The Transformative Role of 3D Culture Models in Triple-Negative Breast Cancer Research
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Three-Dimensional Cell Culture Models
3.1.1. Cell Aggregates and Spheroids
3.1.2. Organoids and Tumoroids
3.1.3. Patient-Derived Xenografts (PDXs) and PDX-Organoids (PDXOs)
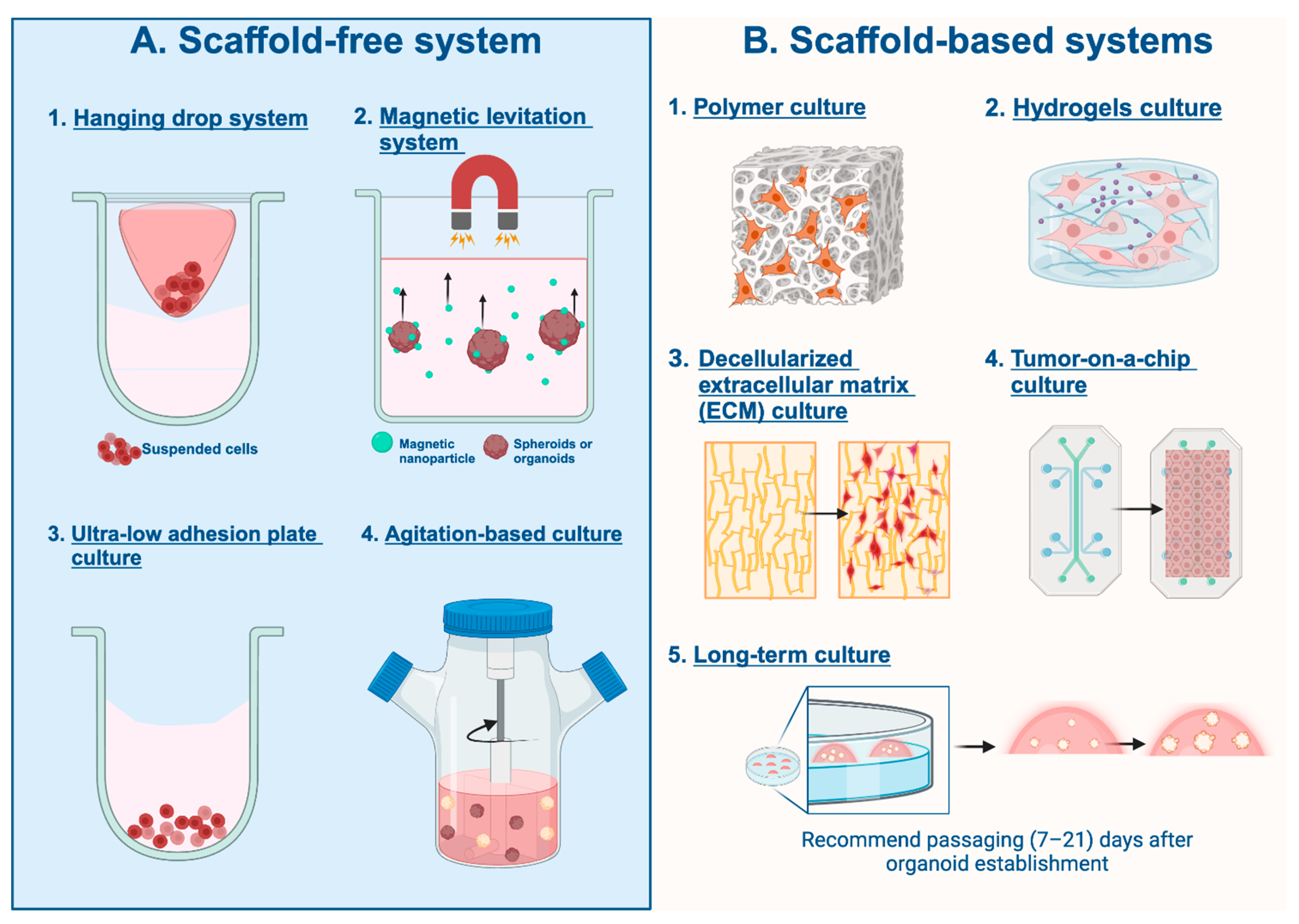
3.2. Three-Dimensional Cell Culturing Techniques
3.2.1. Scaffold-Free System
Hanging Drop System
Magnetic Levitation System
Ultra-Low-Adhesion Plate Culture
Agitation-Based Culture
3.2.2. Scaffold-Based Systems
Polymer Scaffold-Based Culture
Hydrogels Scaffold-Based Culture
Decellularized Extracellular Matrix (ECM) Culture
Tumor-on-a-Chip Culture
Long-Term Culture
3.2.3. 3D Culturing Challenges and Alternative Approaches
3.3. Three-Dimensional Culturing Media and Additives
3.3.1. Base Media for 3D Cell Culture
Plasmax Culture Media
3.3.2. Rho-Protein-Dependent Kinase (ROCK) Inhibitor
3.4. Approaches to Assessing Growth Kinetics, Migration, Morphology, Autophagy, and Cell Death for PDO/PDXO
3.4.1. Live Cell Imaging Techniques and Analysis Instruments
3.4.2. Endpoint Assays
3.5. PDO and PDXO Multi-Omics Analysis
3.5.1. Two-Dimensional vs. Three-Dimensional Culture Multi-Omics Analysis
3.5.2. Omics Analysis Guides Model Selection
3.6. Drug Discovery Using 3D Cancer Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartfeld, S.; Clevers, H. Stem cell-derived organoids and their application for medical research and patient treatment. J. Mol. Med. 2017, 95, 729–738. [Google Scholar] [CrossRef]
- Fujii, M.; Sato, T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat. Mater. 2020, 20, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Kramer, M.; Russo, S.; Naik, P.; Arun, G.; Brophy, K.; Andrews, P.; Fan, C.; Perou, C.M.; Preall, J.; et al. Patient-derived triple negative breast cancer organoids provide robust model systems that recapitulate tumor intrinsic characteristics. Cancer Res. 2022, 82, 1174–1192. [Google Scholar] [CrossRef]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386.e10. [Google Scholar] [CrossRef] [PubMed]
- El Harane, S.; Zidi, B.; El Harane, N.; Krause, K.-H.; Matthes, T.; Preynat-Seauve, O. Cancer Spheroids and Organoids as Novel Tools for Research and Therapy: State of the Art and Challenges to Guide Precision Medicine. Cells 2023, 12, 1001. [Google Scholar] [CrossRef] [PubMed]
- Guillen, K.P.; Fujita, M.; Butterfield, A.J.; Scherer, S.D.; Bailey, M.H.; Chu, Z.; DeRose, Y.S.; Zhao, L.; Cortes-Sanchez, E.; Yang, C.-H.; et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat. Cancer 2022, 3, 232–250. [Google Scholar] [CrossRef]
- Shi, Y.; Guan, Z.; Cai, G.; Nie, Y.; Zhang, C.; Luo, W.; Liu, J. Patient-derived organoids: A promising tool for breast cancer research. Front. Oncol. 2024, 14, 1350935. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Muthuswamy, S.K.; Brugge, J.S. Organoid Cultures for the Study of Mammary Biology and Breast Cancer: The Promise and Challenges. Cold Spring Harb. Perspect. Med. 2023, a041661. [Google Scholar] [CrossRef]
- Huerta-Reyes, M.; Aguilar-Rojas, A. Three-dimensional models to study breast cancer (Review). Int. J. Oncol. 2021, 58, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Han, S.J.; Kwon, S.; Kim, K.S. Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int. 2021, 21, 152. [Google Scholar] [CrossRef] [PubMed]
- Visal, T.H.; Hollander, P.D.; Cristofanilli, M.; Mani, S.A. Circulating tumour cells in the -omics era: How far are we from achieving the ‘singularity’? Br. J. Cancer 2022, 127, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.P.; Grabowska, A.M. Spheroid arrays for high-throughput single-cell analysis of spatial patterns and biomarker expression in 3D. Sci. Rep. 2017, 7, srep41160. [Google Scholar] [CrossRef]
- Huang, Z.; Yu, P.; Tang, J. Characterization of Triple-Negative Breast Cancer MDA-MB-231 Cell Spheroid Model. OncoTargets Ther. 2020, 13, 5395–5405. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.C.; Zboril, E.K.; Olex, A.L.; Leftwich, T.J.; Hairr, N.S.; Byers, H.A.; Valentine, A.D.; Altman, J.E.; Alzubi, M.A.; Grible, J.M.; et al. Discovering Synergistic Compounds with BYL-719 in PI3K Overactivated Basal-like PDXs. Cancers 2023, 15, 1582. [Google Scholar] [CrossRef] [PubMed]
- Rashid, N.S.; Boyd, D.C.; Olex, A.L.; Grible, J.M.; Duong, A.K.; Alzubi, M.A.; Altman, J.E.; Leftwich, T.J.; Valentine, A.D.; Hairr, N.S.; et al. Transcriptomic changes underlying EGFR inhibitor resistance in human and mouse models of basal-like breast cancer. Sci. Rep. 2022, 12, 21248. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.H.; Alzubi, M.A.; Harrell, J.C. Identification of synergistic drug combinations using breast cancer patient-derived xenografts. Sci. Rep. 2020, 10, 1493. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 2020, 13, 4. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, X.; Ding, R.; Yang, L.; Lyu, X.; Zeng, J.; Lei, J.H.; Wang, L.; Bi, J.; Shao, N.; et al. Patient-Derived Organoids Can Guide Personalized-Therapies for Patients with Advanced Breast Cancer. Adv. Sci. 2021, 8, 2101176. [Google Scholar] [CrossRef]
- Takahashi, N.; Higa, A.; Hiyama, G.; Tamura, H.; Hoshi, H.; Dobashi, Y.; Katahira, K.; Ishihara, H.; Takagi, K.; Goda, K.; et al. Construction of in vitro patient-derived tumor models to evaluate anticancer agents and cancer immunotherapy. Oncol. Lett. 2021, 21, 406. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.T.; Alférez, D.G.; Amant, F.; Annibali, D.; Arribas, J.; Biankin, A.V.; Bruna, A.; Budinská, E.; Caldas, C.; Chang, D.K.; et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 2017, 17, 254–268. [Google Scholar] [CrossRef]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, L.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Nayak, B.; Balachander, G.M.; Manjunath, S.; Rangarajan, A.; Chatterjee, K. Tissue mimetic 3D scaffold for breast tumor-derived organoid culture toward personalized chemotherapy. Colloids Surfaces B Biointerfaces 2019, 180, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Chittiboyina, S.; Chang, C.-L.; Lelièvre, S.A.; Savran, C.A. Deterministic culturing of single cells in 3D. Sci. Rep. 2020, 10, 10805. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, Q.; Li, G.; Li, H.; Xu, S.; Pang, D. Cancer organoids: A platform in basic and translational research. Genes Dis. 2024, 11, 614–632. [Google Scholar] [CrossRef]
- Qu, J.; Kalyani, F.S.; Liu, L.; Cheng, T.; Chen, L. Tumor organoids: Synergistic applications, current challenges, and future prospects in cancer therapy. Cancer Commun. 2021, 41, 1331–1353. [Google Scholar] [CrossRef]
- Invrea, F.; Rovito, R.; Torchiaro, E.; Petti, C.; Isella, C.; Medico, E. Patient-derived xenografts (PDXs) as model systems for human cancer. Curr. Opin. Biotechnol. 2020, 63, 151–156. [Google Scholar] [CrossRef]
- Scherer, S.D.; Zhao, L.; Butterfield, A.J.; Yang, C.-H.; Cortes-Sanchez, E.; Guillen, K.P.; Welm, B.E.; Welm, A.L. Breast cancer PDxO cultures for drug discovery and functional precision oncology. STAR Protoc. 2023, 4, 102402. [Google Scholar] [CrossRef]
- Foty, R. A Simple Hanging Drop Cell Culture Protocol for Generation of 3D Spheroids. JoVE (J. Vis. Exp.) 2011, 51, 2720. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, C.; Muñoz-Garcia, J.; Griscom, L.; Cochonneau, D.; Ollivier, E.; Heymann, M.-F.; Vallette, F.M.; Oliver, L.; Heymann, D. Three-dimensional in vitro culture models in oncology research. Cell Biosci. 2022, 12, 155. [Google Scholar] [CrossRef] [PubMed]
- Leung, B.M.; Lesher-Perez, S.C.; Matsuoka, T.; Moraes, C.; Takayama, S. Media additives to promote spheroid circularity and compactness in hanging drop platform. Biomater. Sci. 2015, 3, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.G.; Lee, J.S.; Shim, J.K.; Hur, W. A scaffold-free surface culture of B16F10 murine melanoma cells based on magnetic levitation. Cytotechnology 2016, 68, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, H.; Gage, J.; Leonard, F.; Srinivasan, S.; Souza, G.R.; Dave, B.; Godin, B. Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation. Sci. Rep. 2014, 4, 6468. [Google Scholar] [CrossRef] [PubMed]
- Michl, J.; Park, K.C.; Swietach, P. Evidence-based guidelines for controlling pH in mammalian live-cell culture systems. Commun. Biol. 2019, 2, 144. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.H.; Alzubi, M.A.; Sohal, S.S.; Olex, A.L.; Dozmorov, M.G.; Harrell, J.C. Characterizing the efficacy of cancer therapeutics in patient-derived xenograft models of metastatic breast cancer. Breast Cancer Res. Treat. 2018, 170, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Byerly, J.H.; Port, E.R.; Irie, H.Y. PRKCQ inhibition enhances chemosensitivity of triple-negative breast cancer by regulating Bim. Breast Cancer Res. 2020, 22, 72. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef]
- Gupta, N.; Liu, J.R.; Patel, B.; Solomon, D.E.; Vaidya, B.; Gupta, V. Microfluidics-based 3D cell culture models: Utility in novel drug discovery and delivery research. Bioeng. Transl. Med. 2016, 1, 63–81. [Google Scholar] [CrossRef]
- Abuwatfa, W.H.; Pitt, W.G.; Husseini, G.A. Scaffold-based 3D cell culture models in cancer research. J. Biomed. Sci. 2024, 31, 7. [Google Scholar] [CrossRef]
- Rabie, A.M.I.; Ali, A.S.M.; Al-Zeer, M.A.; Barhoum, A.; El-Hallouty, S.; Shousha, W.G.; Berg, J.; Kurreck, J.; Khalil, A.S.G. Spontaneous Formation of 3D Breast Cancer Tissues on Electrospun Chitosan/Poly(ethylene oxide) Nanofibrous Scaffolds. ACS Omega 2022, 7, 2114–2126. [Google Scholar] [CrossRef] [PubMed]
- De, T.; Goyal, S.; Balachander, G.; Chatterjee, K.; Kumar, P.; Babu, K.G.; Rangarajan, A. A novel ex vivo system using 3D polymer scaffold to culture circulating tumor cells from breast cancer patients exhibits dynamic E-M phenotypes. J. Clin. Med. 2019, 8, 1473. [Google Scholar] [CrossRef] [PubMed]
- Prince, E.; Cruickshank, J.; Ba-Alawi, W.; Hodgson, K.; Haight, J.; Tobin, C.; Wakeman, A.; Avoulov, A.; Topolskaia, V.; Elliott, M.J.; et al. Biomimetic hydrogel supports initiation and growth of patient-derived breast tumor organoids. Nat. Commun. 2022, 13, 1466. [Google Scholar] [CrossRef]
- Badea, M.A.; Balas, M.; Hermenean, A.; Ciceu, A.; Herman, H.; Ionita, D.; Dinischiotu, A. Influence of Matrigel on Single- and Multiple-Spheroid Cultures in Breast Cancer Research. SLAS Discov. Adv. Sci. Drug Discov. 2019, 24, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Wang, H.; Li, G.; Zhao, B. Three-dimensional decellularized tumor extracellular matrices with different stiffness as bioengineered tumor scaffolds. Bioact. Mater. 2021, 6, 2767–2782. [Google Scholar] [CrossRef] [PubMed]
- Iazzolino, G.; Mendibil, U.; Arnaiz, B.; Ruiz-De-Angulo, A.; Azkargorta, M.; Uribe, K.B.; Khatami, N.; Elortza, F.; Olalde, B.; Gomez-Vallejo, V.; et al. Decellularization of xenografted tumors provides cell-specific in vitro 3D environment. Front. Oncol. 2022, 12, 956940. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, J.; Huang, S.; Wu, X.; Xie, X.; Wang, J.; Liu, F.; Zhang, M.; Peng, Z.; Hu, N. Tumor-on-a-chip: From bioinspired design to biomedical application. Microsyst. Nanoeng. 2021, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Lanz, H.L.; Saleh, A.; Kramer, B.; Cairns, J.; Ng, C.P.; Yu, J.; Trietsch, S.J.; Hankemeier, T.; Joore, J.; Vulto, P.; et al. Therapy response testing of breast cancer in a 3D high-throughput perfused microfluidic platform. BMC Cancer 2017, 17, 709. [Google Scholar] [CrossRef]
- Dekkers, J.F.; van Vliet, E.J.; Sachs, N.; Rosenbluth, J.M.; Kopper, O.; Rebel, H.G.; Wehrens, E.J.; Piani, C.; Visvader, J.E.; Verissimo, C.S.; et al. Long-term culture, genetic manipulation and xenotransplantation of human normal and breast cancer organoids. Nat. Protoc. 2021, 16, 1936–1965. [Google Scholar] [CrossRef]
- Urbischek, M.; Rannikmae, H.; Foets, T.; Ravn, K.; Hyvönen, M.; de la Roche, M. Organoid culture media formulated with growth factors of defined cellular activity. Sci. Rep. 2019, 9, 6193. [Google Scholar] [CrossRef]
- Liu, P.; Roberts, S.; Shoemaker, J.T.; Vukasinovic, J.; Tomlinson, D.C.; Speirs, V. Validation of a 3D perfused cell culture platform as a tool for humanised preclinical drug testing in breast cancer using established cell lines and patient-derived tissues. PLoS ONE 2023, 18, e0283044. [Google Scholar] [CrossRef] [PubMed]
- Mazzucchelli, S.; Piccotti, F.; Allevi, R.; Truffi, M.; Sorrentino, L.; Russo, L.; Agozzino, M.; Signati, L.; Bonizzi, A.; Villani, L.; et al. Establishment and Morphological Characterization of Patient-Derived Organoids from Breast Cancer. Biol. Proced. Online 2019, 21, 12. [Google Scholar] [CrossRef]
- Kou, F.; Zhu, B.; Zhou, W.; Lv, C.; Cheng, Y.; Wei, H. Targeted metabolomics in the cell culture media reveals increased uptake of branched amino acids by breast cancer cells. Anal. Biochem. 2021, 624, 114192. [Google Scholar] [CrossRef]
- Msalbi, D.; Jellali, F.; Elloumi-Mseddi, J.; Hakim, B.; Sahli, E.; Aifa, S. Toxicity evaluation of synthetic glucocorticoids against breast cancer cell lines MDA-MB-231, MCF-7 and human embryonic kidney HEK293. Med. Oncol. 2023, 40, 309. [Google Scholar] [CrossRef]
- Ryu, A.H.; Eckalbar, W.L.; Kreimer, A.; Yosef, N.; Ahituv, N. Use antibiotics in cell culture with caution: Genome-wide identification of antibiotic-induced changes in gene expression and regulation. Sci. Rep. 2017, 7, 7533. [Google Scholar] [CrossRef] [PubMed]
- Psilopatis, I.; Mantzari, A.; Vrettou, K.; Theocharis, S. The Role of Patient-Derived Organoids in Triple-Negative Breast Cancer Drug Screening. Biomedicines 2023, 11, 773. [Google Scholar] [CrossRef] [PubMed]
- Divoux, J.; Florent, R.; Jacobs, M.; Lequesne, J.; Grellard, J.-M.; San, C.; Grossi, S.; Kerdja, K.; Clarisse, B.; Boudier, G.; et al. The TRIPLEX study: Use of patient-derived tumor organoids as an innovative tool for precision medicine in triple-negative breast cancer. BMC Cancer 2023, 23, 883. [Google Scholar] [CrossRef]
- Vande Voorde, J.; Ackermann, T.; Pfetzer, N.; Sumpton, D.; Mackay, G.; Kalna, G.; Nixon, C.; Blyth, K.; Gottlieb, E.; Tardito, S. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci. Adv. 2019, 5, eaau7314. [Google Scholar] [CrossRef]
- Ackermann, T.; Tardito, S. Cell Culture Medium Formulation and Its Implications in Cancer Metabolism. Trends Cancer 2019, 5, 329–332. [Google Scholar] [CrossRef]
- Yu, J.; Huang, W. The Progress and Clinical Application of Breast Cancer Organoids. Int. J. Stem Cells 2020, 13, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Bhandary, L.; Whipple, R.A.; Vitolo, M.I.; Charpentier, M.S.; Boggs, A.E.; Chakrabarti, K.R.; Thompson, K.N.; Martin, S.S. ROCK inhibition promotes microtentacles that enhance reattachment of breast cancer cells. Oncotarget 2015, 6, 6251–6266. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.J.; Cook, R.S.; Skala, M.C. Functional Optical Imaging of Primary Human Tumor Organoids: Development of a Personalized Drug Screen. J. Nucl. Med. 2017, 58, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.J.; Cook, R.S.; Sanders, M.E.; Aurisicchio, L.; Ciliberto, G.; Arteaga, C.L.; Skala, M.C. Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res 2014, 74, 5184–5194. [Google Scholar] [CrossRef]
- Cheng, G.; Zielonka, J.; McAllister, D.M.; Mackinnon, A.C., Jr.; Joseph, J.; Dwinell, M.B.; Kalyanaraman, B. Mitochondria-targeted vitamin E analogs inhibit breast cancer cell energy metabolism and promote cell death. BMC Cancer 2013, 13, 285. [Google Scholar] [CrossRef] [PubMed]
- Larson, B.; Malayter, D.; Shure, M. Multiplexed Detection of Cytokine Cancer Biomarkers using Fluorescence RNA In Situ Hybridization and Cellular Imaging; BioTek Applications Notes; Agilent Technologies, Inc.: Santa Clara, CA, USA, 2016; pp. 1–5. [Google Scholar]
- Marqués, G.; Pengo, T.; A Sanders, M. Imaging methods are vastly underreported in biomedical research. eLife 2020, 9, e55133. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Xu, L.; Wei, W.; Cheng, A.; Zhang, L.; Zhang, M.; Wu, G.; Cai, C. CDK16 promotes the progression and metastasis of triple-negative breast cancer by phosphorylating PRC1. J. Exp. Clin. Cancer Res. 2021, 41, 149. [Google Scholar] [CrossRef]
- Hu, W.-Y.; Hu, D.-P.; Xie, L.; Birch, L.A.; Prins, G.S. Isolation of Stem-like Cells from 3-Dimensional Spheroid Cultures. JoVE (J. Vis. Exp.) 2019, 2019, e60357. [Google Scholar] [CrossRef]
- Deben, C.; De La Hoz, E.C.; Le Compte, M.; Van Schil, P.; Hendriks, J.M.; Lauwers, P.; Yogeswaran, S.K.; Lardon, F.; Pauwels, P.; Van Laere, S.; et al. OrBITS: Label-free and time-lapse monitoring of patient derived organoids for advanced drug screening. Cell. Oncol. 2023, 46, 299–314. [Google Scholar] [CrossRef]
- Xie, B.-Y.; Wu, A.-W. Organoid Culture of Isolated Cells from Patient-derived Tissues with Colorectal Cancer. Chin. Med. J. 2016, 129, 2469–2475. [Google Scholar] [CrossRef]
- Smabers, L.P.; Wensink, E.; Verissimo, C.S.; Koedoot, E.; Pitsa, K.-C.; Huismans, M.A.; Barón, C.H.; Doorn, M.; Iersel, L.B.V.-V.; Cirkel, G.A.; et al. Organoids as a biomarker for personalized treatment in metastatic colorectal cancer: Drug screen optimization and correlation with patient response. J. Exp. Clin. Cancer Res. 2024, 43, 61. [Google Scholar] [CrossRef] [PubMed]
- Firestein, R.; Marcinkiewicz, C.; Nie, L.; Chua, H.K.; Velazquez-Quesada, I.; Torelli, M.; Sternberg, M.; Gligorijevic, B.; Shenderova, O.; Schirhagl, R.; et al. Pharmacodynamic Studies of Fluorescent Diamond Carriers of Doxorubicin in Liver Cancer Cells and Colorectal Cancer Organoids. Nanotechnol. Sci. Appl. 2021, 14, 139–159. [Google Scholar] [CrossRef]
- Wiley, L.A.; Beebe, D.C.; Mullins, R.F.; Stone, E.M.; Tucker, B.A. A method for sectioning and immunohistochemical analysis of stem cell–derived 3D organoids. Curr. Protoc. Stem Cell Biol. 2016, 37, 1C–19C. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Gut, G.; Sanchís Calleja, F.; Okamoto, R.; Streib, S.; He, Z.; Zenk, F.; Santel, M.; Seimiya, M.; Holtackers, R.; et al. Morphodynamics of human early brain organoid development. bioRxiv 2023. [Google Scholar] [CrossRef]
- Du, X.; Chen, Z.; Li, Q.; Yang, S.; Jiang, L.; Yang, Y.; Li, Y.; Gu, Z. Organoids revealed: Morphological analysis of the profound next generation in-vitro model with artificial intelligence. Bio-Design Manuf. 2023, 6, 319–339. [Google Scholar] [CrossRef] [PubMed]
- Mukashyaka, P.; Kumar, P.; Mellert, D.J.; Nicholas, S.; Noorbakhsh, J.; Brugiolo, M.; Courtois, E.T.; Anczukow, O.; Liu, E.T.; Chuang, J.H. High-throughput deconvolution of 3D organoid dynamics at cellular resolution for cancer pharmacology with Cellos. Nat. Commun. 2023, 14, 8406. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Vera, Y.M.; Valdés, J.; Hidalgo-Miranda, A.; Cisneros-Villanueva, M.; Marchat, L.A.; Nuñez-Olvera, S.I.; Ramos-Payán, R.; Pérez-Plasencia, C.; Arriaga-Pizano, L.A.; Prieto-Chávez, J.L.; et al. Three-Dimensional Organotypic Cultures Reshape the microRNAs Transcriptional Program in Breast Cancer Cells. Cancers 2022, 14, 2490. [Google Scholar] [CrossRef] [PubMed]
- Gastélum-López, M.d.l.Á.; Aguilar-Medina, M.; García Mata, C.; López-Gutiérrez, J.; Romero-Quintana, G.; Bermúdez, M.; Avendaño-Felix, M.; López-Camarillo, C.; Pérez-Plascencia, C.; Beltrán, A.S.; et al. Organotypic 3D Cell-Architecture Impacts the Expression Pattern of miRNAs–mRNAs Network in Breast Cancer SKBR3 Cells. Non-Coding RNA 2023, 9, 66. [Google Scholar] [CrossRef]
- Koedoot, E.; Wolters, L.; Smid, M.; Stoilov, P.; Burger, G.A.; Herpers, B.; Yan, K.; Price, L.S.; Martens, J.W.M.; Le Dévédec, S.E.; et al. Differential reprogramming of breast cancer subtypes in 3D cultures and implications for sensitivity to targeted therapy. Sci. Rep. 2021, 11, 7259. [Google Scholar] [CrossRef]
- Liu, K.; Newbury, P.A.; Glicksberg, B.S.; Zeng, W.Z.D.; Paithankar, S.; Andrechek, E.R.; Chen, B. Evaluating cell lines as models for metastatic breast cancer through integrative analysis of genomic data. Nat. Commun. 2019, 10, 2138. [Google Scholar] [CrossRef]
- Zou, J.; Shah, O.; Chiu, Y.-C.; Ma, T.; Atkinson, J.M.; Oesterreich, S.; Lee, A.V.; Tseng, G.C. Systems approach for congruence and selection of cancer models towards precision medicine. PLOS Comput. Biol. 2024, 20, e1011754. [Google Scholar] [CrossRef]
- Kondo, J.; Inoue, M. Application of Cancer Organoid Model for Drug Screening and Personalized Therapy. Cells 2019, 8, 470. [Google Scholar] [CrossRef]
- Campaner, E.; Zannini, A.; Santorsola, M.; Bonazza, D.; Bottin, C.; Cancila, V.; Tripodo, C.; Bortul, M.; Zanconati, F.; Schoeftner, S.; et al. Breast Cancer Organoids Model Patient-Specific Response to Drug Treatment. Cancers 2020, 12, 3869. [Google Scholar] [CrossRef]
- He, S.; Fang, Y.; Wu, M.; Zhang, P.; Gao, F.; Hu, H.; Sheng, C.; Dong, G. Enhanced Tumor Targeting and Penetration of Proteolysis-Targeting Chimeras through iRGD Peptide Conjugation: A Strategy for Precise Protein Degradation in Breast Cancer. J. Med. Chem. 2023, 66, 16828–16842. [Google Scholar] [CrossRef]
- Tan, X.; Kong, D.; Tao, Z.; Cheng, F.; Zhang, B.; Wang, Z.; Mei, Q.; Chen, C.; Wu, K. Simultaneous inhibition of FAK and ROS1 synergistically repressed triple-negative breast cancer by upregulating p53 signalling. Biomark. Res. 2024, 12, 13. [Google Scholar] [CrossRef]
- Rowdo, F.P.M.; Xiao, G.; Khramtsova, G.F.; Nguyen, J.; Martini, R.; Stonaker, B.; Boateng, R.; Oppong, J.K.; Adjei, E.K.; Awuah, B.; et al. Patient-derived tumor organoids with p53 mutations, and not wild-type p53, are sensitive to synergistic combination PARP inhibitor treatment. Cancer Lett. 2024, 584, 216608. [Google Scholar] [CrossRef]
- Rao, X.; Qiao, Z.; Yang, Y.; Deng, Y.; Zhang, Z.; Yu, X.; Guo, X. Unveiling Epigenetic Vulnerabilities in Triple-Negative Breast Cancer through 3D Organoid Drug Screening. Pharmaceuticals 2024, 17, 225. [Google Scholar] [CrossRef]
- Önder, C.E.; Moustafa-Oglou, M.; Schröder, S.M.; Hartkopf, A.D.; Koch, A.; Seitz, C.M. Precision Immunotherapy Utilizing Adapter CAR-T Cells (AdCAR-T) in Metastatic Breast Cancer Leads to Target Specific Lysis. Cancers 2024, 16, 168. [Google Scholar] [CrossRef]
- Shu, D.; Shen, M.; Li, K.; Han, X.; Li, H.; Tan, Z.; Wang, Y.; Peng, Y.; Tang, Z.; Qu, C.; et al. Organoids from patient biopsy samples can predict the response of BC patients to neoadjuvant chemotherapy. Ann. Med. 2022, 54, 2581–2597. [Google Scholar] [CrossRef]
- Pellizzari, S.; Bhat, V.; Athwal, H.; Cescon, D.W.; Allan, A.L.; Parsyan, A. PLK4 as a potential target to enhance radiosensitivity in triple-negative breast cancer. Radiat. Oncol. 2024, 19, 24. [Google Scholar] [CrossRef]
- Derouane, F.; Desgres, M.; Moroni, C.; Ambroise, J.; Berlière, M.; Van Bockstal, M.R.; Galant, C.; van Marcke, C.; Vara-Messler, M.; Hutten, S.J.; et al. Metabolic adaptation towards glycolysis supports resistance to neoadjuvant chemotherapy in early triple negative breast cancers. Breast Cancer Res. 2024, 26, 29. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Huang, H.; Wei, Z.; Chen, W.; Li, J.; Yao, Y.; Zhou, J.; Liu, J.; Sun, S.; Xia, W.; et al. Integrated transcriptomics, proteomics, and functional analysis to characterize the tissue-specific small extracellular vesicle network of breast cancer. MedComm 2023, 4, e433. [Google Scholar] [CrossRef] [PubMed]

| Cell Culture Models | Advantages | Challenges | References |
|---|---|---|---|
| 2D cultured cell | Simple setup, well-established protocols, and low-cost maintenance. Easily scale up for high-throughput drug screens and offer a uniform environment for straightforward microscopy and immunostaining analysis. | Monolayer culture (1) alters cellular behavior, gene expression, and intricate cell interactions, (2) leads to discrepancies in drug responses, and (3) lacks modeling of spatial gradients, such as nutrients, oxygen, and signaling molecules. | [24,25] |
| 3D cell aggregates and spheroid | Facilitate cell–cell interactions for studying complex cellular behaviors and signaling pathways. Enables a more accurate representation of cancer progression and drug response compared with 2D. Incorporating extracellular matrix components creates a biomimetic microenvironment conducive to cell adhesion, migration, and differentiation. | Often composed of single cell type, limiting to the reflection of molecular diversity. Variability in cell density, size, and shape complicates standardization, high-throughput screens, and imaging analysis. Non-adherent culture techniques may fail to represent tumor formation in vivo. Requires moderate-cost maintenance, specialized protocols, and complex equipment. | [26] |
| Organoid and PDO | Stem cell- or patient-derived organoids preserve histological, transcriptional, and genetic characteristics of the original tumor, including tumor heterogeneity and mutation patterns during long-term culture. Enables personalized medicine and facilitates drug response studies. Cost-effective compared with animal models and compatibility with emerging technologies. | Lack of standardized protocols for generation and expansion, leading to experiment variabilities. Cellular heterogeneity and uniformity affect maturation and stability, impacting reproducibility and data interpretation. Vascularization absence limits size and organ functions accurately. Requires costly and time-intensive maintenance. | [20,27] |
| PDX | Accurately represents in vivo biology by preserving tumor microenvironments and patient tumor characteristics. Excels in predicting drug responses, aiding therapy development, and enabling long-term studies of tumor behavior. Allows tumor growth and passage to create cohorts or cryopreserving to establish living tissue biobanks. | Technically demanding, requiring specialized equipment, facilities, and expertise in xenotransplantation. Low engraftment success rates, long generation cycles, and costly and time-consuming maintenance. Limited study of tumor–immune interactions due to the use of immunodeficient hosts. | [28,29] |
| PDXO | Faithfully mimics patient tumor traits, enabling predictive drug screening, long-term monitoring, and personalized medicine approaches. Co-culture options facilitate tumor–immune interaction studies. | Technically demanding and costly, requiring specialized equipment, facilities, and expertise in xenotransplantation and organoid culture techniques. Variable culture success rates and potential biases towards aggressive cell lines. Genetic and phenotypic changes over time due to adaptation of the host’s environment. | [30] |
| Additive | Description | References |
|---|---|---|
| Fetal Bovine Serum (FBS) | A universal natural growth supplement of tissue and cell culture media that contains growth-enhancing factors (growth factors, hormones, nutrients, etc.). Note: Smaller molecules within FBS are not fully understood, and their effects on cell cultures are not known and could lead to discrepancies in the reproducibility of cell cultures. FBS can have seasonal batch-to-batch variations which can further contribute to issues in reproducibility. | [30] |
| Advanced DMEM/F12 | Rich in nutritional factors, such as glucose, amino acids, vitamins, zinc, putrescine, hypoxanthine, and thymidine. It is usually coupled with FBS due to its lack of proteins and growth factors. | [5] |
| HEPES | A non-volatile buffer is used to maintain a stable pH while culturing. It is especially useful as a buffering agent when cells are required to be outside of the incubator for extended periods. | [36] |
| GlutaMAX | An exact substitute for L-glutamine, which has been demonstrated to be an effective nutrient for cancer cells to provide more nitrogen and carbon for biosynthetic processes, supporting unchecked proliferation of cancer cells. | [30,54] |
| Hydrocortisone | Helps support cell viability and proliferation, maintain hormone sensitivity, modulate cellular responses, and enhance experimental consistency. Note: Dose-dependent cytotoxic effects have been reported, inhibiting proliferation and inducing cell cycle arrest. | [55] |
| Human Epidermal Growth Factor (hEGF) | hEGF is used to promote cell proliferation by binding to the EGF receptor on the cell surface, maintaining cell viability, and enhancing the responsiveness of 3D models to drugs/therapeutic agents. Note: Although elevated hEGF may enhance proliferation when coupled with BME (Matrigel/Culturex), it has been correlated to hindering the structural integrity of the organoid with gradual sinking and 3D organization loss. | [47] |
| Antibiotics | Antibiotics, such as Pen/Strep and Gentamicin, are used to help prevent cell contamination. They either act by inhibiting cell wall synthesis or interfering with membrane permeability. Note: Regular antibiotic usage may result in bacterial contamination resistance, which might impact cell proliferation and differentiation. It can also drastically change the regulation and expression of genes, which might change the outcomes of research on medication response, cell regulation, and differentiation. | [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bittman-Soto, X.S.; Thomas, E.S.; Ganshert, M.E.; Mendez-Santacruz, L.L.; Harrell, J.C. The Transformative Role of 3D Culture Models in Triple-Negative Breast Cancer Research. Cancers 2024, 16, 1859. https://doi.org/10.3390/cancers16101859
Bittman-Soto XS, Thomas ES, Ganshert ME, Mendez-Santacruz LL, Harrell JC. The Transformative Role of 3D Culture Models in Triple-Negative Breast Cancer Research. Cancers. 2024; 16(10):1859. https://doi.org/10.3390/cancers16101859
Chicago/Turabian StyleBittman-Soto, Xavier S., Evelyn S. Thomas, Madeline E. Ganshert, Laura L. Mendez-Santacruz, and J. Chuck Harrell. 2024. "The Transformative Role of 3D Culture Models in Triple-Negative Breast Cancer Research" Cancers 16, no. 10: 1859. https://doi.org/10.3390/cancers16101859
APA StyleBittman-Soto, X. S., Thomas, E. S., Ganshert, M. E., Mendez-Santacruz, L. L., & Harrell, J. C. (2024). The Transformative Role of 3D Culture Models in Triple-Negative Breast Cancer Research. Cancers, 16(10), 1859. https://doi.org/10.3390/cancers16101859





