Rare Pediatric Cerebellar High-Grade Gliomas Mimic Medulloblastomas Histologically and Transcriptomically and Show p53 Mutations
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Fluorescence In Situ Hybridization (FISH)
2.3. Methylation Profiling
2.4. Identification of Copy Number Variations with 850K Array
2.5. Target Sequencing
2.6. Detection of TERT Promoter Mutation
2.7. Target RNA Sequencing
2.8. Nanostring-Based Affiliation
2.9. Immunohistochemistry
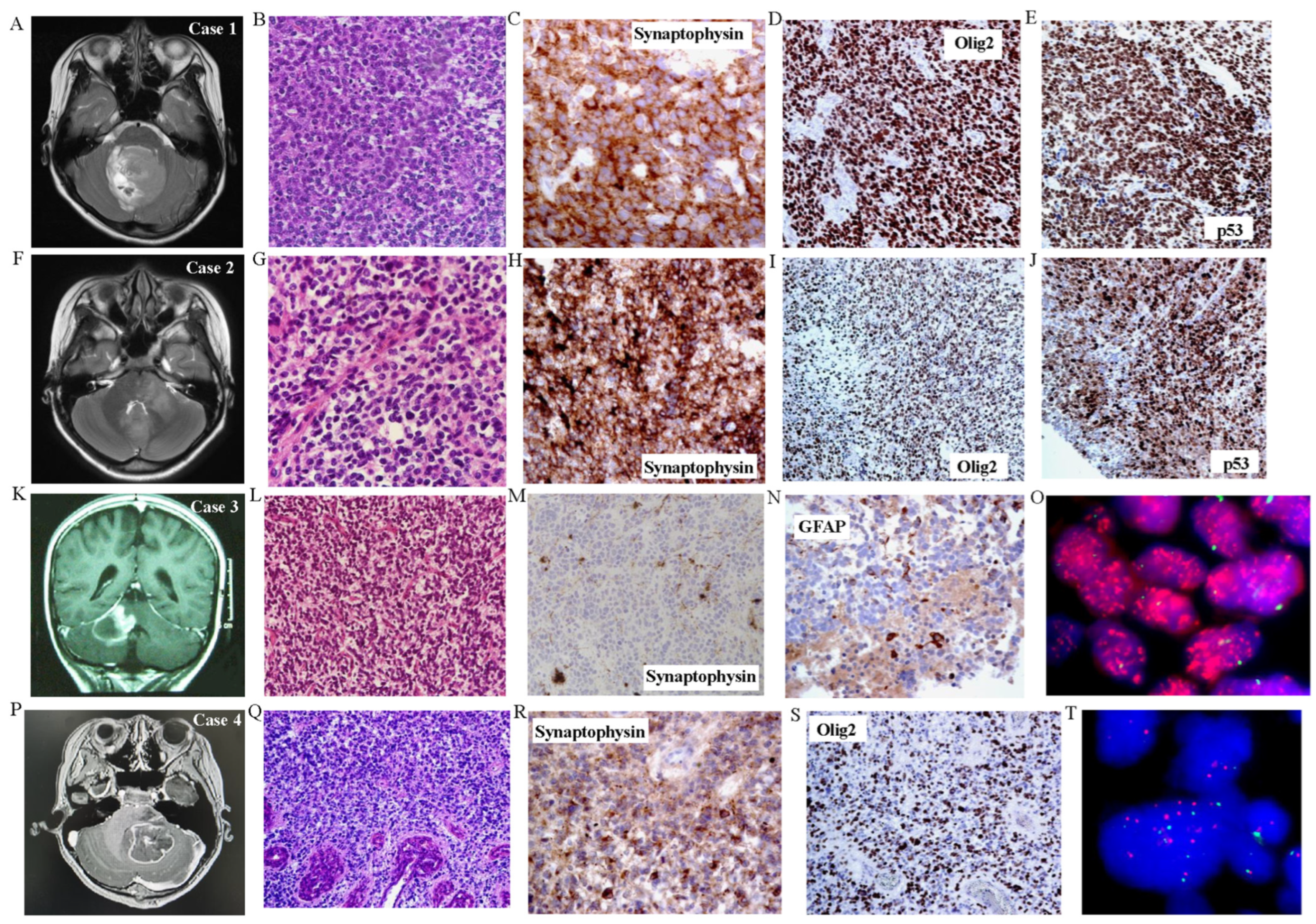
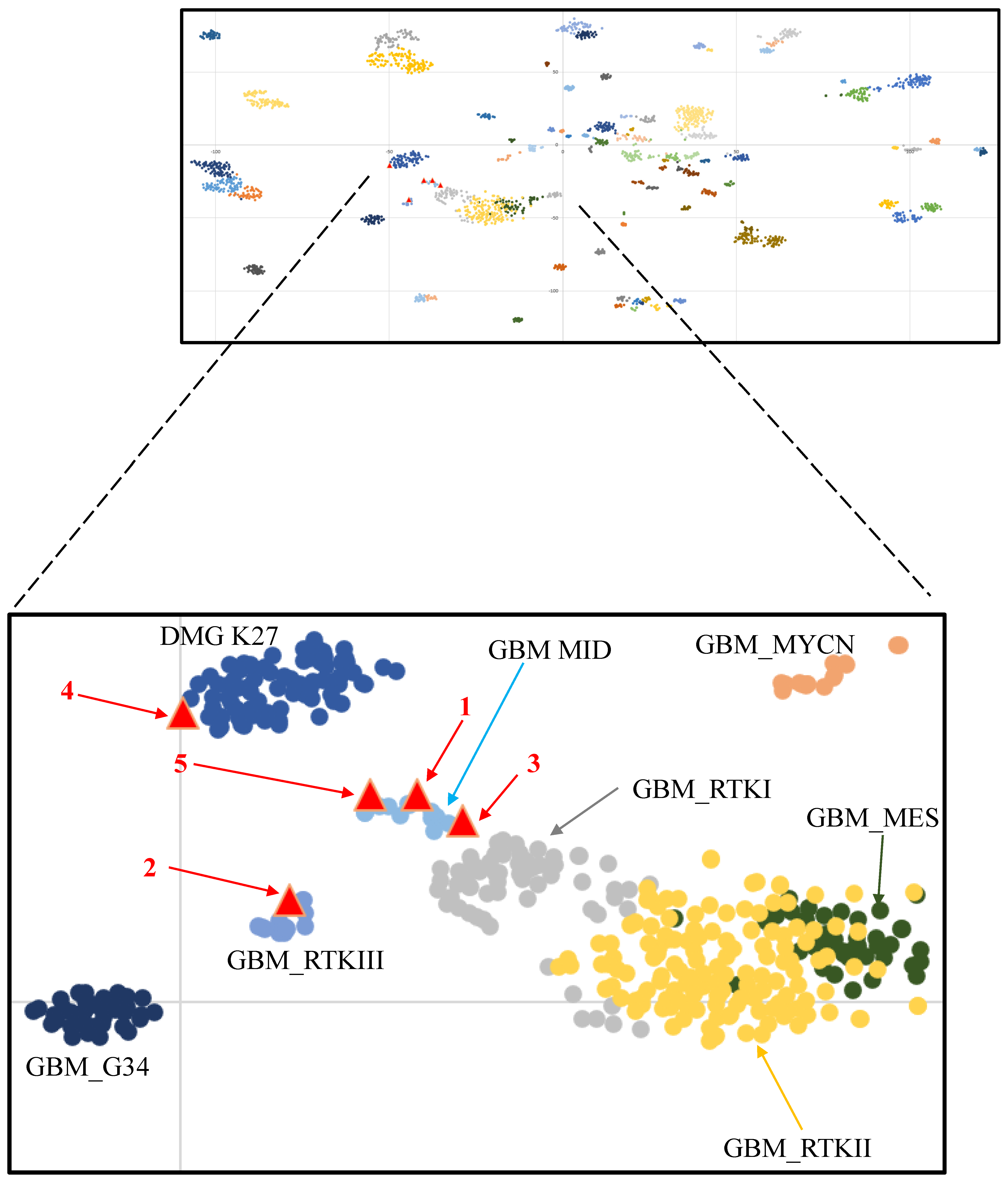
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thorbinson, C.; Kilday, J.P. Childhood Malignant Brain Tumors: Balancing the Bench and Bedside. Cancers 2021, 13, 6099. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; de Blank, P.M.; Kruchko, C.; Petersen, C.M.; Liao, P.; Finlay, J.L.; Stearns, D.S.; Wolff, J.E.; Wolinsky, Y.; Letterio, J.J.; et al. Alex’s Lemonade Stand Foundation Infant and Childhood Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011. Neuro Oncol. 2015, 16 (Suppl. S10), x1–x36. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro Oncol. 2021, 23 (Suppl. S3), iii1–iii105. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Pfister, S.M.; Jones, D.T.W. Pediatric Gliomas: Current Concepts on Diagnosis, Biology, and Clinical Management. J. Clin. Oncol. 2017, 35, 2370–2377. [Google Scholar] [CrossRef] [PubMed]
- Karremann, M.; Rausche, U.; Roth, D.; Kühn, A.; Pietsch, T.; Gielen, G.H.; Warmuth-Metz, M.; Kortmann, R.D.; Straeter, R.; Gnekow, A.; et al. Cerebellar location may predict an unfavourable prognosis in paediatric high-grade glioma. Br. J. Cancer 2013, 109, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, A.; Ryzhova, M.; Hovestadt, V.; Bender, S.; Sturm, D.; Capper, D.; Meyer, J.; Schrimpf, D.; Kool, M.; Northcott, P.A.; et al. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol. 2015, 129, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Mackay, A.; Burford, A.; Molinari, V.; Jones, D.T.W.; Izquierdo, E.; Brouwer-Visser, J.; Giangaspero, F.; Haberler, C.; Pietsch, T.; Jacques, T.S.; et al. Molecular, Pathological, Radiological, and Immune Profiling of Non-brainstem Pediatric High-Grade Glioma from the HERBY Phase II Randomized Trial. Cancer Cell 2018, 33, 829–842.e5. [Google Scholar] [CrossRef]
- Wu, G.; Diaz, A.K.; Paugh, B.S.; Rankin, S.L.; Ju, B.; Li, Y.; Zhu, X.; Qu, C.; Chen, X.; Zhang, J.; et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 2014, 46, 444–450. [Google Scholar]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.A.; Jones, D.T.; Konermann, C.; Pfaff, E.; Tönjes, M.; Sill, M.; Bender, S.; et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef]
- Mackay, A.; Burford, A.; Carvalho, D.; Izquierdo, E.; Fazal-Salom, J.; Taylor, K.R.; Bjerke, L.; Clarke, M.; Vinci, M.; Nandhabalan, M.; et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017, 32, 520–537.e5. [Google Scholar] [CrossRef]
- Reinhardt, A.; Stichel, D.; Schrimpf, D.; Koelsche, C.; Wefers, A.K.; Ebrahimi, A.; Sievers, P.; Huang, K.; Casalini, M.B.; Fernández-Klett, F.; et al. Tumors diagnosed as cerebellar glioblastoma comprise distinct molecular entities. Acta Neuropathol. Commun. 2019, 7, 163. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Ellison, D.W.; Figarella-Branger, D.; Hawkins, C.E.; Louis, D.N.; Perry, A.; Pfister, S.M.; Reifenberger, G.; von Deimling, A. Gliomas, Glioneuronal Tumours, and Neuronal Tumours: WHO Classification of Tumours, 5th ed.; Central Nervous System Tumours, the WHO Classification of Tumours Editorial Board, Chapter 2; IRAC: Lyon, France, 2021; pp. 90–92. [Google Scholar]
- Reinhardt, A.; Stichel, D.; Schrimpf, D.; Sahm, F.; Korshunov, A.; Reuss, D.E.; Koelsche, C.; Huang, K.; Wefers, A.K.; Hovestadt, V.; et al. Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol. 2018, 136, 273–291. [Google Scholar] [CrossRef]
- Buccoliero, A.M.; Giunti, L.; Moscardi, S.; Castiglione, F.; Provenzano, A.; Sardi, I.; Scagnet, M.; Genitori, L.; Caporalini, C. Pediatric High Grade Glioma Classification Criteria and Molecular Features of a Case Series. Genes 2022, 13, 624. [Google Scholar] [CrossRef] [PubMed]
- Gareton, A.; Tauziède-Espariat, A.; Dangouloff-Ros, V.; Roux, A.; Saffroy, R.; Castel, D.; Kergrohen, T.; Fina, F.; Figarella-Branger, D.; Pagès, M.; et al. The histomolecular criteria established for adult anaplastic pilocytic astrocytoma are not applicable to the pediatric population. Acta Neuropathol. 2020, 139, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, A.; Schrimpf, D.; Ryzhova, M.; Sturm, D.; Chavez, L.; Hovestadt, V.; Sharma, T.; Habel, A.; Burford, A.; Jones, C.; et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017, 134, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.R.; Aibaidula, A.; Wang, W.W.; Chan, A.K.; Shi, Z.F.; Zhang, Z.; Chan, D.T.M.; Poon, W.S.; Liu, X.Z.; Li, W.C.; et al. Pediatric low-grade gliomas can be molecularly stratified for risk. Acta Neuropathol. 2018, 136, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Aibaidula, A.; Chan, A.K.; Shi, Z.; Li, Y.; Zhang, R.; Yang, R.; Li, K.K.; Chung, N.Y.; Yao, Y.; Zhou, L.; et al. Adult IDH wild-type lower-grade gliomas should be further stratified. Neuro Oncol. 2017, 19, 1327–1337. [Google Scholar] [CrossRef]
- Horbinski, C.; Hamilton, R.L.; Nikiforov, Y.; Pollack, I.F. Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathol. 2010, 119, 641–649. [Google Scholar] [CrossRef]
- Liu, E.M.; Shi, Z.F.; Li, K.K.; Malta, T.M.; Chung, N.Y.; Chen, H.; Chan, J.Y.; Poon, M.F.; Kwan, J.S.; Chan, D.T.; et al. Molecular landscape of IDH-wild type, pTERT-wild type adult glioblastomas. Brain Pathol. 2022, 32, e13107. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Wong, Q.H.; Li, K.K.; Wang, W.W.; Malta, T.M.; Noushmehr, H.; Grabovska, Y.; Jones, C.; Chan, A.K.; Kwan, J.S.; Huang, Q.J.; et al. Molecular landscape of IDH-mutant primary astrocytoma Grade IV/glioblastomas. Mod. Pathol. 2021, 34, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Shirahata, M.; Ono, T.; Stichel, D.; Schrimpf, D.; Reuss, D.E.; Sahm, F.; Koelsche, C.; Wefers, A.; Reinhardt, A.; Huang, K.; et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018, 136, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.K.; Yao, Y.; Zhang, Z.; Chung, N.Y.; Liu, J.S.; Li, K.K.; Shi, Z.; Chan, D.T.; Poon, W.S.; Zhou, L.; et al. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Mod. Pathol. 2015, 28, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.K.; Li, K.K.; Chung, N.Y.; Chan, J.Y.; Poon, M.F.; Wong, Q.H.; Kwan, J.S.; Poon, W.S.; Chen, H.; Chan, D.T.; et al. Molecular landscapes of longitudinal NF2/22q and non-NF2/22q meningiomas show different life histories. Brain Pathol. 2023, 33, e13120. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.C.; Li, K.K.; Wang, W.W.; Liu, A.P.; Huang, Q.J.; Chan, A.K.; Poon, M.F.; Chung, N.Y.; Wong, Q.H.; Chen, H.; et al. Clinical and mutational profiles of adult medulloblastoma groups. Acta Neuropathol. Commun. 2020, 8, 191. [Google Scholar] [CrossRef]
- Northcott, P.A.; Shih, D.J.; Remke, M.; Cho, Y.J.; Kool, M.; Hawkins, C.; Eberhart, C.G.; Dubuc, A.; Guettouche, T.; Cardentey, Y.; et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012, 123, 615–626. [Google Scholar] [CrossRef]
- Takami, H.; Yoshida, A.; Fukushima, S.; Arita, H.; Matsushita, Y.; Nakamura, T.; Ohno, M.; Miyakita, Y.; Shibui, S.; Narita, Y.; et al. Revisiting TP53 Mutations and Immunohistochemistry—A Comparative Study in 157 Diffuse Gliomas. Brain Pathol. 2015, 25, 256–265. [Google Scholar] [CrossRef]
- Stichel, D.; Ebrahimi, A.; Reuss, D.; Schrimpf, D.; Ono, T.; Shirahata, M.; Reifenberger, G.; Weller, M.; Hänggi, D.; Wick, W.; et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018, 136, 793–803. [Google Scholar] [CrossRef]
- Orr, B.A. Pathology, diagnostics, and classification of medulloblastoma. Brain Pathol. 2020, 30, 664–678. [Google Scholar] [CrossRef]
- Burger, P.C. (Ed.) Medulloblastomas. In Diagnostic Pathology: Neuropathology; Section 1; Amirsys, Inc.: Salt Lake City, UT, USA; pp. 220–233.
- Yachnis, A.T.; Perry, A. Embryonal (Primitive) Neoplasms of the Central Nervous System. In Practical Surgical Pathology; Perry, A., Brat, D.J., Eds.; Churchill Livingstone: Philadelphia, PA, USA, 2010; pp. 165–184. [Google Scholar]
- Korshunov, A.; Chavez, L.; Northcott, P.A.; Sharma, T.; Ryzhova, M.; Jones, D.T.W.; von Deimling, A.; Pfister, S.M.; Kool, M. DNA-methylation profiling discloses significant advantages over NanoString method for molecular classification of medulloblastoma. Acta Neuropathol. 2017, 134, 965–967. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, F.; Donehower, L.A.; Scheurer, M.E.; Creighton, C.J. A pediatric brain tumor atlas of genes deregulated by somatic genomic rearrangement. Nat. Commun. 2021, 12, 937. [Google Scholar] [CrossRef] [PubMed]
- DeSisto, J.; Lucas, J.T., Jr.; Xu, K.; Donson, A.; Lin, T.; Sanford, B.; Wu, G.; Tran, Q.T.; Hedges, D.; Hsu, C.Y.; et al. Comprehensive molecular characterization of pediatric radiation-induced high-grade glioma. Nat. Commun. 2021, 12, 5531. [Google Scholar] [CrossRef] [PubMed]
- Ellison, D.W.; Figarella-Branger, D.; Pfister, S.M.; von Deimling, A.; Wesseling, P. Embryonal Tumours: WHO Classification of Tumours, 5th ed.; Central Nervous System Tumours, the WHO Classification of Tumours Editorial Board, Chapter 4; IRAC: Lyon, France, 2021; pp. 200–219. [Google Scholar]
- Tanboon, J.; Williams, E.A.; Louis, D.N. The Diagnostic Use of Immunohistochemical Surrogates for Signature Molecular Genetic Alterations in Gliomas. J. Neuropathol. Exp. Neurol. 2016, 75, 4–18. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Z.-F.; Li, K.K.-W.; Liu, A.P.-Y.; Chung, N.Y.-F.; Chow, C.; Chen, H.; Kan, N.-C.A.; Zhu, X.-L.; Chan, D.T.-M.; Mao, Y.; et al. Rare Pediatric Cerebellar High-Grade Gliomas Mimic Medulloblastomas Histologically and Transcriptomically and Show p53 Mutations. Cancers 2024, 16, 232. https://doi.org/10.3390/cancers16010232
Shi Z-F, Li KK-W, Liu AP-Y, Chung NY-F, Chow C, Chen H, Kan N-CA, Zhu X-L, Chan DT-M, Mao Y, et al. Rare Pediatric Cerebellar High-Grade Gliomas Mimic Medulloblastomas Histologically and Transcriptomically and Show p53 Mutations. Cancers. 2024; 16(1):232. https://doi.org/10.3390/cancers16010232
Chicago/Turabian StyleShi, Zhi-Feng, Kay Ka-Wai Li, Anthony Pak-Yin Liu, Nellie Yuk-Fei Chung, Chit Chow, Hong Chen, Nim-Chi Amanda Kan, Xian-Lun Zhu, Danny Tat-Ming Chan, Ying Mao, and et al. 2024. "Rare Pediatric Cerebellar High-Grade Gliomas Mimic Medulloblastomas Histologically and Transcriptomically and Show p53 Mutations" Cancers 16, no. 1: 232. https://doi.org/10.3390/cancers16010232
APA StyleShi, Z.-F., Li, K. K.-W., Liu, A. P.-Y., Chung, N. Y.-F., Chow, C., Chen, H., Kan, N.-C. A., Zhu, X.-L., Chan, D. T.-M., Mao, Y., & Ng, H.-K. (2024). Rare Pediatric Cerebellar High-Grade Gliomas Mimic Medulloblastomas Histologically and Transcriptomically and Show p53 Mutations. Cancers, 16(1), 232. https://doi.org/10.3390/cancers16010232





