. Sci. 2023, 24, 4593. [Google Scholar] [CrossRef]
- Li, D.; Jia, H.; Zhang, H.; Lv, M.; Liu, J.; Zhang, Y.; Huang, T.; Huang, B. TLR4 signaling induces the release of microparticles by tumor cells that regulate inflammatory cytokine IL-6 of macrophages via microRNA let-7b. Oncoimmunology 2012, 1, 687–693. [Google Scholar] [CrossRef]
- Nakano, T.; Chen, I.H.; Wang, C.C.; Chen, P.J.; Tseng, H.P.; Huang, K.T.; Hu, T.H.; Li, L.C.; Goto, S.; Cheng, Y.F.; et al. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am. J. Transplant. 2019, 19, 3250–3262. [Google Scholar] [CrossRef]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef]
- Davodabadi, F.; Sajjadi, S.F.; Sarhadi, M.; Mirghasemi, S.; Nadali Hezaveh, M.; Khosravi, S.; Kamali Andani, M.; Cordani, M.; Basiri, M.; Ghavami, S. Cancer chemotherapy resistance: Mechanisms and recent breakthrough in targeted drug delivery. Eur. J. Pharmacol. 2023, 958, 176013. [Google Scholar] [CrossRef] [PubMed]
- Cabral, L.K.D.; Tiribelli, C.; Sukowati, C.H.C. Sorafenib Resistance in Hepatocellular Carcinoma: The Relevance of Genetic Heterogeneity. Cancers 2020, 12, 1576. [Google Scholar] [CrossRef]
- Jaffar Ali, D.; He, C.; Xu, H.; Kumaravel, S.; Sun, B.; Zhou, Y.; Liu, R.; Xiao, Z. Microvesicles mediate sorafenib resistance in liver cancer cells through attenuating p53 and enhancing FOXM1 expression. Life Sci. 2021, 271, 119149. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, W.; Wang, H.; Qiu, G.; Jiang, Z.; Wei, G.; Li, X. Exosomal MiR-744 Inhibits Proliferation and Sorafenib Chemoresistance in Hepatocellular Carcinoma by Targeting PAX2. Med. Sci. Monit. 2019, 25, 7209–7217. [Google Scholar] [CrossRef]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef]
- Sorop, A.; Constantinescu, D.; Cojocaru, F.; Dinischiotu, A.; Cucu, D.; Dima, S.O. Exosomal microRNAs as Biomarkers and Therapeutic Targets for Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 4997. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, T.; Zhu, Z.; Huang, H.; Zhang, L.; Goel, A.; Yang, M.; Wang, X. An integrated workflow for biomarker development using microRNAs in extracellular vesicles for cancer precision medicine. Semin. Cancer Biol. 2021, 74, 134–155. [Google Scholar] [CrossRef]
- Cui, Y.; Xu, H.F.; Liu, M.Y.; Xu, Y.J.; He, J.C.; Zhou, Y.; Cang, S.D. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J. Gastroenterol. 2019, 25, 1890–1898. [Google Scholar] [CrossRef]
- Sorop, A.; Iacob, R.; Iacob, S.; Constantinescu, D.; Chitoiu, L.; Fertig, T.E.; Dinischiotu, A.; Chivu-Economescu, M.; Bacalbasa, N.; Savu, L.; et al. Plasma Small Extracellular Vesicles Derived miR-21-5p and miR-92a-3p as Potential Biomarkers for Hepatocellular Carcinoma Screening. Front. Genet. 2020, 11, 712. [Google Scholar] [CrossRef]
- Cho, H.J.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Sung, S.; Son, J.A.; Kim, S.S.; Cho, S.W.; Jang, J.W.; Nam, S.W.; et al. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020, 9, 5459–5472. [Google Scholar] [CrossRef]
- Cho, H.J.; Eun, J.W.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Kim, S.S.; Cho, S.W.; Cheong, J.Y. Serum Exosomal MicroRNA, miR-10b-5p, as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. J. Clin. Med. 2020, 9, 281. [Google Scholar] [CrossRef]
- Boonkaew, B.; Satthawiwat, N.; Pinjaroen, N.; Chuaypen, N.; Tangkijvanich, P. Circulating Extracellular Vesicle-Derived microRNAs as Novel Diagnostic and Prognostic Biomarkers for Non-Viral-Related Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 16043. [Google Scholar] [CrossRef]
- Bao, L.; Zhao, J.; Dai, X.; Wang, Y.; Ma, R.; Su, Y.; Cui, H.; Niu, J.; Bai, S.; Xiao, Z.; et al. Correlation between miR-23a and onset of hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2014, 38, 318–330. [Google Scholar] [CrossRef]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, L.; Gu, J.; Zhang, H.; Yuan, J.; Lian, Q.; Lv, G.; Wang, S.; Wu, Y.; Yang, Y.T.; et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat. Commun. 2017, 8, 14421. [Google Scholar] [CrossRef]
- Rosenbloom, K.R.; Dreszer, T.R.; Long, J.C.; Malladi, V.S.; Sloan, C.A.; Raney, B.J.; Cline, M.S.; Karolchik, D.; Barber, G.P.; Clawson, H.; et al. ENCODE whole-genome data in the UCSC Genome Browser: Update 2012. Nucleic Acids Res. 2012, 40, D912–D917. [Google Scholar] [CrossRef]
- Tsagakis, I.; Douka, K.; Birds, I.; Aspden, J.L. Long non-coding RNAs in development and disease: Conservation to mechanisms. J. Pathol. 2020, 250, 480–495. [Google Scholar] [CrossRef]
- Tellez-Gabriel, M.; Heymann, D. Exosomal lncRNAs: The newest promising liquid biopsy. Cancer Drug Resist. 2019, 2, 1002–1017. [Google Scholar] [CrossRef]
- Li, B.; Mao, R.; Liu, C.; Zhang, W.; Tang, Y.; Guo, Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018, 197, 122–129. [Google Scholar] [CrossRef]
- Zhuo, C.; Yi, T.; Pu, J.; Cen, X.; Zhou, Y.; Feng, S.; Wei, C.; Chen, P.; Wang, W.; Bao, C.; et al. Exosomal linc-FAM138B from cancer cells alleviates hepatocellular carcinoma progression via regulating miR-765. Aging 2020, 12, 26236–26247. [Google Scholar] [CrossRef]
- Huang, X.; Sun, L.; Wen, S.; Deng, D.; Wan, F.; He, X.; Tian, L.; Liang, L.; Wei, C.; Gao, K.; et al. RNA sequencing of plasma exosomes revealed novel functional long noncoding RNAs in hepatocellular carcinoma. Cancer Sci. 2020, 111, 3338–3349. [Google Scholar] [CrossRef]
- You, L.N.; Tai, Q.W.; Xu, L.; Hao, Y.; Guo, W.J.; Zhang, Q.; Tong, Q.; Zhang, H.; Huang, W.K. Exosomal LINC00161 promotes angiogenesis and metastasis via regulating miR-590-3p/ROCK axis in hepatocellular carcinoma. Cancer Gene Ther. 2021, 28, 719–736. [Google Scholar] [CrossRef]
- Ma, D.; Gao, X.; Liu, Z.; Lu, X.; Ju, H.; Zhang, N. Exosome-transferred long non-coding RNA ASMTL-AS1 contributes to malignant phenotypes in residual hepatocellular carcinoma after insufficient radiofrequency ablation. Cell Prolif. 2020, 53, e12795. [Google Scholar] [CrossRef]
- Wang, D.; Xing, N.; Yang, T.; Liu, J.; Zhao, H.; He, J.; Ai, Y.; Yang, J. Exosomal lncRNA H19 promotes the progression of hepatocellular carcinoma treated with Propofol via miR-520a-3p/LIMK1 axis. Cancer Med. 2020, 9, 7218–7230. [Google Scholar] [CrossRef]
- Pan, G.; Zhang, J.; You, F.; Cui, T.; Luo, P.; Wang, S.; Li, X.; Yuan, Q. ETS Proto-Oncogene 1-activated muskelin 1 antisense RNA drives the malignant progression of hepatocellular carcinoma by targeting miR-22-3p to upregulate ETS Proto-Oncogene 1. Bioengineered 2022, 13, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Haga, H.; Patel, T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J. Cell Sci. 2014, 127, 1585–1594. [Google Scholar]
- Lu, L.; Huang, J.; Mo, J.; Da, X.; Li, Q.; Fan, M.; Lu, H. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol. Biol. Lett. 2022, 27, 17. [Google Scholar] [CrossRef]
- Alzahrani, F.A.; El-Magd, M.A.; Abdelfattah-Hassan, A.; Saleh, A.A.; Saadeldin, I.M.; El-Shetry, E.S.; Badawy, A.A.; Alkarim, S. Potential Effect of Exosomes Derived from Cancer Stem Cells and MSCs on Progression of DEN-Induced HCC in Rats. Stem Cells Int. 2018, 2018, 8058979. [Google Scholar] [CrossRef]
- Kogure, T.; Yan, I.K.; Lin, W.-L.; Patel, T. Extracellular vesicle–mediated transfer of a novel long noncoding RNA TUC339: A mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer 2013, 4, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Ye, P.; Wang, T.; Liu, W.; Li, X.; Tang, L.; Tian, F. Enhancing HOTAIR/MiR-10b drives normal liver stem cells toward a tendency to malignant transformation through inducing epithelial-to-mesenchymal transition. Rejuvenat. Res. 2015, 18, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Zhang, Z.; Xu, Z.; Xia, F.; Yan, Y. A prognostic exosome-related LncRNA risk model correlates with the immune microenvironment in liver cancer. Front. Genet. 2022, 13, 965329. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Baek, G.O.; Son, J.A.; Ahn, H.R.; Yoon, M.K.; Cho, H.J.; Yoon, J.H.; Nam, S.W.; Cheong, J.Y.; Eun, J.W. Early detection of hepatocellular carcinoma via liquid biopsy: Panel of small extracellular vesicle-derived long noncoding RNAs identified as markers. Mol. Oncol. 2021, 15, 2715–2731. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Su, Y.; Liu, X.; Xu, M.; Chen, X.; Zhu, Y.; Guo, Z.; Bai, T.; Dong, L.; Wei, C.; et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J. Cancer 2018, 9, 2631–2639. [Google Scholar] [CrossRef]
- Wang, J.; Pu, J.; Zhang, Y.; Yao, T.; Luo, Z.; Li, W.; Xu, G.; Liu, J.; Wei, W.; Deng, Y. Exosome-transmitted long non-coding RNA SENP3-EIF4A1 suppresses the progression of hepatocellular carcinoma. Aging 2020, 12, 11550–11567. [Google Scholar] [CrossRef]
- Wang, T.; Zhu, H.; Xiao, M.; Zhou, S. Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J. Clin. Lab. Anal. 2021, 35, e23959. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Xiang, Y.; Sheng, J.; Liu, S.; Cui, M.; Zhang, X. Long non-coding RNA CRNDE promotes malignant progression of hepatocellular carcinoma through the miR-33a-5p/CDK6 axis. J. Physiol. Biochem. 2020, 76, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Hua, X.; Shi, J.; Hu, X.; Lui, K.; He, K.; Mai, J.; Lan, T.; Lu, M. LncRNA THEMIS2-211, a tumor-originated circulating exosomal biomarker, promotes the growth and metastasis of hepatocellular carcinoma by functioning as a competing endogenous RNA. FASEB J. 2022, 36, e22238. [Google Scholar] [CrossRef]
- Hou, Y.; Yu, Z.; Tam, N.L.; Huang, S.; Sun, C.; Wang, R.; Zhang, X.; Wang, Z.; Ma, Y.; He, X.; et al. Exosome-related lncRNAs as predictors of HCC patient survival: A prognostic model. Am. J. Transl. Res. 2018, 10, 1648–1662. [Google Scholar]
- Matboli, M.; Labib, M.E.; Nasser, H.E.; El-Tawdi, A.H.F.; Habib, E.K.; Ali-Labib, R. Exosomal miR-1298 and lncRNA-RP11-583F2.2 Expression in Hepato-cellular Carcinoma. Curr. Genom. 2020, 21, 46–55. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, X.; Qi, Q.; Gao, Y.; Wei, Q.; Han, S. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018, 21, 651–659. [Google Scholar] [CrossRef]
- Lu, Y.; Duan, Y.; Xu, Q.; Zhang, L.; Chen, W.; Qu, Z.; Wu, B.; Liu, W.; Shi, L.; Wu, D.; et al. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J. Cell Mol. Med. 2020, 24, 1311–1318. [Google Scholar] [CrossRef]
- Xu, H.; Chen, Y.; Dong, X.; Wang, X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2018, 27, 710–716. [Google Scholar] [CrossRef]
- Lee, Y.R.; Kim, G.; Tak, W.Y.; Jang, S.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Chun, J.M.; Park, S.Y.; et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 2019, 144, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Jia, C.; Tai, Y.; Liang, H.; Zhong, Z.; Xiong, Z.; Deng, M.; Zhang, Q. Serum exosomal long noncoding RNAs lnc-FAM72D-3 and lnc-EPC1-4 as diagnostic biomarkers for hepatocellular carcinoma. Aging 2020, 12, 11843–11863. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lei, Y.; Wu, M.; Li, N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018, 10, 2958. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Chen, K.; Lu, X.; Li, A.; Liu, C.; Wu, B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol. Int. 2021, 15, 444–458. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.I.; Jeong, D.; Ji, S.; Ahn, T.S.; Bae, S.H.; Chin, S.; Chung, J.C.; Kim, H.C.; Lee, M.S.; Baek, M.J. Overexpression of PD-L1 and PD-L2 Is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res. Treat. 2017, 49, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Kogure, T.; Haga, H.; Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 2014, 4, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Wood, J.; Haga, H.; Patel, T. Involvement of Extracellular Vesicle long non-coding RNA (linc-VLDLR) in Tumor Cell Responses to Chemotherapy. Mol. Cancer Res. 2014, 12, 1377–1387. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The landscape of circular RNA in cancer. Cell 2019, 176, 869–881. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, X.; Wu, X.; Guo, H.; Hu, Y.; Tang, F.; Huang, Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015, 16, 148. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Xu, S.; Guo, J. Novel potential tumor biomarkers: Circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J. Clin. Lab. Anal. 2020, 34, e23359. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Li, X.; Jiang, Z.; Xia, Q.; Hu, Y.; Guo, J.; Fu, L. Exosomes and circular RNAs: Promising partners in hepatocellular carcinoma from bench to bedside. Discov. Oncol. 2023, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Zhao, Y.; Yao, J. The Biological Functions and Clinical Values of Exosomal Circular RNAs in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 885214. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Chen, C.; Yang, Q.; Xue, J.; Chen, X.; Sun, B.; Luo, F.; Liu, X.; Xiao, T.; Xu, H.; et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018, 9, 454. [Google Scholar] [CrossRef]
- Su, Y.; Lv, X.; Yin, W.; Zhou, L.; Hu, Y.; Zhou, A.; Qi, F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging 2019, 11, 8183–8203. [Google Scholar] [CrossRef]
- Yu, Y.; Bian, L.; Liu, R.; Wang, Y.; Xiao, X. Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer Cell Int. 2021, 21, 10. [Google Scholar] [CrossRef]
- Liu, C.; Ren, C.; Guo, L.; Yang, C.; Yu, Q. Exosome-mediated circTTLL5 transfer promotes hepatocellular carcinoma malignant progression through miR-136-5p/KIAA1522 axis. Pathol. Res. Pract. 2023, 241, 154276. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Liu, W. Roles of circRNAs in the Tumorigenesis and Metastasis of HCC: A Mini Review. Cancer Manag. Res. 2022, 14, 1847–1856. [Google Scholar] [CrossRef]
- Huang, Y.; Hong, W.; Wei, X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, Q.; Shen, C.; Qian, Y. Circular RNA circ_0003028 regulates cell development through modulating miR-498/ornithine decarboxylase 1 axis in hepatocellular carcinoma. Anticancer Drugs 2023, 34, 507–518. [Google Scholar] [CrossRef]
- Liu, L.; Liao, R.; Wu, Z.; Du, C.; You, Y.; Que, K.; Duan, Y.; Yin, K.; Ye, W. Hepatic stellate cell exosome-derived circWDR25 promotes the progression of hepatocellular carcinoma via the miRNA-4474-3P-ALOX-15 and EMT axes. Biosci. Trends 2022, 16, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Su, Y.; Liu, L.; Wang, S.; Liu, Y.; Wu, J. Circular RNA hsa_circ_0004277 Stimulates Malignant Phenotype of Hepatocellular Carcinoma and Epithelial-Mesenchymal Transition of Peripheral Cells. Front. Cell Dev. Biol. 2021, 8, 585565. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Song, J.; Wang, F.; Chen, B. Exosome-transmitted circ_002136 promotes hepatocellular carcinoma progression by miR-19a-3p/RAB1A pathway. BMC Cancer 2022, 22, 1284. [Google Scholar] [CrossRef]
- Lyu, P.; Zhai, Z.; Hao, Z.; Zhang, H.; He, J. CircWHSC1 serves as an oncogene to promote hepatocellular carcinoma progression. Eur. J. Clin. Investig. 2021, 51, e13487. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, R.; Li, J.; Tang, S.; Li, S.; Tong, Q.; Li, S. Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int. J. Nanomed. 2021, 16, 2803–2818. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, G.; Zhao, Y.; Gao, H.; Li, L.; Yin, Y.; Jiang, J.; Wang, L.; Mang, Y.; Gao, Y.; et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol. Cancer 2023, 22, 55. [Google Scholar] [CrossRef]
- Zhang, P.F.; Gao, C.; Huang, X.Y.; Lu, J.C.; Guo, X.J.; Shi, G.M.; Cai, J.B.; Ke, A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef]
- Hu, Z.Q.; Zhou, S.L.; Li, J.; Zhou, Z.J.; Wang, P.C.; Xin, H.Y.; Mao, L.; Luo, C.B.; Yu, S.Y.; Huang, X.W.; et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology 2020, 72, 906–922. [Google Scholar] [CrossRef]
- Huang, M.; Huang, X.; Huang, N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022, 113, 1968–1983. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Tan, J.; Wu, M.; Fan, W.; Wei, J.; Zhu, B.; Guo, J.; Wang, S.; Zhou, P.; Zhang, H.; et al. High-affinity neoantigens correlate with better prognosis and trigger potent antihepatocellular carcinoma (HCC) activity by activating CD39+CD8+ T cells. Gut 2020, 70, 1965–1977. [Google Scholar] [CrossRef]
- Lu, J.C.; Zhang, P.F.; Huang, X.Y.; Guo, X.J.; Gao, C.; Zeng, H.Y.; Zheng, Y.M.; Wang, S.W.; Cai, J.B.; Sun, Q.M.; et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J. Hematol. Oncol. 2021, 14, 200. [Google Scholar] [CrossRef]
- Chen, B.; Li, Q.; Li, Y.; Li, Q.; Lai, H.; Huang, S.; Li, C.; Li, Y. circTMEM181 upregulates ARHGAP29 to inhibit hepatocellular carcinoma migration and invasion by sponging miR-519a-5p. Hepatol. Res. 2023, 53, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Kong, S.; Wang, F.; Ju, S. CircRNAs: Biogenesis, functions, and role in drug-resistant Tumours. Mol. Cancer 2020, 19, 119. [Google Scholar] [CrossRef]
- Xu, J.; Ji, L.; Liang, Y.; Wan, Z.; Zheng, W.; Song, X.; Gorshkov, K.; Sun, Q.; Lin, H.; Zheng, X.; et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target. Ther. 2020, 5, 298. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Zhang, Y.; Shi, X.; Liu, H.; Zheng, Z.; Han, G.; Rong, D.; Zhang, C.; Tang, W.; Wang, X. CircPAK1 promotes the progression of hepatocellular carcinoma via modulation of YAP nucleus localization by interacting with 14-3-3ζ. J. Exp. Clin. Cancer Res. 2022, 41, 281. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, W.; Zhuo, H.; Zhu, D.; Rong, D.; Sun, J.; Song, J. Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/nuclear factor-kappa B (NF-κB) pathway. Bioengineered 2022, 13, 4786–4797. [Google Scholar] [CrossRef]
- Sun, X.H.; Wang, Y.T.; Li, G.F.; Zhang, N.; Fan, L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020, 20, 226. [Google Scholar] [CrossRef]
- Lin, Y.; Zheng, Z.H.; Wang, J.X.; Zhao, Z.; Peng, T.Y. Tumor Cell-Derived Exosomal Circ-0072088 Suppresses Migration and Invasion of Hepatic Carcinoma Cells Through Regulating MMP-16. Front. Cell Dev. Biol. 2021, 9, 726323. [Google Scholar] [CrossRef]
- Lai, Z.; Wei, T.; Li, Q.; Wang, X.; Zhang, Y.; Zhang, S. Exosomal circFBLIM1 Promotes Hepatocellular Carcinoma Progression and Glycolysis by Regulating the miR-338/LRP6 Axis. Cancer Biother. Radiopharm. 2020, 38, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pei, L.; Yue, Z.; Jia, M.; Wang, H.; Cao, L.L. The Potential of Serum Exosomal has_circ_0028861 as the Novel Diagnostic Biomarker of HBV-Derived Hepatocellular Cancer. Front. Genet. 2021, 12, 703205. [Google Scholar] [CrossRef] [PubMed]
- Lyu, L.; Yang, W.; Yao, J.; Wang, H.; Zhu, J.; Jin, A.; Liu, T.; Wang, B.; Zhou, J.; Fan, J.; et al. The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomark. Med. 2021, 15, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jing, B.; Bai, Y.; Zhang, Y.; Yu, H. Circular RNA circTMEM45A Acts as the Sponge of MicroRNA-665 to Promote Hepatocellular Carcinoma Progression. Mol. Ther. Nucleic Acids 2020, 22, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yu, W.; Wang, Q.; Huang, T.; Ding, Y. CircANTXR1 Contributes to the Malignant Progression of Hepatocellular Carcinoma by Promoting Proliferation and Metastasis. J. Hepatocell. Carcinoma 2021, 8, 1339–1353. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Quan, Y.; Fan, S.; Wang, H.; Liang, J.; Huang, L.; Chen, L.; Liu, Q.; He, P.; Ye, Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020, 475, 119–128. [Google Scholar] [CrossRef]
- Czystowska-Kuzmicz, M.; Whiteside, T.L. The potential role of tumor-derived exosomes in diagnosis, prognosis, and response to therapy in cancer. Expert. Opin. Biol. Ther. 2021, 21, 241–258. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Augello, G.; Cusimano, A.; Cervello, M.; Cusimano, A. Extracellular Vesicle-Related Non-Coding RNAs in Hepatocellular Carcinoma: An Overview. Cancers 2024, 16, 1415. https://doi.org/10.3390/cancers16071415
Augello G, Cusimano A, Cervello M, Cusimano A. Extracellular Vesicle-Related Non-Coding RNAs in Hepatocellular Carcinoma: An Overview. Cancers. 2024; 16(7):1415. https://doi.org/10.3390/cancers16071415
Chicago/Turabian StyleAugello, Giuseppa, Alessandra Cusimano, Melchiorre Cervello, and Antonella Cusimano. 2024. "Extracellular Vesicle-Related Non-Coding RNAs in Hepatocellular Carcinoma: An Overview" Cancers 16, no. 7: 1415. https://doi.org/10.3390/cancers16071415
APA StyleAugello, G., Cusimano, A., Cervello, M., & Cusimano, A. (2024). Extracellular Vesicle-Related Non-Coding RNAs in Hepatocellular Carcinoma: An Overview. Cancers, 16(7), 1415. https://doi.org/10.3390/cancers16071415







