MALAT-1 Is a Key Regulator of Epithelial–Mesenchymal Transition in Cancer: A Potential Therapeutic Target for Metastasis
Abstract
Simple Summary
Abstract
1. Introduction
2. Methodology for Searching the Literature and Study Selection Criteria
3. MALAT-1 Modulates EMT and Promotes Cancer Metastasis, Stemness, and Chemoresistance
3.1. MALAT-1 Induces Cancer Progression and Metastasis by Modulating EMT
3.2. MALAT-1 Promotes Chemoresistance via Modulating EMT
3.3. MALAT-1 Drives Cancer Cells toward More Stem Cell-like Features by Inducing EMT
4. MALAT-1 Regulates EMT by Competitively Inhibiting microRNAs, Enabling Cancer Invasion, Metastasis, and Chemoresistance
| Cancer | MicroRNA | Target Gene | Effect | Mechanism | Cell Lines | In Vivo | References |
|---|---|---|---|---|---|---|---|
| CC | miR-202-3p | Periostin | Downregulate EMT and cancer metastasis | ↑ E-cadherin, ↓ N-cadherin, ↓ vimentin, ↓ Periostin | H8, HeLa, and SiHa | - | [87] |
| CRC | miR-218 | EZH2/CDH1 | Downregulate EMT and chemoresistance | ↑ E-cadherin | T29, SW480, SW620, and FHC | - | [61] |
| EEC | miR-200c | TGF-β | Downregulate EMT and cancer metastasis | ↑ E-cadherin, ↓ ZEB1, ↓ N-cadherin, ↓ β-catenin, ↓ vimentin. | L-952, HEC-1-B, and JEC | BALB/c nude mice | [88] |
| EC | miR-1-3p | CORO1C/TPM3 | Downregulate EMT and cancer metastasis | ↑ E-cadherin, ↓ N-cadherin. | KYSE-510, and TE-7 | BALB/c mice | [89] |
| HNSCC | miR-30a | TGF-β/STAT3 | Downregulate EMT and cancer metastasis | ↓ Twist, ↓ MMP2/9, ↓ STAT3, ↑ E-cadherin, ↓ N-cadherin, ↓ vimentin, | SCC15, SCC25, CAL27, and HaCaT | BALB/c-nu mice | [90] |
| HCC | miR-142-3p | SMAD5 | Downregulate cancer cell growth, migration, and invasion | ↓ vimentin, ↑ E-cadherin, ↓ SMAD5, ↓ Ki-67 | Bel-7402, Huh-7, SMMC-7721, HL7702, and HepG2, | NOD/SCID mouse | [91] |
| miR-125a-3p | FOXM1 | Decrease cell proliferation, migration, and invasion | ↓ FOXM1 | Huh-6, HCCLM3, SK-HEP1, HuH-7, and PLC, L02 | Female athymic nude mice | [92] | |
| miR-22 | SNAI1 | Downregulate EMT | ↓ SNAI1, ↑ E-cadherin | HepG2, Hep3B, HuH7, and PLC/PRF5 | BALB/c nude mice | [93] | |
| Lung adenocarcinoma | miR-429 | RhoA | inhibit cell growth and metastasis | ↓ N-cadherin, ↑ E-cadherin, ↓ vimentin, ↓ Cyclin D1, ↓ MMP-9 | BEAS-2B, HBE, A549, H1299, SPC-A-1, and PG49 HPAEpiC | - | [94] |
| miR-204 | Slug | Downregulate EMT and cancer metastasis | ↑ E-cadherin, ↓ N-cadherin, ↓ vimentin, ↓ Slug | A549, H1299, H460, H446, and BEAS-2B | BALB/c-nu/nu mice | [95] | |
| NPC | miR-124 | Capn4 | Downregulate EMT and cancer metastasis | ↑ E-cadherin, ↓ N-cadherin, ↓ vimentin, ↓ Capn4 | HNEpC, C666-1, HONE-1,5-8F, and CNE-2 | - | [96] |
4.1. MALAT-1 Regulates EMT by Competitively Inhibiting microRNAs in Hepatocellular Carcinoma
4.2. MALAT-1 Regulates EMT via Competitively Inhibiting microRNAs in Lung Cancers
4.3. MALAT-1 Regulates EMT by Competitively Inhibiting microRNAs in Genitourinary Cancers
4.4. MALAT-1 Regulates EMT by Competitively Inhibiting microRNAs in GIT Cancers
4.5. MALAT-1 Regulates EMT by Competitively Inhibiting microRNAs in Head and Neck Cancers
5. MALAT-1 Promotes EMT via Modulation of Different Signaling Pathways
5.1. PI3K/Akt Signaling Pathway
5.2. Wnt/β-Catenin Signaling Pathway
5.3. Other Signaling Pathways
6. Drug Targeting
7. Conclusions and Future Prospective
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ASOs | Antisense oligonucleotides |
| CC | Cervical cancer |
| CDH1 | Cadherin 1 |
| CRC | Colorectal cancer |
| EEC | Endometrioid endometrial carcinoma |
| EMT | Epithelial to mesenchymal transitions |
| ESCC | Esophageal Squamous Cell Carcinoma |
| HCC | Hepatocellular carcinoma |
| HNSCC | Head and neck squamous cell carcinoma |
| lncRNA | Long non-coding RNA |
| MALAT-1 | Metastasis-associated lung adenocarcinoma transcript 1 |
| NEAT2 | Nuclear enriched abundant transcript 2 |
| NPC | Nasopharyngeal carcinoma |
| NSCLC | Non-small cell lung cancer |
| nt | Nucleotides |
| OSCC | Oral squamous cell carcinoma |
| TMZ | Temozolomide |
References
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.X.; Zhu, Q.N.; Zhang, H.B.; Hu, Y.; Wang, G.; Zhu, Y.S. MALAT1: A potential biomarker in cancer. Cancer Manag. Res. 2018, 10, 6757–6768. [Google Scholar] [CrossRef] [PubMed]
- Dueva, R.; Akopyan, K.; Pederiva, C.; Trevisan, D.; Dhanjal, S.; Lindqvist, A.; Farnebo, M. Neutralization of the Positive Charges on Histone Tails by RNA Promotes an Open Chromatin Structure. Cell Chem. Biol. 2019, 26, 1436–1449.e1435. [Google Scholar] [CrossRef] [PubMed]
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019, 26, 2904–2915.e2904. [Google Scholar] [CrossRef] [PubMed]
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent transcription from active promoters. Science 2008, 322, 1849–1851. [Google Scholar] [CrossRef] [PubMed]
- Hartford, C.C.R.; Lal, A. When Long Noncoding Becomes Protein Coding. Mol. Cell Biol. 2020, 40, e00528-19. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lin, J.; Fang, H.; Fang, J.; Li, C.; Chen, W.; Liu, S.; Ondrejka, S.; Gong, Z.; Reu, F.; et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia 2018, 32, 2250–2262. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lin, C.; Dong, H.; Piao, Z.; Jin, C.; Han, H.; Jin, D. Targeting MALAT1 induces DNA damage and sensitize non-small cell lung cancer cells to cisplatin by repressing BRCA1. Cancer Chemother. Pharmacol. 2020, 86, 663–672. [Google Scholar] [CrossRef]
- Anbiyaee, O.; Moalemnia, A.; Ghaedrahmati, F.; Shooshtari, M.K.; Khoshnam, S.E.; Kempisty, B.; Halili, S.A.; Farzaneh, M.; Morenikeji, O.B. The functions of long non-coding RNA (lncRNA)-MALAT-1 in the pathogenesis of renal cell carcinoma. BMC Nephrol. 2023, 24, 380. [Google Scholar] [CrossRef]
- Hou, Z.H.; Xu, X.W.; Fu, X.Y.; Zhou, L.D.; Liu, S.P.; Tan, D.M. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am. J. Physiol. Cell Physiol. 2020, 318, C649–C663. [Google Scholar] [CrossRef]
- Mekky, R.Y.; Ragab, M.F.; Manie, T.; Attia, A.A.; Youness, R.A. MALAT-1: Immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Transl. Oncol. 2023, 31, 101653. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Tang, X.; Tian, X.; Tian, J.; Zhang, Y.; Ma, J.; Xu, H.; Wang, S. LncRNA MALAT1 negatively regulates MDSCs in patients with lung cancer. J. Cancer 2018, 9, 2436–2442. [Google Scholar] [CrossRef] [PubMed]
- Adewunmi, O.; Shen, Y.; Zhang, X.H.; Rosen, J.M. Targeted Inhibition of lncRNA Malat1 Alters the Tumor Immune Microenvironment in Preclinical Syngeneic Mouse Models of Triple-Negative Breast Cancer. Cancer Immunol. Res. 2023, 11, 1462–1479. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wu, Y.; Tan, D.; Meng, H.; Wang, K.; Bai, Y.; Yang, K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 7. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Toden, S.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M.; Boland, C.R.; et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 2014, 35, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Chen, L.; Wang, Y.; Jiang, X.; Xia, H.; Zhuang, Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neurooncol. 2015, 121, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.T.; Shi, D.B.; Wang, Y.W.; Li, X.X.; Xu, Y.; Tripathi, P.; Gu, W.L.; Cai, G.X.; Cai, S.J. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3174–3181. [Google Scholar]
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.M.; Chen, L.M.; Zheng, S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2012, 29, 1810–1816. [Google Scholar] [CrossRef]
- Xu, S.; Sui, S.; Zhang, J.; Bai, N.; Shi, Q.; Zhang, G.; Gao, S.; You, Z.; Zhan, C.; Liu, F.; et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 4881–4891. [Google Scholar]
- Jiao, F.; Hu, H.; Yuan, C.; Wang, L.; Jiang, W.; Jin, Z.; Guo, Z.; Wang, L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol. Rep. 2014, 32, 2485–2492. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Liang, G.; Yuan, B.; Yang, C.; Gao, R.; Zhou, X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015, 36, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Tee, A.E.; Ling, D.; Nelson, C.; Atmadibrata, B.; Dinger, M.E.; Xu, N.; Mizukami, T.; Liu, P.Y.; Liu, B.; Cheung, B.; et al. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget 2014, 5, 1793–1804. [Google Scholar] [CrossRef] [PubMed]
- Glover, A.R.; Zhao, J.T.; Ip, J.C.; Lee, J.C.; Robinson, B.G.; Gill, A.J.; Soon, P.S.; Sidhu, S.B. Long noncoding RNA profiles of adrenocortical cancer can be used to predict recurrence. Endocr. Relat. Cancer 2015, 22, 99–109. [Google Scholar] [CrossRef] [PubMed]
- (UALCAN), The University of Alabama at Birmingham. MALAT-1 Gene Expression across Different Cancers Using Data from The Cancer Genome Atlas (TCGA). Available online: http://ualcan.path.uab.edu/analysis.html (accessed on 20 May 2022).
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wang, J.; Li, J.; Zhao, M.; Huang, S.K.; Gu, Y.Y.; Li, Y.; Sun, X.J.; Yang, L.; Luo, Q.; et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016, 151, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Du, L.; Jiang, X.; Wang, R.; Yan, S.; Xie, Y.; Yan, K.; Wang, Q.; Wang, L.; Zhang, X.; et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget 2016, 7, 78850–78858. [Google Scholar] [CrossRef]
- Huang, S.K.; Luo, Q.; Peng, H.; Li, J.; Zhao, M.; Wang, J.; Gu, Y.Y.; Li, Y.; Yuan, P.; Zhao, G.H.; et al. A Panel of Serum Noncoding RNAs for the Diagnosis and Monitoring of Response to Therapy in Patients with Breast Cancer. Med. Sci. Monit. 2018, 24, 2476–2488. [Google Scholar] [CrossRef]
- He, B.; Zeng, J.; Chao, W.; Chen, X.; Huang, Y.; Deng, K.; Huang, Z.; Li, J.; Dai, M.; Chen, S.; et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget 2017, 8, 41166–41177. [Google Scholar] [CrossRef]
- Eissmann, M.; Gutschner, T.; Hammerle, M.; Gunther, S.; Caudron-Herger, M.; Gross, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zornig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, Y.Q.; You, S.; Zhang, C.M.; Wu, L.; Zhao, W.; Wang, X.M. Long Non-Coding RNA MALAT1 as a Detection and Diagnostic Molecular Marker in Various Human Cancers: A Pooled Analysis Based on 3255 Subjects. Onco Targets Ther. 2020, 13, 5807–5817. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Wang, J.; Zhu, H.; Liu, L.; Jiang, Y. Chronic oxymatrine treatment induces resistance and epithelialmesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells. Oncol. Rep. 2018, 39, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sarkissyan, M.; Ogah, O.; Kim, J.; Vadgama, J.V. Expression of MALAT1 Promotes Trastuzumab Resistance in HER2 Overexpressing Breast Cancers. Cancers 2020, 12, 1918. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lu, X.; Yu, R. lncRNA MALAT1 Promotes EMT Process and Cisplatin Resistance of Oral Squamous Cell Carcinoma via PI3K/AKT/m-TOR Signal Pathway. Onco Targets Ther. 2020, 13, 4049–4061. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Hu, H.; Han, T.; Yuan, C.; Wang, L.; Jin, Z.; Guo, Z.; Wang, L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015, 16, 6677–6693. [Google Scholar] [CrossRef] [PubMed]
- Iderzorig, T.; Kellen, J.; Osude, C.; Singh, S.; Woodman, J.A.; Garcia, C.; Puri, N. Comparison of EMT mediated tyrosine kinase inhibitor resistance in NSCLC. Biochem. Biophys. Res. Commun. 2018, 496, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Dedhar, S.; Kalluri, R.; Thompson, E.W. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J. Cell Biol. 2006, 172, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e1624. [Google Scholar] [CrossRef]
- Jang, M.H.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Park, S.Y. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum. Pathol. 2015, 46, 1267–1274. [Google Scholar] [CrossRef]
- Dong, J.; Hu, Y.; Fan, X.; Wu, X.; Mao, Y.; Hu, B.; Guo, H.; Wen, L.; Tang, F. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 2018, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Jechlinger, M.; Grunert, S.; Beug, H. Mechanisms in epithelial plasticity and metastasis: Insights from 3D cultures and expression profiling. J. Mammary Gland. Biol. Neoplasia 2002, 7, 415–432. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Niessen, K.; Fu, Y.; Chang, L.; Hoodless, P.A.; McFadden, D.; Karsan, A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 2008, 182, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Medici, D.; Hay, E.D.; Olsen, B.R. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol. Biol. Cell 2008, 19, 4875–4887. [Google Scholar] [CrossRef]
- Kokudo, T.; Suzuki, Y.; Yoshimatsu, Y.; Yamazaki, T.; Watabe, T.; Miyazono, K. Snail is required for TGFbeta-induced endothelial-mesenchymal transition of embryonic stem cell-derived endothelial cells. J. Cell Sci. 2008, 121, 3317–3324. [Google Scholar] [CrossRef]
- Patel, M.; Eckburg, A.; Gantiwala, S.; Hart, Z.; Dein, J.; Lam, K.; Puri, N. Resistance to Molecularly Targeted Therapies in Melanoma. Cancers 2021, 13, 1115. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Garside, V.C.; Chang, A.C.; Karsan, A.; Hoodless, P.A. Co-ordinating Notch, BMP, and TGF-beta signaling during heart valve development. Cell Mol. Life Sci. 2013, 70, 2899–2917. [Google Scholar] [CrossRef]
- Micalizzi, D.S.; Farabaugh, S.M.; Ford, H.L. Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J. Mammary Gland. Biol. Neoplasia 2010, 15, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Wells, A.; Yates, C.; Shepard, C.R. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin. Exp. Metastasis 2008, 25, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Tse, J.C.; Kalluri, R. Mechanisms of metastasis: Epithelial-to-mesenchymal transition and contribution of tumor microenvironment. J. Cell Biochem. 2007, 101, 816–829. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Cen, Y.; Chen, J. Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol. Lett. 2018, 15, 2117–2122. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Qin, C.; Jiang, B.; Fang, S.; Pan, X.; Peng, L.; Liu, Z.; Li, W.; Li, Y.; Li, G. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial-mesenchymal transition. Mol. Biosyst. 2016, 12, 952–962. [Google Scholar] [CrossRef] [PubMed]
- Li, L.J.; Chai, Y.; Guo, X.J.; Chu, S.L.; Zhang, L.S. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed. Pharmacother. 2017, 89, 939–948. [Google Scholar] [CrossRef]
- Chen, D.; Liu, L.; Wang, K.; Yu, H.; Wang, Y.; Liu, J.; Guo, Y.; Zhang, H. The role of MALAT-1 in the invasion and metastasis of gastric cancer. Scand. J. Gastroenterol. 2017, 52, 790–796. [Google Scholar] [CrossRef]
- Li, H.; Yuan, X.; Yan, D.; Li, D.; Guan, F.; Dong, Y.; Wang, H.; Liu, X.; Yang, B. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cell Physiol. Biochem. 2017, 42, 1192–1201. [Google Scholar] [CrossRef]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol. Cancer Ther. 2017, 16, 739–751. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Zeng, Y.; Zhang, X.; Hu, Q.; Zheng, J.; Chen, B.; Xie, B.; Zhang, W.M. Implication of epithelial-mesenchymal transition in IGF1R-induced resistance to EGFR-TKIs in advanced non-small cell lung cancer. Oncotarget 2015, 6, 44332–44345. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zhao, Y.; Li, Y.; Zhang, T.; Ma, Y.; Liu, Y. LncRNA MALAT1 Promotes Lung Cancer Proliferation and Gefitinib Resistance by Acting as a miR-200a Sponge. Arch Bronconeumol. 2019, 55, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Q.; Liu, J.; Gao, L.; Liu, J.; Wang, R.; Chu, W.; Zhang, Y.; Tan, G.; Zhao, X.; Lv, B. MicroRNA-200a Targets EGFR and c-Met to Inhibit Migration, Invasion, and Gefitinib Resistance in Non-Small Cell Lung Cancer. Cytogenet. Genome Res. 2015, 146, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhang, G.; Wang, X.; Wang, Y.; Wang, K. Functions and mechanisms of lncRNA MALAT1 in cancer chemotherapy resistance. Biomark. Res. 2023, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Song, X.; Xu, D.; Tiek, D.; Goenka, A.; Wu, B.; Sastry, N.; Hu, B.; Cheng, S.Y. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics 2020, 10, 8721–8743. [Google Scholar] [CrossRef] [PubMed]
- Nassar, D.; Blanpain, C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu. Rev. Pathol. 2016, 11, 47–76. [Google Scholar] [CrossRef]
- Castro-Oropeza, R.; Melendez-Zajgla, J.; Maldonado, V.; Vazquez-Santillan, K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 2018, 41, 585–603. [Google Scholar] [CrossRef]
- Choudhuri, S. Small noncoding RNAs: Biogenesis, function, and emerging significance in toxicology. J. Biochem. Mol. Toxicol. 2010, 24, 195–216. [Google Scholar] [CrossRef]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537–2545. [Google Scholar] [CrossRef] [PubMed]
- Ciafre, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Pallante, P.; Visone, R.; Ferracin, M.; Ferraro, A.; Berlingieri, M.T.; Troncone, G.; Chiappetta, G.; Liu, C.G.; Santoro, M.; Negrini, M.; et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer 2006, 13, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.G.; Sevignani, C.; Ferracin, M.; Felli, N.; Dumitru, C.D.; Shimizu, M.; Cimmino, A.; Zupo, S.; Dono, M.; et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA 2004, 101, 11755–11760. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Aspects Med. 2019, 70, 3–20. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Meng, X.; Li, A.; Yu, B.; Li, S. Interplay between miRNAs and lncRNAs: Mode of action and biological roles in plant development and stress adaptation. Comput. Struct. Biotechnol. J. 2021, 19, 2567–2574. [Google Scholar] [CrossRef]
- Tang, X.J.; Wang, W.; Hann, S.S. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie 2019, 163, 58–72. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Wang, Q.; Zhang, X.; Wang, D.; Tang, H.C.; Meng, X.; Ding, X. Identification of an lncRNAmiRNAmRNA interaction mechanism in breast cancer based on bioinformatic analysis. Mol. Med. Rep. 2017, 16, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.Y.; Wu, K.Y.; Lin, T.Y.; Chen, C.C. The Crosstalk of Long Non-Coding RNA and MicroRNA in Castration-Resistant and Neuroendocrine Prostate Cancer: Their Interaction and Clinical Importance. Int. J. Mol. Sci. 2021, 23, 392. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Liu, C.; Li, H.; Zhang, L.; Luo, G.; Liang, S.; Lu, M. Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol. Lett. 2020, 19, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Urrutia, E.; Bustamante Montes, L.P.; Ladron de Guevara Cervantes, D.; Perez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, Q.; Wang, Y.; Hu, B.; Dong, X.; Zhang, H.; Wang, W. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1/microRNA-202-3p/periostin axis modulates invasion and epithelial-mesenchymal transition in human cervical cancer. J. Cell Physiol. 2019, 234, 14170–14180. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, C.; Chen, R.; Xiong, H.; Qiu, F.; Liu, S.; Zhang, M.; Wang, F.; Wang, Y.; Zhou, X.; et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. 2016, 383, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Dai, Z.; Xia, C.; Jin, L.; Chen, X. Suppression of long non-coding RNA MALAT1 inhibits survival and metastasis of esophagus cancer cells by sponging miR-1-3p/CORO1C/TPM3 axis. Mol. Cell Biochem. 2020, 470, 165–174. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, C.; Zhang, C.; Li, Z.; Zhu, T.; Chen, J.; Ren, Y.; Wang, X.; Zhang, L.; Zhou, X. TGF-beta-induced STAT3 overexpression promotes human head and neck squamous cell carcinoma invasion and metastasis through malat1/miR-30a interactions. Cancer Lett. 2018, 436, 52–62. [Google Scholar] [CrossRef]
- Yu, Q.; Xiang, L.; Chen, Z.; Liu, X.; Ou, H.; Zhou, J.; Yang, D. MALAT1 functions as a competing endogenous RNA to regulate SMAD5 expression by acting as a sponge for miR-142-3p in hepatocellular carcinoma. Cell Biosci. 2019, 9, 39. [Google Scholar] [CrossRef]
- Liu, S.; Qiu, J.; He, G.; Liang, Y.; Wang, L.; Liu, C.; Pan, H. LncRNA MALAT1 acts as a miR-125a-3p sponge to regulate FOXM1 expression and promote hepatocellular carcinoma progression. J. Cancer 2019, 10, 6649–6659. [Google Scholar] [CrossRef]
- Chen, S.; Wang, G.; Tao, K.; Cai, K.; Wu, K.; Ye, L.; Bai, J.; Yin, Y.; Wang, J.; Shuai, X.; et al. Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 cooperates with enhancer of zeste homolog 2 to promote hepatocellular carcinoma development by modulating the microRNA-22/Snail family transcriptional repressor 1 axis. Cancer Sci. 2020, 111, 1582–1595. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zhu, Q.; Zhou, J. Long non-coding RNA MALAT1 interaction with miR-429 regulates the proliferation and EMT of lung adenocarcinoma cells through RhoA. Int. J. Clin. Exp. Pathol. 2019, 12, 419–430. [Google Scholar] [PubMed]
- Li, J.; Wang, J.; Chen, Y.; Li, S.; Jin, M.; Wang, H.; Chen, Z.; Yu, W. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am. J. Cancer Res. 2016, 6, 1099–1107. [Google Scholar] [PubMed]
- Shi, B.; Wang, Y.; Yin, F. MALAT1/miR-124/Capn4 axis regulates proliferation, invasion and EMT in nasopharyngeal carcinoma cells. Cancer Biol. Ther. 2017, 18, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Zhang, Y.; Zhang, S.; Wen, F.; Miao, Y.; Guo, K. MicroRNA-142-3p inhibits cell proliferation and invasion of cervical cancer cells by targeting FZD7. Tumour Biol. 2015, 36, 8065–8073. [Google Scholar] [CrossRef]
- Wu, J.W.; Hu, M.; Chai, J.; Seoane, J.; Huse, M.; Li, C.; Rigotti, D.J.; Kyin, S.; Muir, T.W.; Fairman, R.; et al. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol. Cell 2001, 8, 1277–1289. [Google Scholar] [CrossRef]
- Raychaudhuri, P.; Park, H.J. FoxM1: A master regulator of tumor metastasis. Cancer Res. 2011, 71, 4329–4333. [Google Scholar] [CrossRef]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Hanna, S.; El-Sibai, M. Signaling networks of Rho GTPases in cell motility. Cell Signal 2013, 25, 1955–1961. [Google Scholar] [CrossRef]
- Lin, Y.; Zheng, Y. Approaches of targeting Rho GTPases in cancer drug discovery. Expert. Opin. Drug Discov. 2015, 10, 991–1010. [Google Scholar] [CrossRef]
- An, N.; Zheng, B. MiR-203a-3p Inhibits Pancreatic Cancer Cell Proliferation, EMT, and Apoptosis by Regulating SLUG. Technol. Cancer Res. Treat. 2020, 19, 1533033819898729. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Xiao, G.; Chen, Y.; Deng, Y. LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anticancer. Drugs 2018, 29, 725–735. [Google Scholar] [CrossRef]
- Wu, J.; Weng, Y.; He, F.; Liang, D.; Cai, L. LncRNA MALAT-1 competitively regulates miR-124 to promote EMT and development of non-small-cell lung cancer. Anticancer. Drugs 2018, 29, 628–636. [Google Scholar] [CrossRef]
- Gillan, L.; Matei, D.; Fishman, D.A.; Gerbin, C.S.; Karlan, B.Y.; Chang, D.D. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002, 62, 5358–5364. [Google Scholar] [PubMed]
- Seton-Rogers, S. Oncogenes: All eyes on YAP1. Nat. Rev. Cancer 2014, 14, 514–515. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Ou, C.; Liu, J.; Chen, C.; Zhou, Q.; Yang, S.; Li, G.; Wang, G.; Song, J.; Li, Z.; et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene 2019, 38, 2627–2644. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Lee, C.K. What does Stat3 do? J. Clin. Investig. 2002, 109, 1143–1148. [Google Scholar] [CrossRef]
- Zhao, C.; Yuan, G.; Jiang, Y.; Xu, J.; Ye, L.; Zhan, W.; Wang, J. Capn4 contributes to tumor invasion and metastasis in gastric cancer via activation of the Wnt/beta-catenin/MMP9 signalling pathways. Exp. Cell Res. 2020, 395, 112220. [Google Scholar] [CrossRef]
- Lunardi, A.; Webster, K.A.; Papa, A.; Padmani, B.; Clohessy, J.G.; Bronson, R.T.; Pandolfi, P.P. Role of aberrant PI3K pathway activation in gallbladder tumorigenesis. Oncotarget 2014, 5, 894–900. [Google Scholar] [CrossRef]
- Carnero, A.; Blanco-Aparicio, C.; Renner, O.; Link, W.; Leal, J.F. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr. Cancer Drug Targets 2008, 8, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Carnero, A. The PKB/AKT pathway in cancer. Curr. Pharm. Des. 2010, 16, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Jiang, L.L.; Xu, H.T.; Zhou, D.W.; Li, Z.S. Expression of PI3K/AKT pathway in gastric cancer and its blockade suppresses tumor growth and metastasis. Int. J. Immunopathol. Pharmacol. 2012, 25, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Fujishita, T. Oncogenic Roles of the PI3K/AKT/mTOR Axis. Curr. Top. Microbiol. Immunol. 2017, 407, 153–189. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, L.; Wu, X.; Li, R.; Wen, J.; Sha, J.; Wen, X. The PI3K/AKT pathway in the pathogenesis of prostate cancer. Front. Biosci. 2016, 21, 1084–1091. [Google Scholar] [CrossRef]
- Dai, Q.; Zhang, T.; Li, C. LncRNA MALAT1 Regulates the Cell Proliferation and Cisplatin Resistance in Gastric Cancer via PI3K/AKT Pathway. Cancer Manag. Res. 2020, 12, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, W.; Sun, W.; Zheng, B.; Wang, C.; Luo, Z.; Wang, J.; Yan, W. LncRNA MALAT1 Promotes Cancer Metastasis in Osteosarcoma via Activation of the PI3K-Akt Signaling Pathway. Cell Physiol. Biochem. 2018, 51, 1313–1326. [Google Scholar] [CrossRef]
- Wang, N.; Hou, M.S.; Zhan, Y.; Shen, X.B.; Xue, H.Y. MALAT1 promotes cisplatin resistance in cervical cancer by activating the PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7653–7659. [Google Scholar] [CrossRef]
- Wang, J.; Sun, G. FOXO1-MALAT1-miR-26a-5p Feedback Loop Mediates Proliferation and Migration in Osteosarcoma Cells. Oncol. Res. 2017, 25, 1517–1527. [Google Scholar] [CrossRef]
- Wang, C.; Mao, Z.P.; Wang, L.; Wu, G.H.; Zhang, F.H.; Wang, D.Y.; Shi, J.L. Long non-coding RNA MALAT1 promotes cholangiocarcinoma cell proliferation and invasion by activating PI3K/Akt pathway. Neoplasma 2017, 64, 725–731. [Google Scholar] [CrossRef]
- Jin, Y.; Feng, S.J.; Qiu, S.; Shao, N.; Zheng, J.H. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3176–3184. [Google Scholar] [PubMed]
- Zhou, X.; Liu, S.; Cai, G.; Kong, L.; Zhang, T.; Ren, Y.; Wu, Y.; Mei, M.; Zhang, L.; Wang, X. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci. Rep. 2015, 5, 15972. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Chen, Q.; Wang, Y.; Zhou, Z.; Huang, Y.; Qiu, F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst. 2012, 8, 2289–2294. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.L.; Bamodu, O.A.; Ong, J.R.; Lee, W.H.; Yeh, C.T.; Tsai, J.T. Targeting the Epigenetic Non-Coding RNA MALAT1/Wnt Signaling Axis as a Therapeutic Approach to Suppress Stemness and Metastasis in Hepatocellular Carcinoma. Cells 2020, 9, 1020. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.J.; Zheng, Y.B.; Pan, F.F.; Zhang, F.W.; Zhuang, P.; Fu, W.M. Gallic Acid Suppressed Tumorigenesis by an LncRNA MALAT1-Wnt/beta-Catenin Axis in Hepatocellular Carcinoma. Front. Pharmacol. 2021, 12, 708967. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zheng, S.; Chen, Y.; Liu, T.; Han, X.; Zhang, X.; Shen, T.; Lu, X. TGF-beta1-Induced Upregulation of MALAT1 Promotes Kazakh’s Esophageal Squamous Cell Carcinoma Invasion by EMT. J. Cancer 2020, 11, 6892–6901. [Google Scholar] [CrossRef]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014, 20, 1531–1541. [Google Scholar] [CrossRef]
- Chen, M.; Xia, Z.; Chen, C.; Hu, W.; Yuan, Y. LncRNA MALAT1 promotes epithelial-to-mesenchymal transition of esophageal cancer through Ezh2-Notch1 signaling pathway. Anticancer. Drugs 2018, 29, 767–773. [Google Scholar] [CrossRef]
- Fathi Dizaji, B. Strategies to target long non-coding RNAs in cancer treatment: Progress and challenges. Egypt. J. Med. Human. Genet. 2020, 21, 41. [Google Scholar] [CrossRef]
- Zhang, Z.C.; Tang, C.; Dong, Y.; Zhang, J.; Yuan, T.; Tao, S.C.; Li, X.L. Targeting the long noncoding RNA MALAT1 blocks the pro-angiogenic effects of osteosarcoma and suppresses tumour growth. Int. J. Biol. Sci. 2017, 13, 1398–1408. [Google Scholar] [CrossRef]
- Gong, N.; Teng, X.; Li, J.; Liang, X.J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater. Interfaces 2019, 11, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhong, Z.; Tan, H.Y.; Guo, W.; Zhang, C.; Cheng, C.S.; Wang, N.; Ren, J.; Feng, Y. Suppression of lncRNA MALAT1 by betulinic acid inhibits hepatocellular carcinoma progression by targeting IAPs via miR-22-3p. Clin. Transl. Med. 2020, 10, e190. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Liu, X.; Fu, X.; Zhang, L.; Sui, H.; Zhou, L.; Sun, J.; Cai, J.; Qin, J.; Ren, J.; et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/beta-catenin signal pathway. PLoS ONE 2013, 8, e78700. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xie, Q.; Chen, Z.; Ni, H.; Xia, L.; Zhao, Q.; Chen, Z.; Chen, P. Resveratrol suppresses the invasion and migration of human gastric cancer cells via inhibition of MALAT1-mediated epithelial-to-mesenchymal transition. Exp. Ther. Med. 2019, 17, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Feng, Y.; Zheng, X.; Sun, L.; Wasan, H.S.; Ruan, S.; Shen, M. Resveratrol and Its Analogs: Potent Agents to Reverse Epithelial-to-Mesenchymal Transition in Tumors. Front. Oncol. 2021, 11, 644134. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Jiang, B.; Wang, H.; Ouyang, W.; Chen, X.; Wang, T.; Dong, D.; Yi, S.; Yi, J.; Huang, Y.; et al. Dihydromyricetin induced lncRNA MALAT1-TFEB-dependent autophagic cell death in cutaneous squamous cell carcinoma. J. Cancer 2019, 10, 4245–4255. [Google Scholar] [CrossRef] [PubMed]
- Francois-Moutal, L.; Miranda, V.G.; Mollasalehi, N.; Gokhale, V.; Khanna, M. In Silico Targeting of the Long Noncoding RNA MALAT1. ACS Med. Chem. Lett. 2021, 12, 915–921. [Google Scholar] [CrossRef]
- Abulwerdi, F.A.; Xu, W.; Ageeli, A.A.; Yonkunas, M.J.; Arun, G.; Nam, H.; Schneekloth, J.S., Jr.; Dayie, T.K.; Spector, D.; Baird, N.; et al. Selective Small-Molecule Targeting of a Triple Helix Encoded by the Long Noncoding RNA, MALAT1. ACS Chem. Biol. 2019, 14, 223–235. [Google Scholar] [CrossRef]
- Donlic, A.; Zafferani, M.; Padroni, G.; Puri, M.; Hargrove, A.E. Regulation of MALAT1 triple helix stability and in vitro degradation by diphenylfurans. Nucleic Acids Res. 2020, 48, 7653–7664. [Google Scholar] [CrossRef]
- Miao, S.; Bhunia, D.; Devari, S.; Liang, Y.; Munyaradzi, O.; Rundell, S.; Bong, D. Bifacial PNAs Destabilize MALAT1 by 3’ A-Tail Displacement from the U-Rich Internal Loop. ACS Chem. Biol. 2021, 16, 1600–1609. [Google Scholar] [CrossRef]
- Liu, X.M.; Zhou, J.; Mao, Y.; Ji, Q.; Qian, S.B. Programmable RNA N(6)-methyladenosine editing by CRISPR-Cas9 conjugates. Nat. Chem. Biol. 2019, 15, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Tan, B.; Ma, F.; Zhu, B.; Zhang, L.; Liu, X.; Li, H.; Yang, J.; Cheng, B. A Synthetic Light-switchable System based on CRISPR Cas13a Regulates the Expression of LncRNA MALAT1 and Affects the Malignant Phenotype of Bladder Cancer Cells. Int. J. Biol. Sci. 2019, 15, 1630–1636. [Google Scholar] [CrossRef] [PubMed]
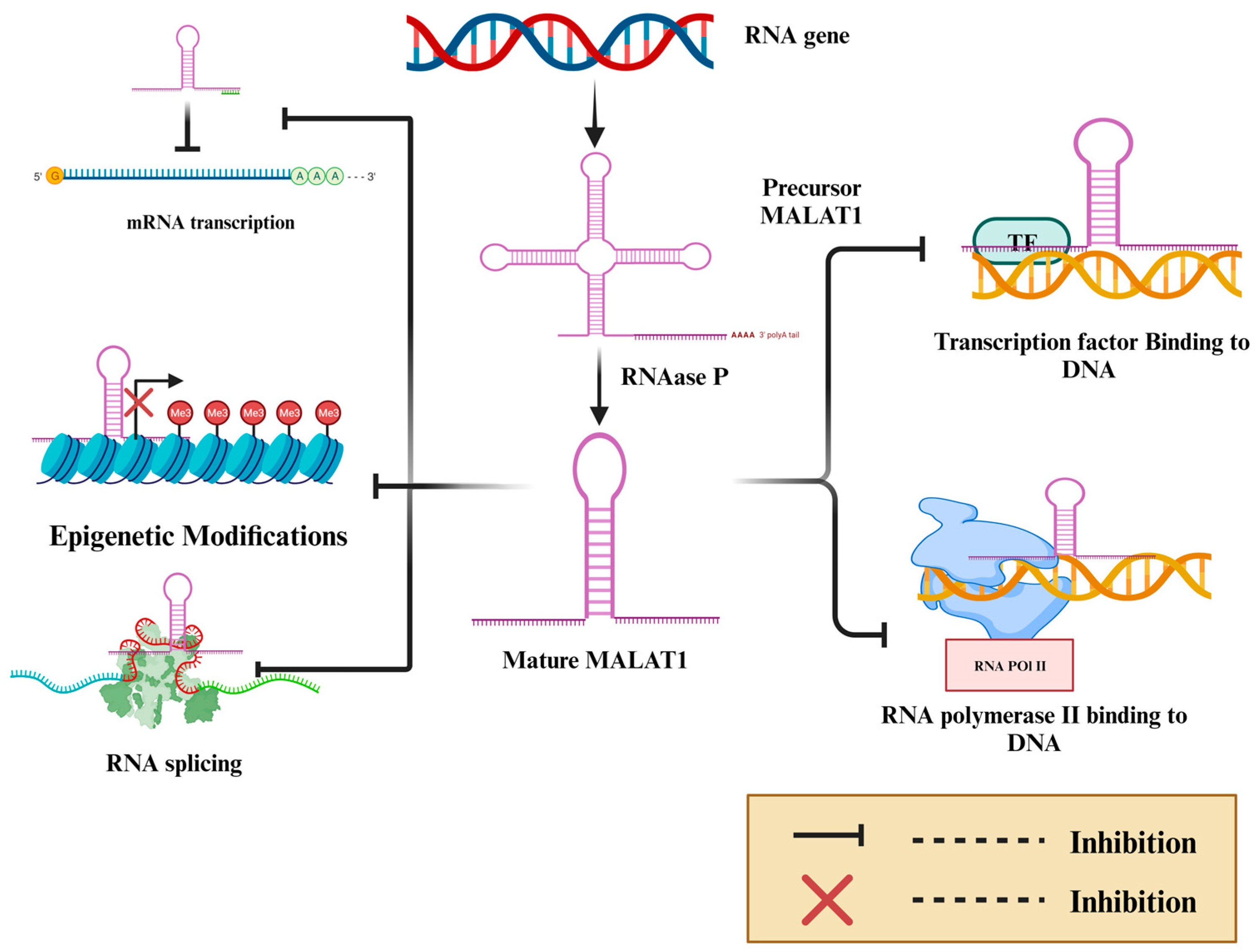
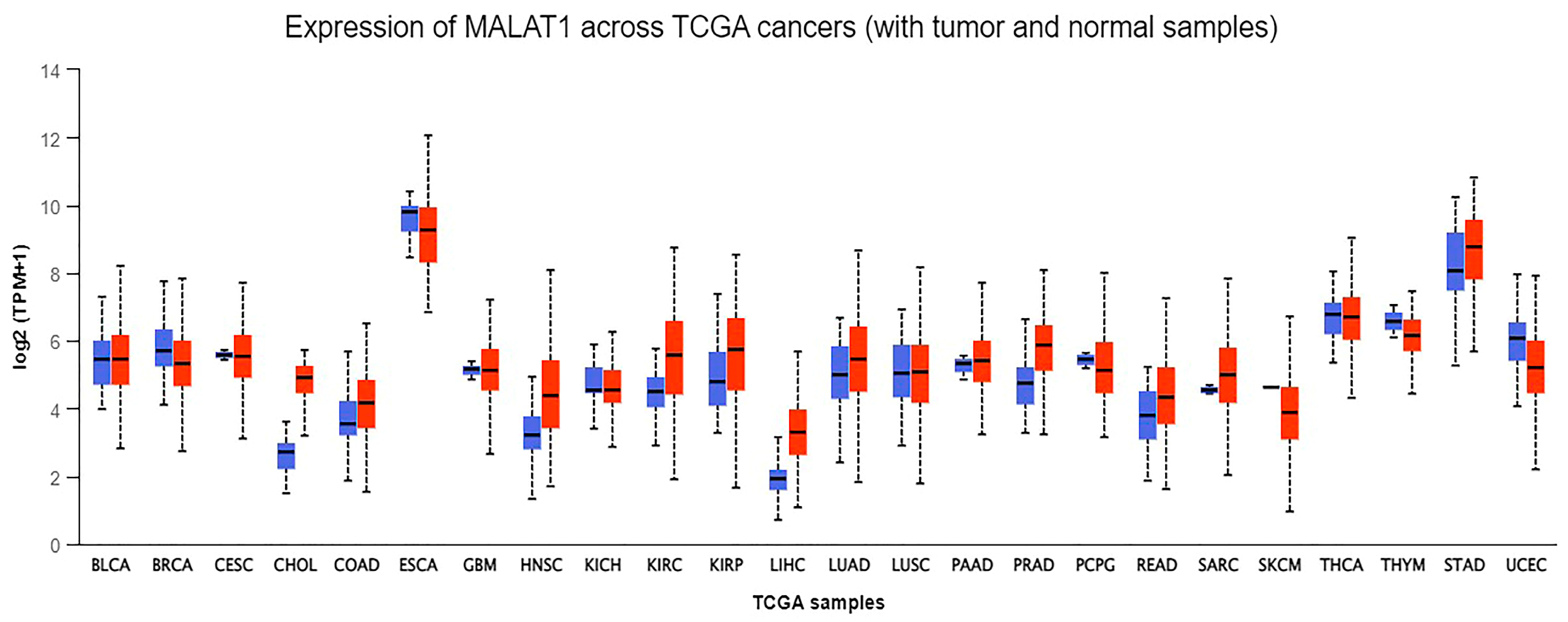
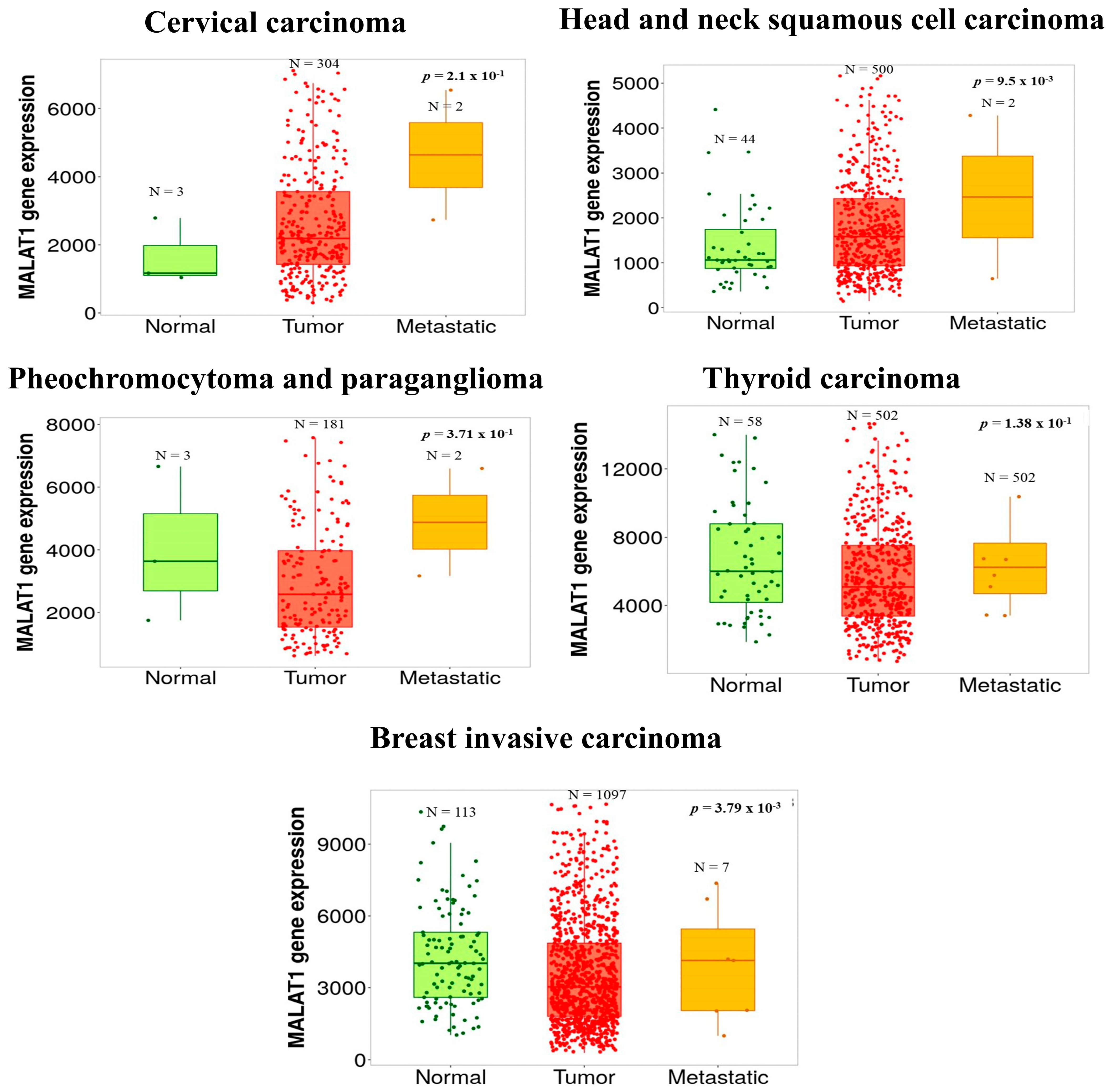
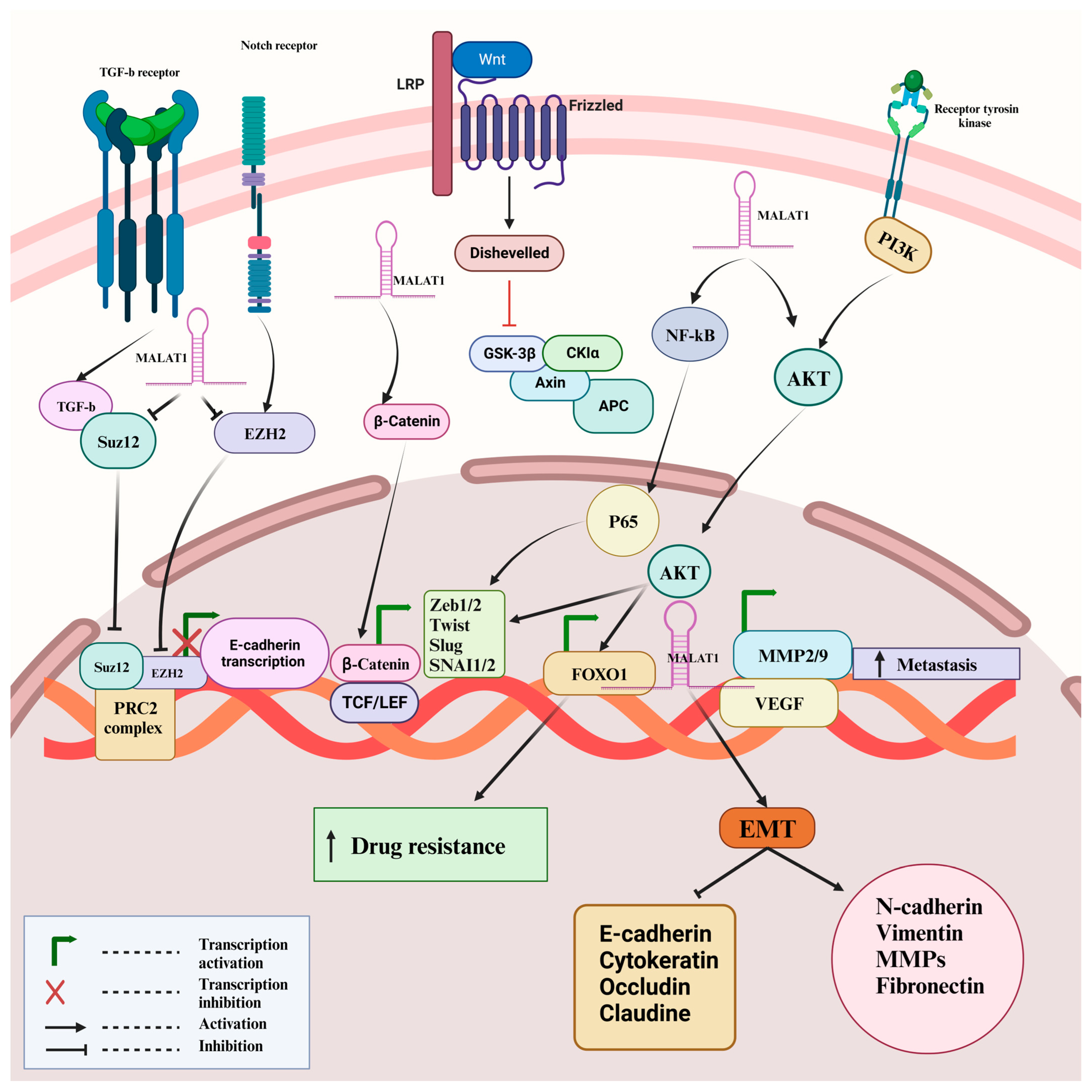
| Cancer Type | Phenotype | Effect | Mechanism | Cell Lines | In Vivo | References |
|---|---|---|---|---|---|---|
| BC | Cancer stem cell-like properties | Modulate stem cell-like properties in BC | ↓ CD133+, ↓ ALDH+, and ↓ Sox2 | MCF7 | - | [55] |
| CC | EMT | Inhibit invasion and metastasis | ↑ E-cadherin, ↑ ZO-1, ↓ vimentin, ↓ β-catenin, and ↓ Snail1 | H8, CC, CaSki, HeLa, SiHa | Female BALB/c nude mice | [56] |
| CRC | Chemoresistance | Reverse EMT and reverse Oxymatrine resistance | ↑ E-cadherin, and ↑ vimentin | HT29, SW480, HT29 Oxymatrine resistant | - | [33] |
| Diffuse large B-cell lymphoma | Chemoresistance | Enhance drug sensitivity by inducing autophagy in DLBCL | ↑ LC3-II, ↑ LC3-I, and ↓ p62 | IM-9I, Ly3, Ly8, Pfeiffer, Farage, Raji, Daud, Ly10, Ly1 | BALB/c-nu/nu nude mice | [57] |
| GC | EMT | Inhibit invasion and metastasis | ↑ E-cadherin, and ↓ vimentin | SGC-7901, BGC823, AGS, MKN4, SGC7901M, SGC7901NM | Female nude mice | [58] |
| Glioblastoma | Chemoresistance | Increase sensitivity to Temozolomide | ↓ MDR1, ↓ MRP5, ↓ LRP, ↓ ZEB, ↑ E-cadherin, ↑ ZO-1, ↓ SMA, and ↓ Fibronectin | U251, U87, U251/TMZ, U87/TMZ | Nude mouse | [59] |
| Lung cancer with brain metastases | EMT | Inhibit invasion and metastasis | ↑ E-cadherin, and ↑ vimentin | H1915-L10, H1915-H10 | Athymic BALB/c-nu/ nu mice | [17] |
| Oral squamous cell carcinoma (OSCC) | Chemoresistance | Reverse EMT and increase Cisplatin chemosensitivity | ↓ p-PI3K, ↓ PI3K ↓ p-AKT, ↓ AKT, ↓ pm-TOR, ↓ mTOR, and ↑ E-cadherin | CAL-27, SCC-9 | BALB/c nude mice | [35] |
| Pancreatic cancer | EMT | promote apoptosis, inhibit tumor invasion and migration | ↑ p21, ↑ p53, ↓ CDC2, ↓ Snail, ↓ Slug, ↑ E-cadherin, ↓ N-cadherin, ↓ vimentin, ↓ MMP-2, and ↓ MMP-9 | BxPC-3, AsPC-1, PANC-1, CFPAC-1, CAPAN-1, SW1990, HS-766T | - | [21] |
| Cancer stem cells like phenotype and chemoresistance | Modulate stem cell-like phenotype and enhance drug sensitivity | ↓ CD133+, and ↓ Sox2 | AsPC-1, CFPAC-1, | BALB/c nude mice | [36] | |
| TNBC | Chemoresistance | Reverse EMT and reverse Trastuzumab resistance | ↓ Snail, ↓ Slug, ↓ Twist, ↓ Nanog | SKBR3, BT474, MCF7, T47D, MDA-MB231, MCF-12A, JIMT1 | Female athymic nude mice | [34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, M.A.; Valinezhad, K.; Adel, E.; Munirathinam, G. MALAT-1 Is a Key Regulator of Epithelial–Mesenchymal Transition in Cancer: A Potential Therapeutic Target for Metastasis. Cancers 2024, 16, 234. https://doi.org/10.3390/cancers16010234
Hussein MA, Valinezhad K, Adel E, Munirathinam G. MALAT-1 Is a Key Regulator of Epithelial–Mesenchymal Transition in Cancer: A Potential Therapeutic Target for Metastasis. Cancers. 2024; 16(1):234. https://doi.org/10.3390/cancers16010234
Chicago/Turabian StyleHussein, Mohamed Ali, Kamyab Valinezhad, Eman Adel, and Gnanasekar Munirathinam. 2024. "MALAT-1 Is a Key Regulator of Epithelial–Mesenchymal Transition in Cancer: A Potential Therapeutic Target for Metastasis" Cancers 16, no. 1: 234. https://doi.org/10.3390/cancers16010234
APA StyleHussein, M. A., Valinezhad, K., Adel, E., & Munirathinam, G. (2024). MALAT-1 Is a Key Regulator of Epithelial–Mesenchymal Transition in Cancer: A Potential Therapeutic Target for Metastasis. Cancers, 16(1), 234. https://doi.org/10.3390/cancers16010234






