Integrating AI and ML in Myelodysplastic Syndrome Diagnosis: State-of-the-Art and Future Prospects
Abstract
:Simple Summary
Abstract
1. Introduction
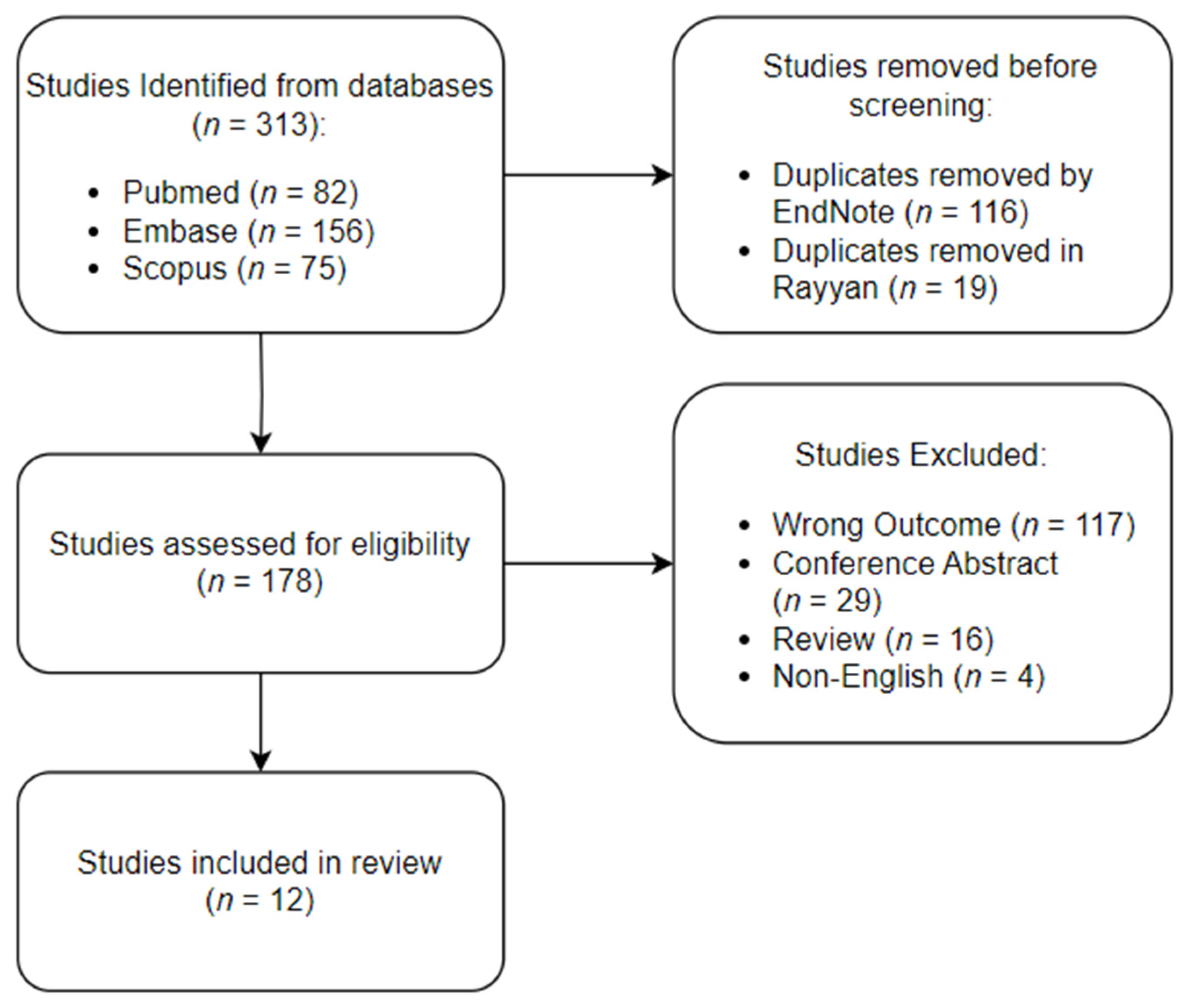
2. Materials and Methods
3. Results
3.1. Diagnosis of MDS Using BM Samples
3.2. Diagnosis of MDS Using PBS
3.3. Diagnosis of MDS Using FC
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ades, L.; Itzykson, R.; Fenaux, P. Myelodysplastic syndromes. Lancet 2014, 383, 2239–2252. [Google Scholar] [CrossRef]
- Muslimani, A.A.; Spiro, T.P.; Chaudhry, A.A.; Daw, H.A. Secondary myelodysplastic syndrome after hydroxychloroquine therapy. Ann. Hematol. 2007, 86, 531–534. [Google Scholar] [CrossRef]
- Beck, D.B.; Ferrada, M.A.; Sikora, K.A.; Ombrello, A.K.; Collins, J.C.; Pei, W.; Balanda, N.; Ross, D.L.; Ospina Cardona, D.; Wu, Z.; et al. Somatic Mutations in UBA1 and Severe Adult-Onset Autoinflammatory Disease. N. Engl. J. Med. 2020, 383, 2628–2638. [Google Scholar] [CrossRef]
- Zeidan, A.M.; Shallis, R.M.; Wang, R.; Davidoff, A.; Ma, X. Epidemiology of myelodysplastic syndromes: Why characterizing the beast is a prerequisite to taming it. Blood Rev. 2019, 34, 1–15. [Google Scholar] [CrossRef]
- Goldberg, S.L.; Chen, E.; Corral, M.; Guo, A.; Mody-Patel, N.; Pecora, A.L.; Laouri, M. Incidence and clinical complications of myelodysplastic syndromes among United States Medicare beneficiaries. J. Clin. Oncol. 2010, 28, 2847–2852. [Google Scholar] [CrossRef]
- Meyers, C.A.; Albitar, M.; Estey, E. Cognitive impairment, fatigue, and cytokine levels in patients with acute myelogenous leukemia or myelodysplastic syndrome. Cancer 2005, 104, 788–793. [Google Scholar] [CrossRef]
- Sekeres, M.A.; Taylor, J. Diagnosis and Treatment of Myelodysplastic Syndromes: A Review. JAMA 2022, 328, 872–880. [Google Scholar] [CrossRef]
- Al-Haidose, A.; Yassin, M.A.; Ahmed, M.N.; Kunhipurayil, H.H.; Al-Harbi, A.A.; Aljaberi, M.A.; Abbasi, S.A.; Kordasti, S.; Abdallah, A.M. Distinct Clinical and Prognostic Features of Myelodysplastic Syndrome in Patients from the Middle East, North Africa, and Beyond: A Systemic Review. J. Clin. Med. 2023, 12, 2832. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Estey, E.; Hasserjian, R.P.; Dohner, H. Distinguishing AML from MDS: A fixed blast percentage may no longer be optimal. Blood 2022, 139, 323–332. [Google Scholar] [CrossRef]
- Steensma, D.P. Does early diagnosis and treatment of myelodysplastic syndromes make a difference? Best Pract. Res. Clin. Haematol. 2019, 32, 101099. [Google Scholar] [CrossRef]
- Al-Antari, M.A. Artificial Intelligence for Medical Diagnostics-Existing and Future AI Technology! Diagnostics 2023, 13, 688. [Google Scholar] [CrossRef]
- Davenport, T.; Kalakota, R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019, 6, 94–98. [Google Scholar] [CrossRef]
- Kumar, Y.; Koul, A.; Singla, R.; Ijaz, M.F. Artificial intelligence in disease diagnosis: A systematic literature review, synthesizing framework and future research agenda. J. Ambient. Intell. Humaniz. Comput. 2023, 14, 8459–8486. [Google Scholar] [CrossRef]
- Undru, T.R.; Uday, U.; Lakshmi, J.T.; Kaliappan, A.; Mallamgunta, S.; Nikhat, S.S.; Sakthivadivel, V.; Gaur, A. Integrating Artificial Intelligence for Clinical and Laboratory Diagnosis—A Review. Maedica 2022, 17, 420–426. [Google Scholar] [CrossRef]
- Clark, J.M.; Sanders, S.; Carter, M.; Honeyman, D.; Cleo, G.; Auld, Y.; Booth, D.; Condron, P.; Dalais, C.; Bateup, S.; et al. Improving the translation of search strategies using the Polyglot Search Translator: A randomized controlled trial. J. Med. Libr. Assoc. 2020, 108, 195–207. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Elshoeibi, A.M.; Ferih, K.; Elsabagh, A.A.; Elsayed, B.; Elhadary, M.; Marashi, M.; Wali, Y.; Al-Rasheed, M.; Al-Khabori, M.; Osman, H.; et al. Applications of Artificial Intelligence in Thrombocytopenia. Diagnostics 2023, 13, 1060. [Google Scholar] [CrossRef]
- Wang, M.; Dong, C.; Gao, Y.; Li, J.; Han, M.; Wang, L. A Deep Learning Model for the Automatic Recognition of Aplastic Anemia, Myelodysplastic Syndromes, and Acute Myeloid Leukemia Based on Bone Marrow Smear. Front. Oncol. 2022, 12, 844978. [Google Scholar] [CrossRef]
- Lee, N.; Jeong, S.; Park, M.J.; Song, W. Deep learning application of the discrimination of bone marrow aspiration cells in patients with myelodysplastic syndromes. Sci. Rep. 2022, 12, 18677. [Google Scholar] [CrossRef]
- Mori, J.; Kaji, S.; Kawai, H.; Kida, S.; Tsubokura, M.; Fukatsu, M.; Harada, K.; Noji, H.; Ikezoe, T.; Maeda, T.; et al. Assessment of dysplasia in bone marrow smear with convolutional neural network. Sci. Rep. 2020, 10, 14734. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, L.; Yin, S.; Wang, H.; Wang, G.; Yuan, J. Differential diagnosis model of hypocellular myelodysplastic syndrome and aplastic anemia based on the medical big data platform. Complexity 2018, 2018, 4824350. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Huang, T.C.; Ye, R.H.; Fang, W.H.; Lai, S.W.; Chang, P.Y.; Liu, W.N.; Kuo, T.Y.; Lee, C.H.; Tsai, W.C.; et al. A Hematologist-Level Deep Learning Algorithm (BMSNet) for Assessing the Morphologies of Single Nuclear Balls in Bone Marrow Smears: Algorithm Development. JMIR Med. Inform. 2020, 8, e15963. [Google Scholar] [CrossRef]
- Acevedo, A.; Merino, A.; Boldu, L.; Molina, A.; Alferez, S.; Rodellar, J. A new convolutional neural network predictive model for the automatic recognition of hypogranulated neutrophils in myelodysplastic syndromes. Comput. Biol. Med. 2021, 134, 104479. [Google Scholar] [CrossRef]
- Kimura, K.; Tabe, Y.; Ai, T.; Takehara, I.; Fukuda, H.; Takahashi, H.; Naito, T.; Komatsu, N.; Uchihashi, K.; Ohsaka, A. A novel automated image analysis system using deep convolutional neural networks can assist to differentiate MDS and AA. Sci. Rep. 2019, 9, 13385. [Google Scholar] [CrossRef]
- Zhu, J.; Lemaire, P.; Mathis, S.; Ronez, E.; Clauser, S.; Jondeau, K.; Fenaux, P.; Ades, L.; Bardet, V. Machine learning-based improvement of MDS-CBC score brings platelets into the limelight to optimize smear review in the hematology laboratory. BMC Cancer 2022, 22, 972. [Google Scholar] [CrossRef]
- Clichet, V.; Lebon, D.; Chapuis, N.; Zhu, J.; Bardet, V.; Marolleau, J.P.; Garcon, L.; Caulier, A.; Boyer, T. Artificial intelligence to empower diagnosis of myelodysplastic syndromes by multiparametric flow cytometry. Haematologica 2023, 108, 2435–2443. [Google Scholar] [CrossRef]
- Duetz, C.; Van Gassen, S.; Westers, T.M.; van Spronsen, M.F.; Bachas, C.; Saeys, Y.; van de Loosdrecht, A.A. Computational flow cytometry as a diagnostic tool in suspected-myelodysplastic syndromes. Cytom. A 2021, 99, 814–824. [Google Scholar] [CrossRef]
- Herbig, M.; Jacobi, A.; Wobus, M.; Weidner, H.; Mies, A.; Krater, M.; Otto, O.; Thiede, C.; Weickert, M.T.; Gotze, K.S.; et al. Machine learning assisted real-time deformability cytometry of CD34+ cells allows to identify patients with myelodysplastic syndromes. Sci. Rep. 2022, 12, 870. [Google Scholar] [CrossRef]
- Li, J.L.; Wang, Y.F.; Ko, B.S.; Li, C.C.; Tang, J.L.; Lee, C.C. Learning a Cytometric Deep Phenotype Embedding for Automatic Hematological Malignancies Classification. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 2019, 1733–1736. [Google Scholar] [CrossRef]
- Dao, K.T. Myelodysplastic Syndromes: Updates and Nuances. Med. Clin. N. Am. 2017, 101, 333–350. [Google Scholar] [CrossRef]
- Mantripragada, V.P.; Piuzzi, N.S.; George, J.; Bova, W.; Ng, M.; Boehm, C.; Muschler, G.F. Reliable assessment of bone marrow and bone marrow concentrates using automated hematology analyzer. Regen. Med. 2019, 14, 639–646. [Google Scholar] [CrossRef]
- Percival, M.E.; Lai, C.; Estey, E.; Hourigan, C.S. Bone marrow evaluation for diagnosis and monitoring of acute myeloid leukemia. Blood Rev. 2017, 31, 185–192. [Google Scholar] [CrossRef]
- Piuzzi, N.S.; Hussain, Z.B.; Chahla, J.; Cinque, M.E.; Moatshe, G.; Mantripragada, V.P.; Muschler, G.F.; LaPrade, R.F. Variability in the Preparation, Reporting, and Use of Bone Marrow Aspirate Concentrate in Musculoskeletal Disorders: A Systematic Review of the Clinical Orthopaedic Literature. J. Bone Jt. Surg. Am. 2018, 100, 517–525. [Google Scholar] [CrossRef]
- Barrett, J.; Saunthararajah, Y.; Molldrem, J. Myelodysplastic syndrome and aplastic anemia: Distinct entities or diseases linked by a common pathophysiology? Semin. Hematol. 2000, 37, 15–29. [Google Scholar] [CrossRef]
- DeZern, A.E.; Churpek, J.E. Approach to the diagnosis of aplastic anemia. Blood Adv. 2021, 5, 2660–2671. [Google Scholar] [CrossRef]
- DeZern, A.E.; Sekeres, M.A. The challenging world of cytopenias: Distinguishing myelodysplastic syndromes from other disorders of marrow failure. Oncologist 2014, 19, 735–745. [Google Scholar] [CrossRef]
- Durrani, J.; Maciejewski, J.P. Idiopathic aplastic anemia vs hypocellular myelodysplastic syndrome. Hematol. Am. Soc. Hematol. Educ. Program. 2019, 2019, 97–104. [Google Scholar] [CrossRef]
- Keel, S.B.; Scott, A.; Sanchez-Bonilla, M.; Ho, P.A.; Gulsuner, S.; Pritchard, C.C.; Abkowitz, J.L.; King, M.C.; Walsh, T.; Shimamura, A. Genetic features of myelodysplastic syndrome and aplastic anemia in pediatric and young adult patients. Haematologica 2016, 101, 1343–1350. [Google Scholar] [CrossRef]
- Mohammed, E.A.; Mohamed, M.M.; Far, B.H.; Naugler, C. Peripheral blood smear image analysis: A comprehensive review. J. Pathol. Inform. 2014, 5, 9. [Google Scholar] [CrossRef]
- Gupta, G.; Singh, R.; Kotasthane, D.S.; Kotasthane, V.D. Myelodysplastic syndromes/neoplasms: Recent classification system based on World Health Organization Classification of Tumors—International Agency for Research on Cancer for Hematopoietic and Lymphoid Tissues. J. Blood Med. 2010, 1, 171–182. [Google Scholar] [CrossRef]
- Hast, R.; Nilsson, I.; Widell, S.; Ost, A. Diagnostic significance of dysplastic features of peripheral blood polymorphs in myelodysplastic syndromes. Leuk. Res. 1989, 13, 173–178. [Google Scholar] [CrossRef]
- Parmentier, S.; Schetelig, J.; Lorenz, K.; Kramer, M.; Ireland, R.; Schuler, U.; Ordemann, R.; Rall, G.; Schaich, M.; Bornhauser, M.; et al. Assessment of dysplastic hematopoiesis: Lessons from healthy bone marrow donors. Haematologica 2012, 97, 723–730. [Google Scholar] [CrossRef]
- Widell, S.; Hellstrom-Lindberg, E.; Kock, Y.; Lindberg, M.; Ost, A.; Hast, R. Peripheral blood neutrophil morphology reflects bone marrow dysplasia in myelodysplastic syndromes. Am. J. Hematol. 1995, 49, 115–120. [Google Scholar] [CrossRef]
- Bento, L.C.; Correia, R.P.; Pitangueiras Mangueira, C.L.; De Souza Barroso, R.; Rocha, F.A.; Bacal, N.S.; Marti, L.C. The Use of Flow Cytometry in Myelodysplastic Syndromes: A Review. Front. Oncol. 2017, 7, 270. [Google Scholar] [CrossRef]
- Oelschlaegel, U.; Oelschlaeger, L.; von Bonin, M.; Kramer, M.; Sockel, K.; Mohr, B.; Wagenfuehr, L.; Kroschinsky, F.; Bornhaeuser, M.; Platzbecker, U. Comparison of five diagnostic flow cytometry scores in patients with myelodysplastic syndromes: Diagnostic power and prognostic impact. Cytom. B Clin. Cytom. 2023, 104, 141–150. [Google Scholar] [CrossRef]
- Pembroke, J.S.; Joseph, J.E.; Smith, S.; Parker, A.J.C.; Jiang, W.; Sewell, W.A. Comparison of flow cytometry with other modalities in the diagnosis of myelodysplastic syndrome. Int. J. Lab. Hematol. 2022, 44, 313–319. [Google Scholar] [CrossRef]
- van de Loosdrecht, A.A.; Alhan, C.; Bene, M.C.; Della Porta, M.G.; Drager, A.M.; Feuillard, J.; Font, P.; Germing, U.; Haase, D.; Homburg, C.H.; et al. Standardization of flow cytometry in myelodysplastic syndromes: Report from the first European LeukemiaNet working conference on flow cytometry in myelodysplastic syndromes. Haematologica 2009, 94, 1124–1134. [Google Scholar] [CrossRef]
- Vardiman, J.W.; Harris, N.L.; Brunning, R.D. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 2002, 100, 2292–2302. [Google Scholar] [CrossRef]
- El Alaoui, Y.; Elomri, A.; Qaraqe, M.; Padmanabhan, R.; Yasin Taha, R.; El Omri, H.; El Omri, A.; Aboumarzouk, O. A Review of Artificial Intelligence Applications in Hematology Management: Current Practices and Future Prospects. J. Med. Internet Res. 2022, 24, e36490. [Google Scholar] [CrossRef]
- Elsayed, B.; Elshoeibi, A.M.; Elhadary, M.; Ferih, K.; Elsabagh, A.A.; Rahhal, A.; Abu-Tineh, M.; Afana, M.S.; Abdulgayoom, M.; Yassin, M. Applications of Artificial Intelligence in Philadelphia-Negative Myeloproliferative Neoplasms. Diagnostics 2023, 13, 1123. [Google Scholar] [CrossRef]
- Ferih, K.; Elsayed, B.; Elshoeibi, A.M.; Elsabagh, A.A.; Elhadary, M.; Soliman, A.; Abdalgayoom, M.; Yassin, M. Applications of Artificial Intelligence in Thalassemia: A Comprehensive Review. Diagnostics 2023, 13, 1551. [Google Scholar] [CrossRef]
- Elhadary, M.; Elsabagh, A.A.; Ferih, K.; Elsayed, B.; Elshoeibi, A.M.; Kaddoura, R.; Akiki, S.; Ahmed, K.; Yassin, M. Applications of Machine Learning in Chronic Myeloid Leukemia. Diagnostics 2023, 13, 1330. [Google Scholar] [CrossRef]
- Elsabagh, A.A.; Elhadary, M.; Elsayed, B.; Elshoeibi, A.M.; Ferih, K.; Kaddoura, R.; Alkindi, S.; Alshurafa, A.; Alrasheed, M.; Alzayed, A.; et al. Artificial intelligence in sickle disease. Blood Rev. 2023, 61, 101102. [Google Scholar] [CrossRef]
- Bleeker, S.E.; Moll, H.A.; Steyerberg, E.W.; Donders, A.R.; Derksen-Lubsen, G.; Grobbee, D.E.; Moons, K.G. External validation is necessary in prediction research: A clinical example. J. Clin. Epidemiol. 2003, 56, 826–832. [Google Scholar] [CrossRef]
- Cabitza, F.; Campagner, A.; Soares, F.; Garcia de Guadiana-Romualdo, L.; Challa, F.; Sulejmani, A.; Seghezzi, M.; Carobene, A. The importance of being external. methodological insights for the external validation of machine learning models in medicine. Comput. Methods Programs Biomed. 2021, 208, 106288. [Google Scholar] [CrossRef]
- Konig, I.R.; Malley, J.D.; Weimar, C.; Diener, H.C.; Ziegler, A.; German Stroke Study Collaboration. Practical experiences on the necessity of external validation. Stat. Med. 2007, 26, 5499–5511. [Google Scholar] [CrossRef]
- Yagi, R.; Goto, S.; Katsumata, Y.; MacRae, C.A.; Deo, R.C. Importance of external validation and subgroup analysis of artificial intelligence in the detection of low ejection fraction from electrocardiograms. Eur. Heart J. Digit. Health 2022, 3, 654–657. [Google Scholar] [CrossRef]
| Study | Method | Outcome | Advantages | Disadvantages |
|---|---|---|---|---|
| Wang, M. et al. [19] | BMS | Diagnosing MDS and distinguishing it from AA and AML |
|
|
| Lee, N. et al. [20] | BMS | Detection of dysplastic erythrocytes, granulocytes, megakaryocytes, and blasts |
|
|
| Mori, J. et al. [21] | BMS | Diagnosing MDS using hypogranulated dysplastic neutrophils |
|
|
| Wu, J. et al. [22] | BMS and PBS | Diagnosing hypocellular MDS and distinguishing it from AA |
|
|
| Wu, Y. et al. [23] | BMS | Detection of elevated blasts to diagnose MDS |
|
|
| Acevedo, A. et al. [24] | PBS | Detection of hypogranulated dysplastic neutrophils to diagnose MDS |
|
|
| Kimura, K. et al. [25] | PBS | Diagnosing MDS and distinguishing it from AA |
|
|
| Zhu, J. et al. [26] | PBS | Diagnosing MDS using CBC and immature platelet fraction |
|
|
| Clichet, V. et al. [27] | FC | Diagnosing MDS using MFC |
|
|
| Duetz, C. et al. [28] | FC | Diagnosing MDS in suspected patients using FC |
|
|
| Herbig, M. et al. [29] | FC | Diagnosing MDS using RT-DC |
|
|
| Li, J. L. et al. [30] | FC | Diagnosing MDS and distinguishing it from AML using FC |
|
|
| Study | Data Source | Outcomes |
Model Utilized | Validation | AUC | ACC | SEN | SPE |
|---|---|---|---|---|---|---|---|---|
| Wang, M. et al. [19] | American Society of Hematology image bank and Hospital BMS samples (AA, AML, MDS) | Diagnosing MDS | CNN | Internal | 0.985 | 0.914 | 0.992 | 0.881 |
| External | 0.942 | 0.921 | 0.886 | 0.938 | ||||
| Distinguishing MDS from AA and AML | CNN | Internal | 0.968 | 0.929 | 0.857 | 0.967 | ||
| External | 0.948 | 0.915 | 0.887 | 0.929 | ||||
| Lee, N. et al. [20] | Hospital BMS (MDS and healthy controls) | Detecting dysplastic erythrocytes | CNN | Internal | 0.972 | 0.988 | 0.790 | 0.992 |
| Detecting dysplastic granulocytes | CNN | Internal | 0.996 | 0.993 | 0.900 | 0.999 | ||
| Detecting dysplastic megakaryocytes | CNN | Internal | 0.971 | 0.931 | 0.899 | 0.948 | ||
| Detecting blasts | CNN | Internal | 0.973 | 0.932 | 0.831 | 0.951 | ||
| Mori, J. et al. [21] | Hospital BMS (MDS, “other hematological diseases”) | Diagnosing MDS using severe dysplasia (DG-3) | CNN | Internal | 0.944 | 0.972 | 0.910 | 0.977 |
| Diagnosing MDS using dysplasia and severe dysplasia | CNN | Internal | 0.921 | 0.982 | 0.852 | 0.989 | ||
| Wu, J. et al. [22] | Hospital BMS and PBS (Hypo-MDS, AA) | Diagnosing hypocellular MDS and distinguishing it from AA | Decision tree | Internal | 0.800 | 0.805 | 0.765 | 0.837 |
| Wu, Y. et al. [23] | Hospital BMS (MDS, multiple myeloma, MPD, AA, lymphoma) | Detecting > 5% blasts | CNN: BMSnet | Internal | 0.948 | NR | NR | NR |
| Acevedo, A. et al. [24] | Hospital PBS samples (MDS and healthy controls) | Detecting hypogranulated dysplastic neutrophils | CNN: model M1 | Internal | 0.982 | 0.949 | 0.955 | 0.943 |
| Kimura, K. et al. [25] | Hospital PBS data (MDS, MPN, AML, ALL, multiple myeloma, multiple lymphoma) | Diagnosing MDS and distinguishing it from AA | CNN with Xgboost | Internal | 0.990 | >0.900 | 0.962 | 1.000 |
| Zhu, J. et al. [26] | Hospital PBS (MDS and non-MDS controls) | Diagnosing MDS | CART | Internal | NR | NR | 0.845 | 0.978 |
| Clichet, V. et al. [27] | Hospital MFC data (MDS) | Diagnosing MDS | Elasticnet (LinearR) | External | 0.935 | NR | 0.918 | 0.925 |
| Duetz, C. et al. [28] | Hospital FC data (MDS, healthy controls, non-neoplastic cytopenia) | Diagnosing MDS in suspected patients | Random forest | Internal | 0.964 | NR | 0.850 | 0.950 |
| External | NR | NR | 0.970 | 0.950 | ||||
| Herbig, M. et al. [29] | University Hospital RT-DC data (MDS, AML, CML, AA) | Predicting MDS | Random forest | Internal | 0.950 | 0.910 | 0.860 | 1.000 |
| Li, J. L. et al. [30] | Hospital FC data (AML, MDS, normal) | Classification of MDS vs. Normal | LogR using AGF-P | Internal | 0.956 | 0.960 | NR | NR |
| Classification of MDS vs. AML | LogR using AGF-P | Internal | 0.911 | 0.875 | NR | NR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elshoeibi, A.M.; Badr, A.; Elsayed, B.; Metwally, O.; Elshoeibi, R.; Elhadary, M.R.; Elshoeibi, A.; Attya, M.A.; Khadadah, F.; Alshurafa, A.; et al. Integrating AI and ML in Myelodysplastic Syndrome Diagnosis: State-of-the-Art and Future Prospects. Cancers 2024, 16, 65. https://doi.org/10.3390/cancers16010065
Elshoeibi AM, Badr A, Elsayed B, Metwally O, Elshoeibi R, Elhadary MR, Elshoeibi A, Attya MA, Khadadah F, Alshurafa A, et al. Integrating AI and ML in Myelodysplastic Syndrome Diagnosis: State-of-the-Art and Future Prospects. Cancers. 2024; 16(1):65. https://doi.org/10.3390/cancers16010065
Chicago/Turabian StyleElshoeibi, Amgad Mohamed, Ahmed Badr, Basel Elsayed, Omar Metwally, Raghad Elshoeibi, Mohamed Ragab Elhadary, Ahmed Elshoeibi, Mohamed Amro Attya, Fatima Khadadah, Awni Alshurafa, and et al. 2024. "Integrating AI and ML in Myelodysplastic Syndrome Diagnosis: State-of-the-Art and Future Prospects" Cancers 16, no. 1: 65. https://doi.org/10.3390/cancers16010065
APA StyleElshoeibi, A. M., Badr, A., Elsayed, B., Metwally, O., Elshoeibi, R., Elhadary, M. R., Elshoeibi, A., Attya, M. A., Khadadah, F., Alshurafa, A., Alhuraiji, A., & Yassin, M. (2024). Integrating AI and ML in Myelodysplastic Syndrome Diagnosis: State-of-the-Art and Future Prospects. Cancers, 16(1), 65. https://doi.org/10.3390/cancers16010065





