Novel Imaging Approaches for Glioma Classification in the Era of the World Health Organization 2021 Update: A Scoping Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
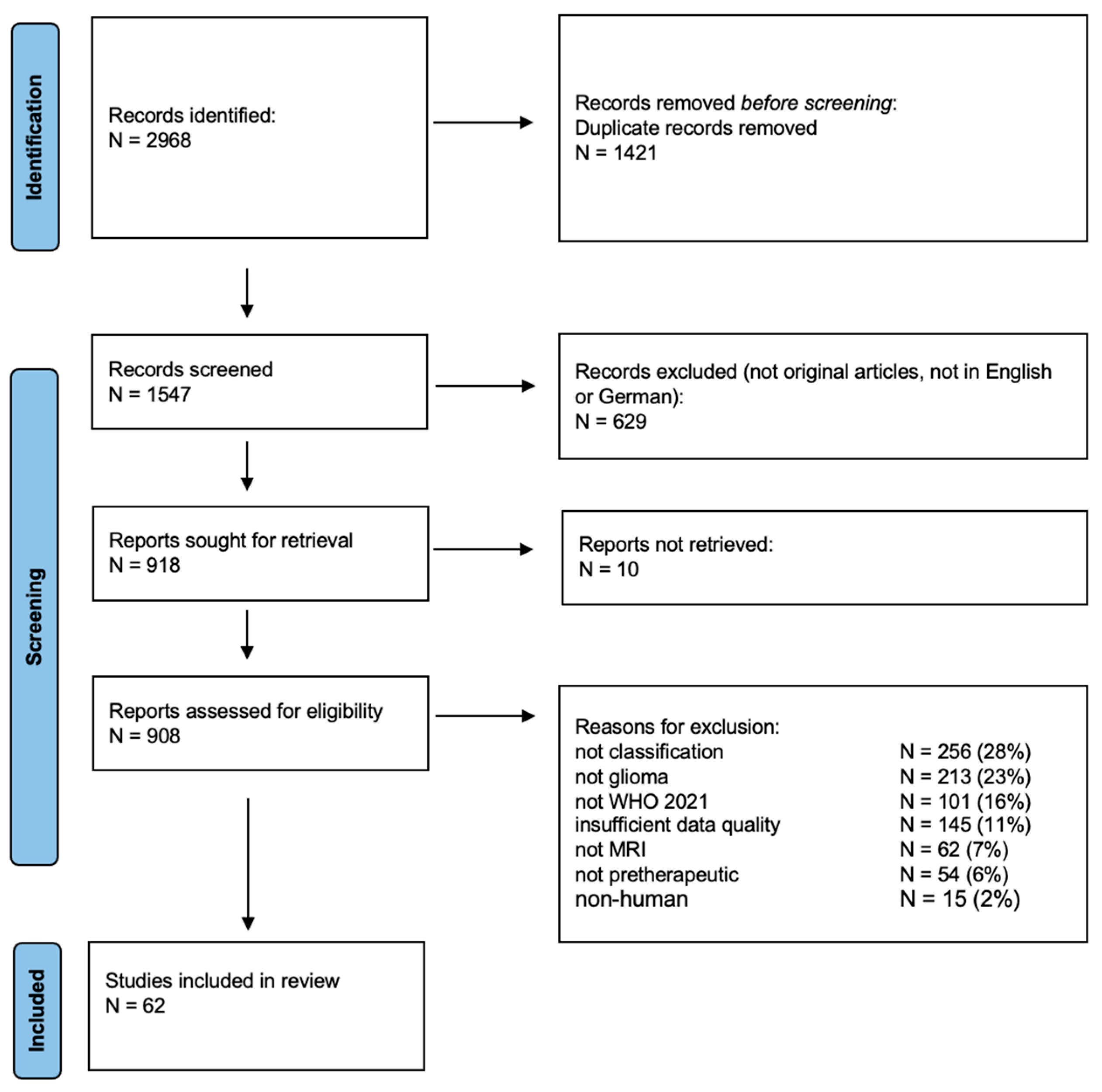
2.1. Search Strategy
2.2. Study Selection and Eligibility Criteria
3. Results
3.1. Bibliographic Results and Eligibility Criteria
3.2. Patient Population
3.3. Data Sources
3.4. Imaging Sequences
3.5. Molecular Subgroups
3.6. Algorithms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Cluceru, J.; Interian, Y.; Phillips, J.J.; Molinaro, A.M.; Luks, T.L.; Alcaide-Leon, P.; Olson, M.P.; Nair, D.; LaFontaine, M.; Shai, A.; et al. Improving the noninvasive classification of glioma genetic subtype with deep learning and diffusion-weighted imaging. Neuro-Oncol. 2022, 24, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Haldar, D.; Kazerooni, A.F.; Arif, S.; Familiar, A.; Madhogarhia, R.; Khalili, N.; Bagheri, S.; Anderson, H.; Shaikh, I.S.; Mahtabfar, A.; et al. Unsupervised machine learning using K-means identifies radiomic subgroups of pediatric low-grade gliomas that correlate with key molecular markers. Neoplasia 2023, 36, 100869. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Gong, J.; Su, X.; Chen, N.; Li, S.; Yang, X.; Zhang, S.; Huang, Z.; Hu, W.; Gong, Q.; et al. MRI characteristics of H3 G34-mutant diffuse hemispheric gliomas and possible differentiation from IDH-wild-type glioblastomas in adolescents and young adults. J. Neurosurg. Pediatr. 2023, 33, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.L.; Lai, M.Y.; Li, S.Q.; Ye, K.L.; Li, L.Z.; Hu, Q.J.; Ai, R.Y.; Zhou, J.F.; Li, J.; Zhen, J.J.; et al. Radiomics features based on MRI predict BRAF V600E mutation in pediatric low-grade gliomas: A non-invasive method for molecular diagnosis. Clin. Neurol. Neurosurg. 2022, 222, 107478. [Google Scholar] [CrossRef] [PubMed]
- Vafaeikia, P.; Wagner, M.W.; Hawkins, C.; Tabori, U.; Ertl-Wagner, B.B.; Khalvati, F. MRI-Based End-To-End Pediatric Low-Grade Glioma Segmentation and Classification. Can. Assoc. Radiol. J. 2024, 75, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Tak, D.; Ye, Z.; Zapaischykova, A.; Zha, Y.; Boyd, A.; Vajapeyam, S.; Chopra, R.; Hayat, H.; Prabhu, S.P.; Liu, K.X.; et al. Noninvasive Molecular Subtyping of Pediatric Low-Grade Glioma with Self-Supervised Transfer Learning. Radiol. Artif. Intell. 2024, 6, e230333. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Hassan, L.; Katib, Y. A texture-based method for predicting molecular markers and survival outcome in lower grade glioma. Appl. Intell. 2023, 53, 24724–24738. [Google Scholar] [CrossRef]
- Lu, J.; Li, X.; Li, H. A radiomics feature-based nomogram to predict telomerase reverse transcriptase promoter mutation status and the prognosis of lower-grade gliomas. Clin. Radiol. 2022, 77, E560–E567. [Google Scholar] [CrossRef]
- Sun, C.; Fan, L.; Wang, W.; Wang, W.; Liu, L.; Duan, W.; Pei, D.; Zhan, Y.; Zhao, H.; Sun, T.; et al. Radiomics and Qualitative Features From Multiparametric MRI Predict Molecular Subtypes in Patients With Lower-Grade Glioma. Front. Oncol. 2021, 11, 756828. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, S.; Sun, Q.; Wang, W.; Duan, W.; Wang, L.; Ding, T.; Pei, D.; Sun, C.; Wang, W.; et al. Predicting 1p/19q co-deletion status from magnetic resonance imaging using deep learning in adult-type diffuse lower-grade gliomas: A discovery and validation study. Lab. Investig. 2022, 102, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, L.; Luo, T.; Nie, C.; Fan, R.; Li, D.; Yang, R.; Zhou, C.; Li, Q.; Hu, X.; et al. Cyclin-Dependent Kinase Inhibitor 2A/B Homozygous Deletion Prediction and Survival Analysis. Brain Sci. 2023, 13, 548. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.T.; Su, C.Q.; Lin, J.; Xia, Z.W.; Lu, S.S.; Hong, X.N. T2-FLAIR mismatch sign and machine learning-based multiparametric MRI radiomics in predicting IDH mutant 1p/19q non-co-deleted diffuse lower-grade gliomas. Clin. Radiol. 2024, 79, e750–e758. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Yan, X.; Qu, Y.; Wen, H.; Zou, X.; Liu, X.; Lu, M.; Mo, J.; Wen, Z. Amide proton transfer weighted and diffusion weighted imaging based radiomics classification algorithm for predicting 1p/19q co-deletion status in low grade gliomas. BMC Med. Imaging 2024, 24, 85. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Zhang, Y.; Mao, Q.; Ju, Y.; Liu, Y.; Su, Z.; Lei, Y.; Ren, Y. Deep learning-based prediction of H3K27M alteration in diffuse midline gliomas based on whole-brain MRI. Cancer Med. 2023, 12, 17139–17148. [Google Scholar] [CrossRef] [PubMed]
- Khalid, F.; Goya-Outi, J.; Escobar, T.; Dangouloff-Ros, V.; Grigis, A.; Philippe, C.; Boddaert, N.; Grill, J.; Frouin, V.; Frouin, F. Multimodal MRI radiomic models to predict genomic mutations in diffuse intrinsic pontine glioma with missing imaging modalities. Front. Med. 2023, 10, 1071447. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Yang, N.; Gu, G.C.; Wang, X.Y.; Jiang, Z.; Li, T.; Zhang, X.R.; Ma, L.F.; Zhang, P.; Liao, H.E.; et al. Diffusion MRI is valuable in brainstem glioma genotyping with quantitative measurements of white matter tracts. Eur. Radiol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Xiao, X.; Gu, G.; Wang, X.; Zhang, X.; Wang, Y.; Pan, C.; Zhang, P.; Ma, L.; Zhang, L.; et al. Diffusion MRI-based connectomics features improve the noninvasive prediction of H3K27M mutation in brainstem gliomas. Radiother. Oncol. 2023, 186, 109789. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Zhang, Z.; Chen, Y.; Zhao, Y.; Sun, Q.; Wang, W.; Zheng, H.; Liang, D.; Cheng, J.; Yan, J.; et al. Imaging phenotypes from MRI for the prediction of glioma immune subtypes from RNA sequencing: A multicenter study. Mol. Oncol. 2023, 17, 629–646. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, Z.M.; Wang, Z.Y.; Qian, X.S.; Yao, Z.G.; Cheng, C.C.; Zhou, Z.Y.; Gao, F.; Dai, Y.K. Using radiomics based on multicenter magnetic resonance images to predict isocitrate dehydrogenase mutation status of gliomas. Quant. Imaging Med. Surg. 2023, 13, 2143–2155. [Google Scholar] [CrossRef]
- Lu, J.; Xu, W.; Chen, X.; Wang, T.; Li, H. Noninvasive prediction of IDH mutation status in gliomas using preoperative multiparametric MRI radiomics nomogram: A mutlicenter study. Magn. Reson. Imaging 2023, 104, 72–79. [Google Scholar] [CrossRef]
- Sha, Y.; Yan, Q.; Tan, Y.; Wang, X.; Zhang, H.; Yang, G. Prediction of the Molecular Subtype of IDH Mutation Combined with MGMT Promoter Methylation in Gliomas via Radiomics Based on Preoperative MRI. Cancers 2023, 15, 1440. [Google Scholar] [CrossRef] [PubMed]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.J.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Nandoe Tewarie, R.; Lycklama, G.J.; De Witt Hamer, P.C.; Eijgelaar, R.S.; et al. Combined molecular subtyping, grading, and segmentation of glioma using multi-task deep learning. Neuro-Oncol. 2023, 25, 279–289. [Google Scholar] [CrossRef]
- Zhang, H.; Fan, X.; Zhang, J.; Wei, Z.; Feng, W.; Hu, Y.; Ni, J.; Yao, F.; Zhou, G.; Wan, C.; et al. Deep-learning and conventional radiomics to predict IDH genotyping status based on magnetic resonance imaging data in adult diffuse glioma. Front. Oncol. 2023, 13, 1143688. [Google Scholar] [CrossRef] [PubMed]
- Foltyn-Dumitru, M.; Schell, M.; Sahm, F.; Kessler, T.; Wick, W.; Bendszus, M.; Rastogi, A.; Brugnara, G.; Vollmuth, P. Advancing noninvasive glioma classification with diffusion radiomics: Exploring the impact of signal intensity normalization. Neurooncol. Adv. 2024, 6, vdae043. [Google Scholar] [CrossRef] [PubMed]
- Santinha, J.; Katsaros, V.; Stranjalis, G.; Liouta, E.; Boskos, C.; Matos, C.; Viegas, C.; Papanikolaou, N. Development of End-to-End AI-Based MRI Image Analysis System for Predicting IDH Mutation Status of Patients with Gliomas: Multicentric Validation. J. Imaging Inf. Med. 2024, 37, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; LaMontagne, P.; Shimony, J.; Marcus, D.S.; Sotiras, A. MRI-based classification of IDH mutation and 1p/19q codeletion status of gliomas using a 2.5D hybrid multi-task convolutional neural network. Neuro-Oncol. Adv. 2023, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- He, A.; Wang, P.; Zhu, A.; Liu, Y.; Chen, J.; Liu, L. Predicting IDH Mutation Status in Low-Grade Gliomas Based on Optimal Radiomic Features Combined with Multi-Sequence Magnetic Resonance Imaging. Diagnostics 2022, 12, 2995. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, T.; Ohka, F.; Aoki, K.; Suzuki, H.; Motomura, K.; Yamaguchi, J.; Maeda, S.; Kibe, Y.; Shimizu, H.; Natsume, A.; et al. Easy-to-use machine learning system for the prediction of IDH mutation and 1p/19q codeletion using MRI images of adult-type diffuse gliomas. Brain Tumor Pathol. 2023, 40, 85–92. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, R.; Gao, J.; Tang, Y.; Xu, X.; Kan, Y.; Cao, X.; Wen, Z.; Liu, Z.; Cui, S.; et al. A novel MRI-based deep learning networks combined with attention mechanism for predicting CDKN2A/B homozygous deletion status in IDH-mutant astrocytoma. Eur. Radiol. 2023, 34, 391–399. [Google Scholar] [CrossRef]
- Kihira, S.; Derakhshani, A.; Leung, M.; Mahmoudi, K.; Bauer, A.; Zhang, H.; Polson, J.; Arnold, C.; Tsankova, N.M.; Hormigo, A.; et al. Multi-Parametric Radiomic Model to Predict 1p/19q Co-Deletion in Patients with IDH-1 Mutant Glioma: Added Value to the T2-FLAIR Mismatch Sign. Cancers 2023, 15, 1037. [Google Scholar] [CrossRef]
- Gemini, L.; Tortora, M.; Giordano, P.; Prudente, M.E.; Villa, A.; Vargas, O.; Giugliano, M.F.; Somma, F.; Marchello, G.; Chiaramonte, C.; et al. Vasari Scoring System in Discerning between Different Degrees of Glioma and IDH Status Prediction: A Possible Machine Learning Application? J. Imaging 2023, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Karami, G.; Pascuzzo, R.; Figini, M.; Del Gratta, C.; Zhang, H.; Bizzi, A. Combining Multi-Shell Diffusion with Conventional MRI Improves Molecular Diagnosis of Diffuse Gliomas with Deep Learning. Cancers 2023, 15, 482. [Google Scholar] [CrossRef] [PubMed]
- Pei, D.; Guan, F.; Hong, X.; Liu, Z.; Wang, W.; Qiu, Y.; Duan, W.; Wang, M.; Sun, C.; Wang, W.; et al. Radiomic features from dynamic susceptibility contrast perfusion-weighted imaging improve the three-class prediction of molecular subtypes in patients with adult diffuse gliomas. Eur. Radiol. 2023, 33, 3455–3466. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.W.; Park, K.S.; Park, J.E.; Ahn, S.S.; Park, I.; Kim, H.S.; Chang, J.H.; Lee, S.K.; Kim, S.H. Qualitative and Quantitative Magnetic Resonance Imaging Phenotypes May Predict CDKN2A/B Homozygous Deletion Status in Isocitrate Dehydrogenase-Mutant Astrocytomas: A Multicenter Study. Korean J. Radiol. 2023, 24, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Sohn, B.; Park, K.; Ahn, S.S.; Park, Y.W.; Choi, S.H.; Kang, S.G.; Kim, S.H.; Chang, J.H.; Lee, S.K. Dynamic contrast-enhanced MRI radiomics model predicts epidermal growth factor receptor amplification in glioblastoma, IDH-wildtype. J. Neuro-Oncol. 2023, 164, 341–351. [Google Scholar] [CrossRef]
- Buz-Yalug, B.; Turhan, G.; Cetin, A.I.; Dindar, S.S.; Danyeli, A.E.; Yakicier, C.; Pamir, M.N.; Ozduman, K.; Dincer, A.; Ozturk-Isik, E. Identification of IDH and TERTp mutations using dynamic susceptibility contrast MRI with deep learning in 162 gliomas. Eur. J. Radiol. 2024, 170, 111257. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Yu, Y.; Chang, J.; Chu, Y.H.; Yu, W.; Hsu, Y.C.; Patrick, L.A.; Liu, M.; Yue, Q. Convolutional neural network to predict IDH mutation status in glioma from chemical exchange saturation transfer imaging at 7 Tesla. Front. Oncol. 2023, 13, 1134626. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Togao, O.; Yamashita, K.; Momosaka, D.; Kikuchi, Y.; Kuga, D.; Yuhei, S.; Fujioka, Y.; Narutomi, F.; Obara, M.; et al. Comparison of diagnostic performance of radiologist- and AI-based assessments of T2-FLAIR mismatch sign and quantitative assessment using synthetic MRI in the differential diagnosis between astrocytoma, IDH-mutant and oligodendroglioma, IDH-mutant and 1p/19q-codeleted. Neuroradiology 2024, 66, 333–341. [Google Scholar] [CrossRef]
- Huo, X.; Wang, Y.; Ma, S.; Zhu, S.; Wang, K.; Ji, Q.; Chen, F.; Wang, L.; Wu, Z.; Li, W. Multimodal MRI-based radiomic nomogram for predicting telomerase reverse transcriptase promoter mutation in IDH-wildtype histological lower-grade gliomas. Medicine 2023, 102, e36581. [Google Scholar] [CrossRef]
- Zhang, H.; Ouyang, Y.; Zhang, H.; Zhang, Y.; Su, R.; Zhou, B.; Yang, W.; Lei, Y.; Huang, B. Sub-region based radiomics analysis for prediction of isocitrate dehydrogenase and telomerase reverse transcriptase promoter mutations in diffuse gliomas. Clin. Radiol. 2024, 79, e682–e691. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, H.; Zhang, Y.; Zhou, B.; Wu, L.; Yang, W.; Lei, Y.; Huang, B. Multiparametric MRI-based fusion radiomics for predicting telomerase reverse transcriptase (TERT) promoter mutations and progression-free survival in glioblastoma: A multicentre study. Neuroradiology 2024, 66, 81–92. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, B.; Zhang, H.; Zhang, Y.; Lei, Y.; Huang, B. Peritumoral Radiomics for Identification of Telomerase Reverse Transcriptase Promoter Mutation in Patients With Glioblastoma Based on Preoperative MRI. Can. Assoc. Radiol. J. 2024, 75, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, R.; Li, T.; Huang, L.; Tang, C.; Li, Y.; Zeng, Z. MRI radiomics model for predicting TERT promoter mutation status in glioblastoma. Brain Behav. 2023, 13, e3324. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, R.R.; Li, T.; Tang, C.Y.; Li, Y.; Zeng, Z.S. Multi-parameter MRI based radiomics nomogram for predicting telomerase reverse transcriptase promoter mutation and prognosis in glioblastoma. Front. Neurol. 2023, 14, 1266658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, H.; Zhang, Y.; Zhou, B.; Wu, L.; Lei, Y.; Huang, B. Deep Learning Radiomics for the Assessment of Telomerase Reverse Transcriptase Promoter Mutation Status in Patients With Glioblastoma Using Multiparametric MRI. J. Magn. Reson. Imaging 2023, 58, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xu, X.; Zhang, W.; Zhang, L.; Wen, M.; Gao, J.; Yang, J.; Kan, Y.; Yang, X.; Wen, Z.; et al. A fusion model integrating magnetic resonance imaging radiomics and deep learning features for predicting alpha-thalassemia X-linked intellectual disability mutation status in isocitrate dehydrogenase-mutant high-grade astrocytoma: A multicenter study. Quant. Imaging Med. Surg. 2024, 14, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Musigmann, M.; Nacul, N.G.; Kasap, D.N.; Heindel, W.; Mannil, M. Use Test of Automated Machine Learning in Cancer Diagnostics. Diagnostics 2023, 13, 2315. [Google Scholar] [CrossRef] [PubMed]
- Nacul Mora, N.G.; Akkurt, B.H.; Kasap, D.; Blomer, D.; Heindel, W.; Mannil, M.; Musigmann, M. Comparison of MRI Sequences to Predict ATRX Status Using Radiomics-Based Machine Learning. Diagnostics 2023, 13, 2216. [Google Scholar] [CrossRef]
- Zhong, S.; Ren, J.X.; Yu, Z.P.; Peng, Y.D.; Yu, C.W.; Deng, D.; Xie, Y.; He, Z.Q.; Duan, H.; Wu, B.; et al. Predicting glioblastoma molecular subtypes and prognosis with a multimodal model integrating convolutional neural network, radiomics, and semantics. J. Neurosurg. Pediatr. 2022, 139, 305–314. [Google Scholar] [CrossRef]
- Ma, C.; Wang, L.; Song, D.; Gao, C.; Jing, L.; Lu, Y.; Liu, D.; Man, W.; Yang, K.; Meng, Z.; et al. Multimodal-based machine learning strategy for accurate and non-invasive prediction of intramedullary glioma grade and mutation status of molecular markers: A retrospective study. BMC Med. 2023, 21, 198. [Google Scholar] [CrossRef] [PubMed]
- Usuzaki, T.; Takahashi, K.; Inamori, R.; Morishita, Y.; Shizukuishi, T.; Takagi, H.; Ishikuro, M.; Obara, T.; Takase, K. Identifying key factors for predicting O6-Methylguanine-DNA methyltransferase status in adult patients with diffuse glioma: A multimodal analysis of demographics, radiomics, and MRI by variable Vision Transformer. Neuroradiology 2024, 66, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xu, Q.Q.; Shi, N.; Dong, L.N.; Zhu, H.; Xu, K. A multitask classification framework based on vision transformer for predicting molecular expressions of glioma. Eur. J. Radiol. 2022, 157, 110560. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xu, Y.; Ye, M.; Li, Y.; Sun, Y.; Liang, J.; Lu, J.; Wang, Z.; Zhu, Z.; Zhang, X.; et al. Predicting MGMT Promoter Methylation in Diffuse Gliomas Using Deep Learning with Radiomics. J. Clin. Med. 2022, 11, 3445. [Google Scholar] [CrossRef] [PubMed]
- Doniselli, F.M.; Pascuzzo, R.; Agro, M.; Aquino, D.; Anghileri, E.; Farinotti, M.; Pollo, B.; Paterra, R.; Cuccarini, V.; Moscatelli, M.; et al. Development of A Radiomic Model for MGMT Promoter Methylation Detection in Glioblastoma Using Conventional MRI. Int. J. Mol. Sci. 2023, 25, 138. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, Z.; Pan, H.; Cao, X.; Kan, Y.; Wen, Z.; Chen, S.; Wen, M.; Zhang, L. Preoperative Discrimination of CDKN2A/B Homozygous Deletion Status in Isocitrate Dehydrogenase-Mutant Astrocytoma: A Deep Learning-Based Radiomics Model Using MRI. J. Magn. Reson. Imaging 2024, 59, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Mao, Y.; Xi, F.; Li, Y.; Luo, Y.; Ma, J. Development of a nomogram based on radiomics and semantic features for predicting chromosome 7 gain/chromosome 10 loss in IDH wild-type histologically low-grade gliomas. Front. Oncol. 2023, 13, 1196614. [Google Scholar] [CrossRef]
- Liu, Y.H.; Wang, P.; Wang, S.Y.; Zhang, H.P.; Song, Y.; Yan, X.; Gao, Y. Heterogeneity matching and IDH prediction in adult-type diffuse gliomas: A DKI-based habitat analysis. Front. Oncol. 2023, 13, 1202170. [Google Scholar] [CrossRef]
- Liu, J.; Cong, C.; Zhang, J.; Qiao, J.; Guo, H.; Wu, H.; Sang, Z.; Kang, H.; Fang, J.; Zhang, W. Multimodel habitats constructed by perfusion and/or diffusion MRI predict isocitrate dehydrogenase mutation status and prognosis in high-grade gliomas. Clin. Radiol. 2024, 79, e127–e136. [Google Scholar] [CrossRef]
- Deng, S.G.; Zhu, Y.Q. Prediction of Glioma Grade by Tumor Heterogeneity Radiomic Analysis Based on Multiparametric MRI. Int. J. Comput. Intell. Syst. 2023, 16, 51. [Google Scholar] [CrossRef]
- Hagiwara, A.; Tatekawa, H.; Yao, J.; Raymond, C.; Everson, R.; Patel, K.; Mareninov, S.; Yong, W.H.; Salamon, N.; Pope, W.B.; et al. Visualization of tumor heterogeneity and prediction of isocitrate dehydrogenase mutation status for human gliomas using multiparametric physiologic and metabolic MRI. Sci. Rep. 2022, 12, 1078. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Strasser, B.; Neuberger, U.; Vollmuth, P.; Bendszus, M.; Wick, W.; Dietrich, J.; Batchelor, T.T.; Cahill, D.P.; Andronesi, O.C. Deep learning super-resolution magnetic resonance spectroscopic imaging of brain metabolism and mutant isocitrate dehydrogenase glioma. Neuro-Oncol. Adv. 2022, 4, vdac071. [Google Scholar] [CrossRef] [PubMed]
- Griessmair, M.; Delbridge, C.; Ziegenfeuter, J.; Bernhardt, D.; Gempt, J.; Schmidt-Graf, F.; Kertels, O.; Thomas, M.; Meyer, H.S.; Zimmer, C.; et al. Imaging the WHO 2021 Brain Tumor Classification: Fully Automated Analysis of Imaging Features of Newly Diagnosed Gliomas. Cancers 2023, 15, 2355. [Google Scholar] [CrossRef] [PubMed]
- Nalawade, S.S.; Yu, F.F.; Bangalore Yogananda, C.G.; Murugesan, G.K.; Shah, B.R.; Pinho, M.C.; Wagner, B.C.; Xi, Y.; Mickey, B.; Patel, T.R.; et al. Brain tumor IDH, 1p/19q, and MGMT molecular classification using MRI-based deep learning: An initial study on the effect of motion and motion correction. J. Med. Imaging 2022, 9, 016001. [Google Scholar] [CrossRef]
- Foltyn-Dumitru, M.; Schell, M.; Rastogi, A.; Sahm, F.; Kessler, T.; Wick, W.; Bendszus, M.; Brugnara, G.; Vollmuth, P. Impact of signal intensity normalization of MRI on the generalizability of radiomic-based prediction of molecular glioma subtypes. Eur. Radiol. 2023, 34, 2782–2790. [Google Scholar] [CrossRef]
| Topic | Search Term | Syntax |
|---|---|---|
| Glioma | Glioma OR brain tumor OR brain neoplasms OR tumor of the brain OR brain cancer | AND |
| Algorithm | radiomic* OR imaging genomic* OR radiogenomic* OR machine learning OR deep learning OR support vector machine OR artificial intelligence | AND |
| Imaging technique | Magnetic Resonance Imaging OR magnetic resonance tomograph OR MRI OR MR imaging OR magnetic resonance brain imaging | AND |
| Subjects | Animals | NOT |
| Date | 2022/01/01:3000/12/31 | AND |
| Population | N (Studies) | % (All Studies) | Number of Patients per Study | |
|---|---|---|---|---|
| Mean | Stdev | |||
| Gliomas (all subtypes) | 48 | 77% | 337 | 445 |
| LGG | 10 | 16% | 223 | 146 |
| DMG | 4 | 6% | 141 | 66 |
| adult | 57 | 92% | 317 | 414 |
| pediatric | 5 | 8% | 185 | 118 |
| Data Source | N (Studies) | % (All Studies) | Number of Patients per Study | ||
|---|---|---|---|---|---|
| Mean | Stdev | p * | |||
| local | 48 | 77% | 207 | 152 | |
| public | 4 | 6% | 812 | 1225 | 0.001 |
| local and public | 10 | 16% | 581 | 461 | 0.003 |
| Sequences | N (Studies) | % (All Studies) | Best Sequence | N | % |
|---|---|---|---|---|---|
| T2 | 52 | 84% | combination | 45 | 73% |
| T1CE | 48 | 77% | not applicable | 11 | 17% |
| T2-FLAIR | 36 | 58% | ADC | 2 | 3% |
| T1 | 35 | 56% | T1CE | 1 | 2% |
| DWI (ADC, DTI, DKI) | 17 | 27% | T1CE, ADC | 1 | 2% |
| PWI (DCE/DSC) | 4 | 6% | T2 | 2 | 3% |
| T2*/SWI | 1 | 2% | |||
| CEST | 2 | 3% | |||
| SyMRI | 1 | 2% |
| Molecular Subgroup | N of Studies | % (All Studies) | AUC | |||
|---|---|---|---|---|---|---|
| Min | Max | Mean AUC | Stdev | |||
| IDH1/2 | 28 | 45% | 0.7 | 0.98 | 0.87 | 0.07 |
| 1p/19q codel | 13 | 21% | 0.6 | 0.98 | 0.84 | 0.11 |
| TERT | 9 | 15% | 0.7 | 0.95 | 0.86 | 0.08 |
| ATRX | 6 | 10% | 0.67 | 0.95 | 0.83 | 0.11 |
| H3K27 | 5 | 8% | 0.89 | 0.92 | 0.9 | 0.01 |
| MGMT | 5 | 8% | 0.57 | 0.98 | 0.85 | 0.16 |
| P53 | 4 | 7% | 0.77 | 0.97 | 0.85 | 0.09 |
| CDKN2A/B | 4 | 7% | 0.82 | 0.95 | 0.88 | 0.07 |
| BRAF | 4 | 7% | 0.73 | 0.87 | 0.79 | 0.07 |
| EGFR | 1 | 2% | 0.8 | |||
| chr7/10 | 1 | 2% | 0.85 | |||
| Mean AUC | p (DL > ML) | p (DL < ML) |
|---|---|---|
| IDH | 0.009 | |
| TERT | 0.01 | |
| ATRX | 0.01 | |
| MGMT | 0.01 | |
| CDKN2A/B | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richter, V.; Ernemann, U.; Bender, B. Novel Imaging Approaches for Glioma Classification in the Era of the World Health Organization 2021 Update: A Scoping Review. Cancers 2024, 16, 1792. https://doi.org/10.3390/cancers16101792
Richter V, Ernemann U, Bender B. Novel Imaging Approaches for Glioma Classification in the Era of the World Health Organization 2021 Update: A Scoping Review. Cancers. 2024; 16(10):1792. https://doi.org/10.3390/cancers16101792
Chicago/Turabian StyleRichter, Vivien, Ulrike Ernemann, and Benjamin Bender. 2024. "Novel Imaging Approaches for Glioma Classification in the Era of the World Health Organization 2021 Update: A Scoping Review" Cancers 16, no. 10: 1792. https://doi.org/10.3390/cancers16101792
APA StyleRichter, V., Ernemann, U., & Bender, B. (2024). Novel Imaging Approaches for Glioma Classification in the Era of the World Health Organization 2021 Update: A Scoping Review. Cancers, 16(10), 1792. https://doi.org/10.3390/cancers16101792







