Clinical Biomarkers of Tumour Radiosensitivity and Predicting Benefit from Radiotherapy: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Literature Search
2.2. Exclusion Criteria
2.3. Definitions
2.4. Gene Lists
3. Results
3.1. Radiosensitivity Signatures Developed In Vitro
3.1.1. Radiosensitivity Index (RSI) and Genomic-Adjusted Radiation Dose (GARD)
3.1.2. 31-Gene Radiosensitivity Signature
3.1.3. Interferon-Related DNA Damage Resistance Signature
3.2. Radiosensitivity Signatures Developed In Vivo
3.2.1. Post-Operative Radiation Therapy Outcomes Score (Decipher PORTOS)
3.2.2. Adjuvant Radiotherapy Intensification Classifier (ARTIC)
3.2.3. Other In Vivo Derived Radiosensitivity Signatures
3.3. Breast Cancer Prognostic Signatures
3.3.1. MammaPrint
3.3.2. Danish Breast Cancer Cooperative Group Radiotherapy Profile (DBCG-RT)
3.3.3. Oncotype DCIS and Oncotype Dx
3.3.4. Prosigna PAM-50
3.3.5. Profile for the Omission of Local Adjuvant Radiation (POLAR)
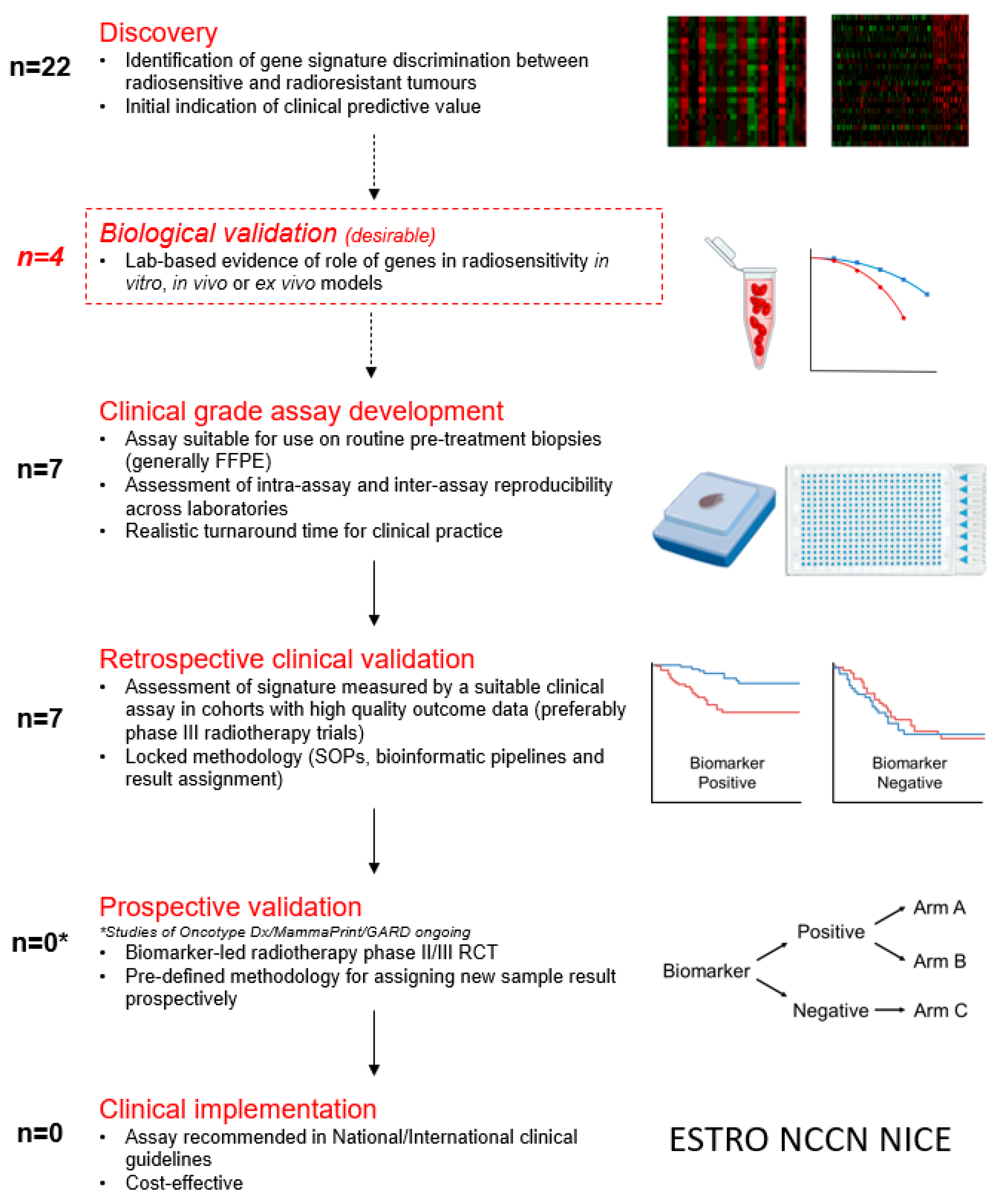
4. Discussion
5. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Borras, J.M.; Lievens, Y.; Dunscombe, P.; Coffey, M.; Malicki, J.; Corral, J.; Gasparotto, C.; Defourny, N.; Barton, M.; Verhoeven, R.; et al. The Optimal Utilization Proportion of External Beam Radiotherapy in European Countries: An ESTRO-HERO Analysis. Radiother. Oncol. 2015, 116, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Chandra, R.A.; Keane, F.K.; Voncken, F.E.M.; Thomas, C.R. Contemporary Radiotherapy: Present and Future. Lancet 2021, 398, 171–184. [Google Scholar] [CrossRef]
- Boustani, J.; Grapin, M.; Laurent, P.; Apetoh, L.; Mirjolet, C. The 6th R of Radiobiology: Reactivation of Anti-Tumor Immune Response. Cancers 2019, 11, 680. [Google Scholar] [CrossRef] [PubMed]
- Steel, G.G.; McMillan, T.J.; Peacock, J.H. The 5Rs of Radiobiology. Int. J. Radiat. Biol. 1989, 56, 1045–1048. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.; Giaccia, A. Radiobiology for the Radiologist, 8th ed.; Wolters Kluwer: Philadelphia, PA, USA, 2019; p. 624. [Google Scholar]
- West, C.M.; Davidson, S.E.; Roberts, S.A.; Hunter, R.D. The Independence of Intrinsic Radiosensitivity as a Prognostic Factor for Patient Response to Radiotherapy of Carcinoma of the Cervix. Br. J. Cancer 1997, 76, 1184–1190. [Google Scholar] [CrossRef]
- Walker, A.K.; Karaszi, K.; Valentine, H.; Strauss, V.Y.; Choudhury, A.; McGill, S.; Wen, K.; Brown, M.D.; Ramani, V.; Bhattarai, S.; et al. MRE11 as a Predictive Biomarker of Outcome after Radiation Therapy in Bladder Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Kalinsky, K.; Barlow, W.E.; Gralow, J.R.; Meric-Bernstam, F.; Albain, K.S.; Hayes, D.F.; Lin, N.U.; Perez, E.A.; Goldstein, L.J.; Chia, S.K.L.; et al. 21-Gene Assay to Inform Chemotherapy Benefit in Node-Positive Breast Cancer. N. Engl. J. Med. 2021, 385, 2336–2347. [Google Scholar] [CrossRef] [PubMed]
- Betts, G.N.J.; Eustace, A.; Patiar, S.; Valentine, H.R.; Irlam, J.; Ramachandran, A.; Merve, A.; Homer, J.J.; Möller-Levet, C.; Buffa, F.M.; et al. Prospective Technical Validation and Assessment of Intra-Tumour Heterogeneity of a Low Density Array Hypoxia Gene Profile in Head and Neck Squamous Cell Carcinoma. Eur. J. Cancer 2013, 49, 156–165. [Google Scholar] [CrossRef]
- Forker, L.J.; Choudhury, A.; Kiltie, A.E. Biomarkers of Tumour Radiosensitivity and Predicting Benefit from Radiotherapy. Clin. Oncol. 2015, 27, 561–569. [Google Scholar] [CrossRef]
- UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, 523–531. [CrossRef]
- Torres-Roca, J.; Erho, N.; Vergara, I.; Davicioni, E.; Jenkins, R.B.; Den, R.B.; Dicker, A.P.; Eschrich, S.A. A Molecular Signature of Radiosensitivity (RSI) is an RT-Specific Biomarker in Prostate Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2014, 90, S157. [Google Scholar] [CrossRef]
- Wushou, A.; Jiang, Y.; Hou, J.; Liu, Y.; Guo, X.; Shao, Z. Development of Triple-Negative Breast Cancer Radiosensitive Gene Signature and Validation Based on Transcriptome Analysis. Breast Cancer Res. Treat. 2015, 154, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.G.; Chang, S.L.; Spratt, D.E.; Erho, N.; Yu, M.; Ashab, H.A.; Alshalalfa, M.; Speers, C.; Tomlins, S.A.; Davicioni, E.; et al. Development and Validation of a 24-Gene Predictor of Response to Postoperative Radiotherapy in Prostate Cancer: A Matched, Retrospective Analysis. Lancet Oncol. 2016, 17, 1612–1620. [Google Scholar] [CrossRef] [PubMed]
- Rakovitch, E.; Nofech-Mozes, S.; Hanna, W.; Sutradhar, R.; Baehner, F.L.; Miller, D.P.; Fong, C.; Gu, S.; Tuck, A.; Sengupta, S.; et al. Multigene Expression Assay and Benefit of Radiotherapy After Breast Conservation in Ductal Carcinoma in Situ. J. Natl. Cancer Inst. 2017, 109, djw256. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zeng, Q.; Li, Y.; Zhang, X.; Ma, J.; Suto, M.J.; Xu, B.; Yi, N. Development of a Radiosensitivity Gene Signature for Patients with Soft Tissue Sarcoma. Oncotarget 2017, 8, 27428–27439. [Google Scholar] [CrossRef]
- Tang, Z.; Zeng, Q.; Li, Y.; Zhang, X.; Suto, M.J.; Xu, B.; Yi, N. Predicting Radiotherapy Response for Patients with Soft Tissue Sarcoma by Developing a Molecular Signature. Oncol. Rep. 2017, 38, 2814–2824. [Google Scholar] [CrossRef][Green Version]
- Zhou, J.; Wu, X.; Li, G.; Gao, X.; Zhai, M.; Chen, W.; Hu, H.; Tang, Z. Prediction of Radiosensitive Patients with Gastric Cancer by Developing Gene Signature. Int. J. Oncol. 2017, 51, 1067–1076. [Google Scholar] [CrossRef]
- Chen, L.; Wen, Y.; Zhang, J.; Sun, W.; Lui, V.W.Y.; Wei, Y.; Chen, F.; Wen, W. Prediction of Radiotherapy Response with a 5-microRNA Signature-Based Nomogram in Head and Neck Squamous Cell Carcinoma. Cancer Med. 2018, 7, 726–735. [Google Scholar] [CrossRef]
- Jang, B.; Kim, I.A. A Radiosensitivity Gene Signature and PD-L1 Predict the Clinical Outcomes of Patients with Lower Grade Glioma in TCGA. Radiother. Oncol. 2018, 128, 245–253. [Google Scholar] [CrossRef]
- Sjöström, M.; Staaf, J.; Edén, P.; Wärnberg, F.; Bergh, J.; Malmström, P.; Fernö, M.; Niméus, E.; Fredriksson, I. Identification and Validation of Single-Sample Breast Cancer Radiosensitivity Gene Expression Predictors. Breast Cancer Res. 2018, 20, 64. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.R.; Seagle, B.L.; Kocherginsky, M.; Donnelly, E.D.; Shahabi, S.; Strauss, J.B. 21-Gene Recurrence Score Assay Predicts Benefit of Post-Mastectomy Radiotherapy in T1-2 N1 Breast Cancer. Clin. Cancer Res. 2018, 24, 3878–3887. [Google Scholar] [CrossRef]
- Ji, Y.; Jiang, Q.; Jian, G.; Sun, H.; Wang, Y.; Qin, H.; Zhang, S.; Cao, J.; Tang, Z. Developing a Radiosensitivity Gene Signature for Caucasian Patients with Breast Cancer. Oncol. Rep. 2018, 40, 1695–1705. [Google Scholar] [CrossRef]
- Cui, Y.; Li, B.; Pollom, E.L.; Horst, K.C.; Li, R. Integrating Radiosensitivity and Immune Gene Signatures for Predicting Benefit of Radiotherapy in Breast Cancer. Clin. Cancer Res. 2018, 24, 4754–4762. [Google Scholar] [CrossRef]
- Sjöström, M.; Chang, S.L.; Fishbane, N.; Davicioni, E.; Zhao, S.G.; Hartman, L.; Holmberg, E.; Feng, F.Y.; Speers, C.W.; Pierce, L.J.; et al. Clinicogenomic Radiotherapy Classifier Predicting the Need for Intensified Locoregional Treatment after Breast-Conserving Surgery for Early-Stage Breast Cancer. J. Clin. Oncol. 2019, 37, 3340–3349. [Google Scholar] [CrossRef]
- Mohammadi, H.; Prince, A.; Figura, N.B.; Peacock, J.S.; Fernandez, D.C.; Montejo, M.E.; Chon, H.S.; Wenham, R.M.; Eschrich, S.A.; Torres-Roca, J.F.; et al. Using the Radiosensitivity Index (RSI) to Predict Pelvic Failure in Endometrial Cancer Treated with Adjuvant Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2020, 106, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.; Kim, I.A. A Radiosensitivity Gene Signature and PD-L1 Status Predict Clinical Outcome of Patients with Glioblastoma Multiforme in the Cancer Genome Atlas Dataset. Cancer. Res. Treat. 2020, 52, 530–542. [Google Scholar] [CrossRef]
- Thiruthaneeswaran, N.; Bibby, B.A.S.; Pereira, R.; More, E.; Denley, H.; Henry, A.; Wylie, J.; Hoskin, P.; Bristow, R.; Choudhury, A.; et al. OC-1031: The Radiosensitivity Index Predicts Benefit from HDR Brachytherapy in High-Risk Prostate Cancer. Radiother. Oncol. 2020, 152, S1086–S1087. [Google Scholar] [CrossRef]
- Nishiwada, S.; Sho, M.; Cui, Y.; Yamamura, K.; Akahori, T.; Nakagawa, K.; Nagai, M.; Nakamura, K.; Takagi, T.; Ikeda, N.; et al. A Gene Expression Signature for Predicting Response to Neoadjuvant Chemoradiotherapy in Pancreatic Ductal Adenocarcinoma. Int. J. Cancer 2021, 148, 769–779. [Google Scholar] [CrossRef]
- Fitzal, F.; Filipits, M.; Fesl, C.; Rudas, M.; Greil, R.; Balic, M.; Moinfar, F.; Herz, W.; Dubsky, P.; Bartsch, R.; et al. PAM-50 Predicts Local Recurrence After Breast Cancer Surgery in Postmenopausal Patients with ER+/HER2- Disease: Results from 1204 Patients in the Randomized ABCSG-8 Trial. Br. J. Surg. 2021, 108, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Shen, M.; Du, Z.; Cao, J.; Tian, Y.; Zeng, P.; Tang, Z. Developing ZNF Gene Signatures Predicting Radiosensitivity of Patients with Breast Cancer. J. Oncol. 2021, 2021, 9255494. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, M.; Qiao, Q.; Wang, Y. Integrating Intrinsic Radiosensitivity and Immune Status for Predicting Benefits of Radiotherapy in Head and Neck Squamous Cell Carcinoma. Med. Sci. Monit. 2021, 27, e932126. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Guo, Y.; Shui, Y.; Li, J.; Jiang, B.; Wei, Q. Combination of Radiosensitivity Gene Signature and PD-L1 Status Predicts Clinical Outcome of Patients with Locally Advanced Head and Neck Squamous Cell Carcinoma: A Study Based on the Cancer Genome Atlas Dataset. Front. Mol. Biosci. 2021, 8, 775562. [Google Scholar] [CrossRef]
- Kim, K.H.; Chang, J.S.; Byun, H.K.; Kim, Y.B. A Novel Gene Signature Associated with Poor Response to Chemoradiotherapy in Patients with Locally Advanced Cervical Cancer. J. Gynecol. Oncol. 2022, 33, e7. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yan, D.; Bai, L.; Geng, R.; Zhao, X.; Li, H.; Dong, Y.; Cao, J.; Tang, Z.; Liu, S. An 11-Gene Signature Based on Treatment Responsiveness Predicts Radiation Therapy Survival Benefit among Breast Cancer Patients. Front. Oncol. 2022, 11, 816053. [Google Scholar] [CrossRef]
- Wu, S.; Xu, J.; Li, G.; Jin, X. Integrating Radiosensitivity Gene Signature Improves Glioma Outcome and Radiotherapy Response Prediction. Medicina 2022, 58, 1327. [Google Scholar] [CrossRef] [PubMed]
- Sjöström, M.; Fyles, A.; Liu, F.; McCready, D.; Shi, W.; Rey-McIntyre, K.; Chang, S.L.; Feng, F.Y.; Speers, C.W.; Pierce, L.J.; et al. Development and Validation of a Genomic Profile for the Omission of Local Adjuvant Radiation in Breast Cancer. J. Clin. Oncol. 2023, 41, 1533–1540. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, Q.; Li, Z.; Xia, X.; Zhang, W.; Qin, Y.; Wu, D.; Ren, C. Exploration of the Radiosensitivity-Related Prognostic Risk Signature in Patients with Glioma: Evidence from Microarray Data. J. Transl. Med. 2023, 21, 618. [Google Scholar] [CrossRef]
- Weichselbaum, R.R.; Ishwaran, H.; Yoon, T.; Nuyten, D.S.A.; Baker, S.W.; Khodarev, N.; Su, A.W.; Shaikh, A.Y.; Roach, P.; Kreike, B.; et al. An Interferon-Related Gene Signature for DNA Damage Resistance is a Predictive Marker for Chemotherapy and Radiation for Breast Cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 18490–18495. [Google Scholar] [CrossRef]
- Eschrich, S.A.; Fulp, W.J.; Pawitan, Y.; Foekens, J.A.; Smid, M.; Martens, J.W.M.; Echevarria, M.; Kamath, V.; Lee, J.; Harris, E.E.; et al. Validation of a Radiosensitivity Molecular Signature in Breast Cancer. Clin. Cancer Res. 2012, 18, 5134–5143. [Google Scholar] [CrossRef]
- Drukker, C.A.; Elias, S.G.; Nijenhuis, M.V.; Wesseling, J.; Bartelink, H.; Elkhuizen, P.; Fowble, B.; Whitworth, P.W.; Patel, R.R.; de Snoo, F.A.; et al. Gene Expression Profiling to Predict the Risk of Locoregional Recurrence in Breast Cancer: A Pooled Analysis. Breast Cancer Res. Treat. 2014, 148, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Tramm, T.; Mohammed, H.; Myhre, S.; Kyndi, M.; Alsner, J.; Børresen-Dale, A.; Sørlie, T.; Frigessi, A.; Overgaard, J. Development and Validation of a Gene Profile Predicting Benefit of Postmastectomy Radiotherapy in Patients with High-Risk Breast Cancer: A Study of Gene Expression in the DBCG82bc Cohort. Clin. Cancer Res. 2014, 20, 5272–5280. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Li, P.; Zhang, Q.; Yang, Z.; Fu, S. A Radiosensitivity Gene Signature in Predicting Glioma Prognostic via EMT Pathway. Oncotarget 2014, 5, 4683–4693. [Google Scholar] [CrossRef] [PubMed]
- Torres-Roca, J.F.; Eschrich, S.; Zhao, H.; Bloom, G.; Sung, J.; McCarthy, S.; Cantor, A.B.; Scuto, A.; Li, C.; Zhang, S.; et al. Prediction of Radiation Sensitivity using a Gene Expression Classifier. Cancer Res. 2005, 65, 7169–7176. [Google Scholar] [CrossRef] [PubMed]
- Eschrich, S.; Zhang, H.; Zhao, H.; Boulware, D.; Lee, J.; Bloom, G.; Torres-Roca, J.F. Systems Biology Modeling of the Radiation Sensitivity Network: A Biomarker Discovery Platform. Int. J. Radiat. Oncol. Biol. Phys. 2009, 75, 497–505. [Google Scholar] [CrossRef]
- Eschrich, S.A.; Pramana, J.; Zhang, H.; Zhao, H.; Boulware, D.; Lee, J.; Bloom, G.; Rocha-Lima, C.; Kelley, S.; Calvin, D.P.; et al. A Gene Expression Model of Intrinsic Tumor Radiosensitivity: Prediction of Response and Prognosis after Chemoradiation. Int. J. Radiat. Oncol. Biol. Phys. 2009, 75, 489–496. [Google Scholar] [CrossRef]
- Scott, J.G.; Berglund, A.; Schell, M.J.; Mihaylov, I.; Fulp, W.J.; Yue, B.; Welsh, E.; Caudell, J.J.; Ahmed, K.; Strom, T.S.; et al. A Genome-Based Model for Adjusting Radiotherapy Dose (GARD): A Retrospective, Cohort-Based Study. Lancet Oncol. 2017, 18, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G.; Sedor, G.; Ellsworth, P.; Scarborough, J.A.; Ahmed, K.A.; Oliver, D.E.; Eschrich, S.A.; Kattan, M.W.; Torres-Roca, J.F. Pan-Cancer Prediction of Radiotherapy Benefit using Genomic-Adjusted Radiation Dose (GARD): A Cohort-Based Pooled Analysis. Lancet Oncol. 2021, 22, 1221–1229. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, S.C.; Kim, S.J.; Park, C.H.; Jeung, H.; Kim, Y.B.; Ahn, J.B.; Chung, H.C.; Rha, S.Y. Identification of a Radiosensitivity Signature Using Integrative Metaanalysis of Published Microarray Data for NCI-60 Cancer Cells. BMC Genom. 2012, 13, 348. [Google Scholar] [CrossRef]
- Van’t Veer, L.J.; Dai, H.; Van De Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T. Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed]
- van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.M.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Ismaila, N.; Allison, K.H.; Barlow, W.E.; Collyar, D.E.; Damodaran, S.; Henry, N.L.; Jhaveri, K.; Kalinsky, K.; Kuderer, N.M.; et al. Biomarkers for Adjuvant Endocrine and Chemotherapy in Early-Stage Breast Cancer: ASCO Guideline Update. J. Clin. Oncol. 2022, 40, 1816–1837. [Google Scholar] [CrossRef] [PubMed]
- Mittempergher, L.; Delahaye, L.J.M.J.; Witteveen, A.T.; Spangler, J.B.; Hassenmahomed, F.; Mee, S.; Mahmoudi, S.; Chen, J.; Bao, S.; Snel, M.H.J.; et al. MammaPrint and BluePrint Molecular Diagnostics using Targeted RNA Next-Generation Sequencing Technology. J. Mol. Diagn. 2019, 21, 808–823. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- National Institute for Health and Care Excellence. Tumour Profiling Tests to Guide Adjuvant Chemotherapy Decisions in Early Breast Cancer. Diagnostics Guidance (DG34); National Institute for Health and Care Excellence: London, UK, 2018. [Google Scholar]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E.; Dees, E.C.; Goetz, M.P.; Olson, J.A.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef]
- Solin, L.J.; Gray, R.; Baehner, F.L.; Butler, S.M.; Hughes, L.L.; Yoshizawa, C.; Cherbavaz, D.B.; Shak, S.; Page, D.L.; Sledge, G.W., Jr.; et al. A Multigene Expression Assay to Predict Local Recurrence Risk for Ductal Carcinoma In Situ of the Breast. J. Natl. Cancer Inst. 2013, 105, 701–710. [Google Scholar] [CrossRef]
- Yuan, Y.; Van Dyke, A.L.; Kurian, A.W.; Negoita, S.; Petkov, V.I. Oncotype DX DCIS use and Clinical Utility: A SEER Population-Based Study. J. Clin. Oncol. 2019, 37, e12046. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular Portraits of Human Breast Tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Wallden, B.; Storhoff, J.; Nielsen, T.; Dowidar, N.; Schaper, C.; Ferree, S.; Liu, S.; Leung, S.; Geiss, G.; Snider, J.; et al. Development and Verification of the PAM50-Based Prosigna Breast Cancer Gene Signature Assay. BMC Med. Genom. 2015, 8, 54–56. [Google Scholar] [CrossRef]
- Effects of Radiotherapy and of Differences in the Extent of Surgery for Early Breast Cancer on Local Recurrence and 15-Year Survival: An Overview of the Randomised Trials. Lancet 2005, 366, 2087–2106. [CrossRef] [PubMed]
- Coindre, J.M.; Terrier, P.; Bui, N.B.; Bonichon, F.; Collin, F.; Le Doussal, V.; Mandard, A.M.; Vilain, M.O.; Jacquemier, J.; Duplay, H.; et al. Prognostic Factors in Adult Patients with Locally Controlled Soft Tissue Sarcoma. A Study of 546 Patients from the French Federation of Cancer Centers Sarcoma Group. J. Clin. Oncol. 1996, 14, 869–877. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.D.; Overton, I.M.; McMahon, S.J. Validation of In Vitro Trained Transcriptomic Radiosensitivity Signatures in Clinical Cohorts. Cancers 2023, 15, 3504. [Google Scholar] [CrossRef] [PubMed]
- Eggener, S.E.; Bryan Rumble, R.; Armstrong, A.J.; Morgan, T.M.; Crispino, T.; Cornford, P.; van Theodorus, D.K.; Grignon, D.J.; Rai, A.J.; Agarwal, N.; et al. Molecular Biomarkers in Localized Prostate Cancer: ASCO Guideline. J. Clin. Oncol. 2020, 38, 1474–1494. [Google Scholar] [CrossRef] [PubMed]
- Venet, D.; Dumont, J.E.; Detours, V. Most Random Gene Expression Signatures are significantly Associated with Breast Cancer Outcome. PLoS Comput. Biol. 2011, 7, e1002240. [Google Scholar] [CrossRef] [PubMed]
- Harris, B.H.L.; Barberis, A.; West, C.M.L.; Buffa, F.M. Gene Expression Signatures as Biomarkers of Tumour Hypoxia. Clin. Oncol. 2015, 27, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Doudna, D. CRISPR Technology: A Decade of Genome Editing is Only the Beginning. Science 2023, 379, eadd8643. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.D.; Overton, I.M.; McMahon, S.J. RadSigBench: A Framework for Benchmarking Functional Genomics Signatures of Cancer Cell Radiosensitivity. Brief Bioinform. 2022, 23, bbab561. [Google Scholar] [CrossRef] [PubMed]
- Mistry, H.B. Radiosensitivity Index is Not Fit to Be Used for Dose Adjustments: A Pan-Cancer Analysis. Clin. Oncol. (R. Coll. Radiol.) 2023, 35, 565–570. [Google Scholar] [CrossRef]
- Torres-Roca, J.F.; Grass, G.D.; Scott, J.G.; Eschrich, S.A. Towards Data Driven RT Prescription: Integrating Genomics into RT Clinical Practice. Semin. Radiat. Oncol. 2023, 33, 221–231. [Google Scholar] [CrossRef]
- Creelan, B.; Eschrich, S.A.; Fulp, W.J.; Torres-Roca, J. A Gene Expression Platform to Predict Benefit from Adjuvant External Beam Radiation in Resected Non-Small Cell Lung Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2014, 90, S76–S77. [Google Scholar] [CrossRef]
- Strom, T.; Torres-Roca, J.F.; Parekh, A.; Naghavi, A.O.; Caudell, J.J.; Oliver, D.E.; Messina, J.L.; Khushalani, N.I.; Zager, J.S.; Sarnaik, A.; et al. Regional Radiation Therapy Impacts Outcome for Node-Positive Cutaneous Melanoma. J. Natl. Compr. Cancer Netw. 2017, 15, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Grass, G.D.; Azizi, M.; Ahmed, K.A.; Yoder, G.S.J.; Welsh, E.A.; Fulp, W.J.; Dhillon, J.; Torres-Roca, J.F.; Giuliano, A.R.; et al. Intrinsic Radiosensitivity, Genomic-Based Radiation Dose and Patterns of Failure of Penile Cancer in Response to Adjuvant Radiation Therapy. Rep. Pract. Oncol. Radiother. 2019, 24, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Strom, T.; Hoffe, S.E.; Fulp, W.; Frakes, J.; Coppola, D.; Springett, G.M.; Malafa, M.P.; Harris, C.L.; Eschrich, S.A.; Torres-Roca, J.; et al. Radiosensitivity Index Predicts for Survival with Adjuvant Radiation in Resectable Pancreatic Cancer. Radiother. Oncol. 2015, 117, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, S.; Serafini, M.S.; Carenzo, A.; Canevari, S.; Brakenhoff, R.H.; René Leemans, C.; Nauta, I.H.; Hoebers, F.; van den Hout, M.F.; Scheckenbach, K.; et al. Clinical Validity of a Prognostic Gene Expression Cluster-Based Model in Human Papillomavirus–Positive Oropharyngeal Carcinoma. JCO Precis. Oncol. 2021, 5, 1666–1676. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Yang, L.; More, E.; Irlam-Jones, J.J.; Valentine, H.R.; Hoskin, P.; Choudhury, A.; West, C.M. Developing Tumor Radiosensitivity Signatures Using LncRNAs. Radiat. Res. 2021, 195, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Unraveling Your Genome. Individualizing Radiation Therapy. Available online: https://www.cvergenx.com/ (accessed on 1 April 2024).
- Genomically Guided Radiation Therapy in the Management of Triple Negative Breast Cancer. Available online: https://clinicaltrials.gov/study/NCT05528133 (accessed on 1 April 2024).
- Murray Brunt, A.; Haviland, J.S.; Wheatley, D.A.; Sydenham, M.A.; Alhasso, A.; Bloomfield, D.J.; Chan, C.; Churn, M.; Cleator, S.; Coles, C.E.; et al. Hypofractionated Breast Radiotherapy for 1 Week Versus 3 Weeks (FAST-Forward): 5-Year Efficacy and Late Normal Tissue Effects Results from a Multicentre, Non-Inferiority, Randomised, Phase 3 Trial. Lancet 2020, 395, 1613–1626. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.Q.; Shubeck, S.; Howard, F.M.; Chen, N.; Nanda, R.; Huo, D. Evaluation of Multigene Assays as Predictors for Response to Neoadjuvant Chemotherapy in Early-Stage Breast Cancer Patients. NPJ Breast Cancer 2023, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- The PRECISION Trial (Profiling Early Breast Cancer for Radiotherapy Omission): A Phase II Study of Breast-Conserving Surgery without Adjuvant Radiotherapy for Favorable-Risk Breast Cancer. Available online: https://www.clinicaltrials.gov/study/NCT02653755 (accessed on 1 April 2024).
- Jagsi, R.; Griffith, K.A.; Harris, E.E.; Wright, J.L.; Recht, A.; Taghian, A.G.; Lee, L.; Moran, M.S.; Small, W.; Johnstone, C.; et al. Omission of Radiotherapy after Breast-Conserving Surgery for Women with Breast Cancer with Low Clinical and Genomic Risk: 5-Year Outcomes of IDEA. J. Clin. Oncol. 2024, 42, 390–398. [Google Scholar] [CrossRef]
- Parulekar, W.R.; Berrang, T.; Kong, I.; Rakovitch, E.; Theberge, V.; Gelmon, K.A.; Chia, S.K.L.; Bellon, J.R.; Jagsi, R.; Ho, A.Y.; et al. Cctg MA.39 Tailor RT: A Randomized Trial of Regional Radiotherapy in Biomarker Low-Risk Node-Positive Breast Cancer (NCT03488693). J. Clin. Oncol. 2019, 37, TPS602. [Google Scholar] [CrossRef]
- White, J.R.; Anderson, S.J.; Eleanor, E.H.; Mamounas, E.P.; Stover, D.G.; Ganz, P.A.; Jagsi, R.; Cecchini, R.S.; Bergom, C.; Theberge, V.; et al. NRG-BR007: A Phase III Trial Evaluating De-Escalation of Breast Radiation (DEBRA) Following Breast-Conserving Surgery (BCS) of Stage 1, Hormone Receptor+, HER2-, RS ≤18 Breast Cancer. J. Clin. Oncol. 2022, 40, TPS613. [Google Scholar] [CrossRef]
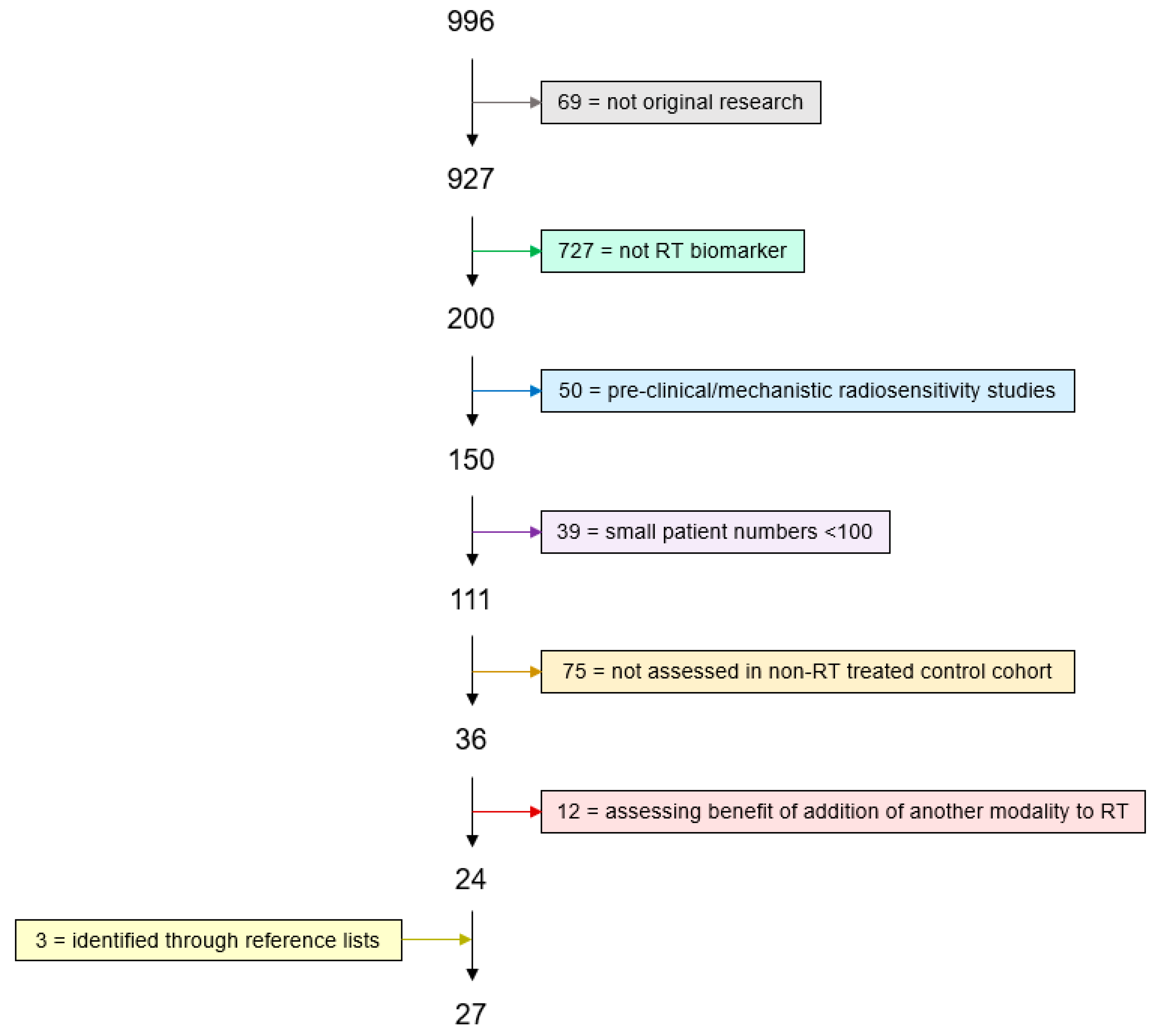
| Not original research article (reviews, letters, comments) |
| Not radiotherapy biomarkers (irrelevant, diagnostic markers, prognostic markers, chemotherapy benefit, immunotherapy benefit, normal tissue toxicity) |
| Pre-clinical or mechanistic studies of radiosensitivity |
| Assessed in a small number of patients (<100) |
| Not assessed as predictive marker (no non-radiotherapy-treated control cohort) |
| Predicting benefit of addition of another modality to radiotherapy (hypoxia modification, concurrent chemotherapy) |
| Signature | Derivation | Proposed Biological Mechanism | Tumour Type | Cohort | N | Cohort Type | Prognosis/Prediction | Endpoint | Assay/Tissue | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 10-gene signature (RSI) | in vitro | Radiosensitivity | Prostate | Moffitt | 618 | Validation | Prediction | Distant metastasis | Microarray/FF | Torres-Roca et al., 2014 [13]. |
| 3-gene signature | in vivo | Cell adhesion molecules | TNBC | Shanghai | 32 | Training | - | - | Microarray/FF | Wushou et al., 2015 [14]. |
| Shanghai | 166 | Validation | Prediction | RFS | ||||||
| 24-gene signature (PORTOS) | in vivo | DNA damage repair and radiation response | Prostate | Mayo Clinic | 196 | Training | Prediction | Distant metastasis | Microarray/FFPE | Zhao et al., 2016 [15]. |
| Pooled | 330 | Validation | Prediction | Distant metastasis | ||||||
| 7-gene signature (Oncotype Dx DCIS) | in vitro | Proliferation | DCIS | Ontario DCIS | 1260 | Validation | Prediction | LRR | qRT-PCR/FFPE | Rakovitch et al., 2017 [16]. |
| 26-gene signature | in vivo | Radiosensitivity | STS | TCGA | 253 | Training/Validation | Prediction | OS | RNA-Seq/FF | Tang et al., 2017 [17]. |
| 65-gene signature | in vivo | Radiosensitivity | STS | TCGA | 218 | Training/Validation | Prediction | OS | RNA-Seq/FF | Tang et al., 2017 [18]. |
| 11-gene signature | in vivo | Radiosensitivity | Gastric | TCGA | 371 | Training/Validation | Prediction | OS | RNA-Seq/FF | Zhou et al., 2017 [19]. |
| 5-miRNA signature | in vivo | Not stated | HNSCC | TCGA | 553 | Training | - | - | miRNA-Seq/FF | Chen et al., 2018 [20]. |
| TCGA | 154 | Training | Prognosis | OS | ||||||
| TCGA | 153 | Validation | Prognosis | OS | ||||||
| TCGA | 509 | Validation | Prediction | OS | ||||||
| 31-gene signature + PD-L1 | in vivo | Cell junction and adhesion | Glioma | TCGA | 511 | Validation | Prediction | OS | RNA-Seq/FF | Jang et al., 2018 [21]. |
| 10-gene signature (RSI) | in vitro | Radiosensitivity | Breast | Sweden | 307 | Validation | Prediction | LR | Nanostring/FF | Sjöström et al., 2018 [22]. |
| 16-gene signature (Oncotype Dx) * | in vitro | Prognostic signature | Breast | NCDB | 7332 | Validation | Prediction | OS | RT-PCR/FFPE | Goodman et al., 2018 [23]. |
| SEER | 3087 | Validation | Prediction | OS | ||||||
| 30-gene signature | in vivo | Radiosensitivity | Breast | TCGA | 700 | Training/Validation | Prediction | OS | RNA-Seq/FF | Ji et al., 2018 [24]. |
| 34-gene signature | in vivo | Radiosensitivity | Breast | GSE30682 | 343 | Training | Prognosis | LRFS | Microarray/FF | Cui et al., 2018 [25]. |
| NKI | 319 | Validation | Prognosis | RFS | ||||||
| GSE2034 | 286 | Validation | Prognosis | RFS | ||||||
| METABRIC | 262 | Validation | Prediction | DSS | ||||||
| 27-gene signature (ARTIC) | in vivo | Not stated | Breast | GSE30692 | 343 | Training | Prognosis | LR | Microarray/FF | Sjöström et al., 2019 [26]. |
| NKI | 228 | Training | Prognosis | LR | Microarray/FF | |||||
| GSE103746 | 106 | Training | Prognosis | LR | Microarray/FF | |||||
| SweBCG91-RT | 748 | Validation | Prediction | LRR | Microarray/FFPE | |||||
| 10-gene signature (RSI) | in vitro | Radiosensitivity | Endometrial | Moffitt | 204 | Validation | Prediction | Pelvic control | Microarray/FFPE | Mohammadi et al., 2020 [27]. |
| 30-gene signature | in vitro | Cell junction and adhesion | GBM | TCGA | 399 | Validation | Prediction | OS | RNA-Seq/FF | Jang et al., 2020 [28]. |
| 10-gene signature (RSI) | in vitro | Radiosensitivity | Prostate | Manchester | 386 | Validation | Prediction | PFS | Microarray/FFPE | Thiruthaneeswaran et al., 2020 [29]. |
| 10-gene signature (RSI) | in vitro | Radiosensitivity | PDAC | TCGA | 182 | Training | Prognosis | OS | RNA-Seq/FF | Nishiwada et al., 2021 [30]. |
| ICGC | 94 | Validation | Prognosis | OS | ||||||
| Nara | 145 | Training | Prognosis | OS | RT-PCR/FFPE | |||||
| Kumamoto | 112 | Validation | Prognosis | OS | ||||||
| pre-NACRT cohort | 56 | Validation | Prediction | Pathological response | ||||||
| 46-gene signature (PAM-50) | in vivo | Prognostic signature | Breast | ABCSG-8 | 1204 | Validation | Prediction | LR | Nanostring/FF | Fitzal et al., 2021 [31]. |
| 4-gene signature | in vivo | Transcriptional regulation | Breast | TCGA | 976 | Training | Prediction | OS | RNA-Seq/FF | Yan et al., 2021 [32]. |
| METABRIC | 1798 | Training/Validation | Prediction | OS | Microarray/FF | |||||
| 3-gene signature | in vivo | Radiosensitivity/immune status | HNSCC | TCGA | 236 | Training | Prediction | OS | RNA-Seq/FF | Sun et al., 2021 [33]. |
| TCGA | 156 | Validation | Prediction | OS | ||||||
| 31-gene signature | in vitro | Cell junction and adhesion | HNSCC | TCGA | 288 | Validation | Prediction | OS | RNA-Seq/FF | Dai et al., 2021 [34]. |
| 185-gene signature | in vivo | Various | Cervix | TCGA | 9 | Derivation | - | PFS | RNA-Seq/FFPE | Kim et al., 2022 [35]. |
| TCGA | 273 | Validation | Prediction | PFS | RNA-Seq/FF | |||||
| 11-gene signature | in vivo | Not stated | Breast | TCGA | 937 | Training | Prediction | PFS | RNA-Seq/FF | Shen et al., 2022 [36]. |
| E-TABM-158 | 130 | Validation | Prediction | PFS | ||||||
| 12-gene signature | in vivo | Radiosensitivity | Glioma | CGGA | 748 | Validation | Prediction | OS | RNA-Seq/FF | Wu et al., 2022 [37]. |
| TCGA | 647 | Validation | Prediction | OS | ||||||
| 16-gene signature (POLAR) | in vivo | Not stated | Breast | SweBCG91-RT | 243 | Training | Prediction | LRR | Microarray/FFPE | Sjöström et al., 2023 [38]. |
| SweBCG91-RT | 354 | Validation | Prediction | LRR | ||||||
| Princess Margaret | 132 | Validation | Prediction | LRR | Microarray/FF | |||||
| 9-gene signature | in vitro/in vivo | Not stated | Glioma/GBM | GSE7696 | 84 | Derivation | - | - | RNA-Seq/FF | Zhang et al., 2023 [39]. |
| TCGA -GBM | 152 | Training/Validation | Prediction | PFS | RNA-Seq/FF | |||||
| TCGA-low grade glioma | 616 | Training/Validation | Prediction | PFS | RNA-Seq/FF | |||||
| SMU-NFH | 31 | Validation | Prediction | PFS | RNA-Seq/FFPE | |||||
| CGGA | 501 | Validation | Prediction | OS | RNA-Seq/FF |
| Biomarker | Derivation | Proposed Biological Mechanism | Tumour Type | Cohort | n | Cohort Type | Prognosis/Prediction | Endpoint | Clinical Assay | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 7-gene signature | in vitro | IFN-related DNA damage resistance | Breast | Multiple (meta-analysis) | 1573 | Validation | Prediction | LRC | Microarray/FF | Weichselbaum et al., 2008 [40]. |
| 10-gene signature (RSI) | in vitro | Radiosensitivity | Breast | Karolinska | 159 | Validation | Prediction | RFS | Microarray/FF | Eschrich et al., 2012 [41]. |
| Erasmus | 344 | Validation | Prediction | MFS | ||||||
| 70-gene signature (Mammaprint) * | in vivo | Prognostic signature | Breast | NKI | 1053 | Validation | Prediction | LRR | Microarray/FFPE | Drukker et al., 2014 [42]. |
| 7-gene signature (DBCG-RT) | in vivo | Not stated | Breast | DBCG82bc | 191 | Training | Prediction | LRR | Microarray/FF | Tramm et al., 2014 [43]. |
| DBCG82bc | 112 | Validation | Prediction | LRR | qRT-PCR/FFPE | |||||
| 31-gene signature | in vitro | Radiosensitivity | Glioma | GSE16011 | 263 | Validation | Prediction | OS | RNA-Seq/FF | Meng et al., 2014 [44]. |
| TCGA | 463 | Validation | Prognosis | OS | Microarray/FF |
| Gene | Signatures (n) | Function | References |
|---|---|---|---|
| ACTN1 | 2 | F-actin cross-linking protein which is thought to anchor actin to a variety of intracellular structures. This is a bundling protein. | Meng et al., 2014 [44], Kim et al., 2022 [35]. |
| ANLN | 2 | Required for cytokinesis. | Zhao et al., 2016 [15], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| BAG1 | 2 | Acts as a nucleotide-exchange factor promoting the release of ADP from the HSP70 and HSC70 proteins thereby triggering client/substrate protein release. | Goodman et al., 2018 (Oncotype Dx) [23], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| BCL2 | 2 | Regulates cell death by controlling the mitochondrial membrane permeability. Appears to function in a feedback loop system with caspases. | Goodman et al., 2018 (Oncotype Dx) [23], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| BTG3 | 2 | Overexpression impairs serum-induced cell cycle progression from the G0/G1 to S phase. | Cui et al., 2018 [25], Sjöström et al., 2019 [26]. |
| CCNB1 | 4 | Essential for the control of the cell cycle at the G2/M (mitosis) transition. | Goodman et al., 2018 (Oncotype Dx) [23], Cui et al., 2018 [25], Sjöström et al., 2019 [26], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| CDC5L * | 2 | DNA-binding protein involved in cell cycle control. May act as a transcription activator. Plays a role in pre-mRNA splicing as core component of precatalytic, catalytic and post-catalytic spliceosomal complexes. | Tang et al., 2017 [17], Tang et al., 2017 [18]. |
| CENPF | 2 | Required for kinetochore function and chromosome segregation in mitosis. | Sjöström et al., 2019 [26], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| CKB | 2 | Reversibly catalyses the transfer of phosphate between ATP and various phosphogens. | Fitzal et al., 2021 (Prosigna PAM-50) [31], Shen et al., 2022 [36]. |
| CLGN | 2 | Functions during spermatogenesis as a chaperone for a range of client proteins that are important for sperm adhesion onto the egg zona pellucida and for subsequent penetration of the zona pellucida. | Cui et al., 2018 [25], Sun et al., 2021 [33]. |
| DAG1 | 2 | The dystroglycan complex is involved in a number of processes including laminin and basement membrane assembly, sarcolemmal stability, cell survival, peripheral nerve myelination, nodal structure, cell migration, and epithelial polarisation. | Meng et al., 2014 [44], Ji et al., 2018 [24]. |
| DRAM1 | 2 | Lysosomal modulator of autophagy that plays a central role in p53/TP53-mediated apoptosis. | Zhao et al., 2016 [15], Tang et al., 2017 [17] |
| DTL | 2 | Substrate-specific adapter of a DCX (DDB1-CUL4-X-box) E3 ubiquitin-protein ligase complex required for cell cycle control, DNA damage response and translesion DNA synthesis. | Drukker et al., 2014 (MammaPrint) [42], Zhao et al., 2016 [15]. |
| ERBB2 | 2 | Protein tyrosine kinase that is part of several cell surface receptor complexes. | Goodman et al., 2018 (Oncotype Dx) [23], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| ESR1 | 2 | Nuclear hormone receptor. | Goodman et al., 2018 (Oncotype Dx) [23], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| GNG11 | 2 | G-protein transmembrane signalling. | Zhao et al., 2016 [15], Sjöström et al., 2023 (POLAR) [38]. |
| HCLS1 | 2 | Substrate of the antigen receptor-coupled tyrosine kinase. Plays a role in antigen receptor signalling for both clonal expansion and deletion in lymphoid cells. May also be involved in the regulation of gene expression. | Meng et al., 2014 [44], Zhao et al., 2016 [15]. |
| KNTC2 | 2 | Acts as a component of the essential kinetochore-associated NDC80 complex, which is required for chromosome segregation and spindle checkpoint activity. | Drukker et al., 2014 (MammaPrint) [42], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| KPNA2 | 2 | Functions in nuclear protein import as an adapter protein for nuclear receptor KPNB1. | Cui et al., 2018 [25], Sjöström et al., 2023 (POLAR) [38]. |
| KRT14 | 2 | The nonhelical tail domain is involved in promoting KRT5-KRT14 filaments to self-organise into large bundles. | Zhao et al., 2016 [15], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| KRT15 | 2 | The keratins are intermediate filament proteins responsible for the structural integrity of epithelial cells and are subdivided into cytokeratins and hair keratins. | Sun et al., 2021 [33], Kim et al., 2022 [35]. |
| MDM2 | 2 | E3 ubiquitin-protein ligase that mediates ubiquitination of p53/TP53, leading to its degradation by the proteasome. | Cui et al., 2018 [25], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| MELK | 2 | Serine/threonine-protein kinase involved in various processes such as cell cycle regulation, self-renewal of stem cells, apoptosis and splicing regulation. | Drukker et al., 2014 (MammaPrint) [42], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| MKI67 | 2 | Required to maintain individual mitotic chromosomes dispersed in the cytoplasm following nuclear envelope disassembly | Goodman et al., 2018 (Oncotype Dx) [23], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| MMD | 2 | Involved in the dynamics of lysosomal membranes associated with microglial activation following brain lesion. | Cui et al., 2018 [25], Kim et al., 2022 [35]. |
| MMP11 | 4 | May play an important role in the progression of epithelial malignancies. | Goodman et al., 2018 (Oncotype Dx) [23], Fitzal et al., 2021 (Prosigna PAM-50) [31], Kim et al., 2022 [35], Sjöström et al., 2023 (POLAR) [38]. |
| MORF4L2 | 2 | Component of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histone H4 and H2A. | Kim et al., 2022 [35], Shen et al., 2022 [36] |
| MX1 | 2 | Interferon-induced dynamin-like GTPase with antiviral activity against a wide range of RNA viruses and some DNA viruses. | Weichselbaum et al., 2008 [40], Kim et al., 2022 [35]. |
| NAT1 | 2 | Participates in the detoxification of a plethora of hydrazine and arylamine drugs. | Tang et al., 2017 [17], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| ORC6L | 2 | Component of the origin recognition complex (ORC) that binds origins of replication. | Drukker et al., 2014 (MammaPrint) [42], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| PGR | 2 | The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. | Goodman et al., 2018 (Oncotype Dx) [23], Fitzal et al., 2021 (Prosigna PAM-50) [31]. |
| PLK2 | 2 | Tumour suppressor serine/threonine-protein kinase involved in synaptic plasticity, centriole duplication and G1/S phase transition. | Zhao et al., 2016 [15], Zhang et al., 2023 [39]. |
| POSTN | 2 | Induces cell attachment and spreading and plays a role in cell adhesion. | Kim et al., 2022 [35], Zhang et al., 2023 [39]. |
| PYGB | 2 | Glycogen phosphorylase that regulates glycogen mobilisation. | Meng et al., 2014 [44], Cui et al., 2018 [25]. |
| RGS4 | 2 | Inhibits signal transduction by increasing the GTPase activity of G protein alpha subunits thereby driving them into their inactive GDP-bound form. | Tang et al., 2017 [17], Kim et al., 2022 [35]. |
| SCUBE2 | 2 | SHH long-range signalling by binding to the dually lipid-modified SHH (ShhNp) and by promoting ShhNp mobilisation, solubilisation and release from the cell membrane. | Drukker et al., 2014 (MammaPrint) [42], Goodman et al., 2018 (Oncotype Dx) [23]. |
| STAT1 | 2 | Signal transducer and transcription activator that mediates cellular responses to interferons (IFNs), cytokine KITLG/SCF and other cytokines and other growth factors | Weichselbaum et al., 2008 [40], Eschrich et al., 2012 [41]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bleaney, C.W.; Abdelaal, H.; Reardon, M.; Anandadas, C.; Hoskin, P.; Choudhury, A.; Forker, L. Clinical Biomarkers of Tumour Radiosensitivity and Predicting Benefit from Radiotherapy: A Systematic Review. Cancers 2024, 16, 1942. https://doi.org/10.3390/cancers16101942
Bleaney CW, Abdelaal H, Reardon M, Anandadas C, Hoskin P, Choudhury A, Forker L. Clinical Biomarkers of Tumour Radiosensitivity and Predicting Benefit from Radiotherapy: A Systematic Review. Cancers. 2024; 16(10):1942. https://doi.org/10.3390/cancers16101942
Chicago/Turabian StyleBleaney, Christopher W., Hebatalla Abdelaal, Mark Reardon, Carmel Anandadas, Peter Hoskin, Ananya Choudhury, and Laura Forker. 2024. "Clinical Biomarkers of Tumour Radiosensitivity and Predicting Benefit from Radiotherapy: A Systematic Review" Cancers 16, no. 10: 1942. https://doi.org/10.3390/cancers16101942
APA StyleBleaney, C. W., Abdelaal, H., Reardon, M., Anandadas, C., Hoskin, P., Choudhury, A., & Forker, L. (2024). Clinical Biomarkers of Tumour Radiosensitivity and Predicting Benefit from Radiotherapy: A Systematic Review. Cancers, 16(10), 1942. https://doi.org/10.3390/cancers16101942






