AI-Powered Segmentation of Invasive Carcinoma Regions in Breast Cancer Immunohistochemical Whole-Slide Images
Abstract
:Simple Summary
Abstract
1. Introduction
2. Method
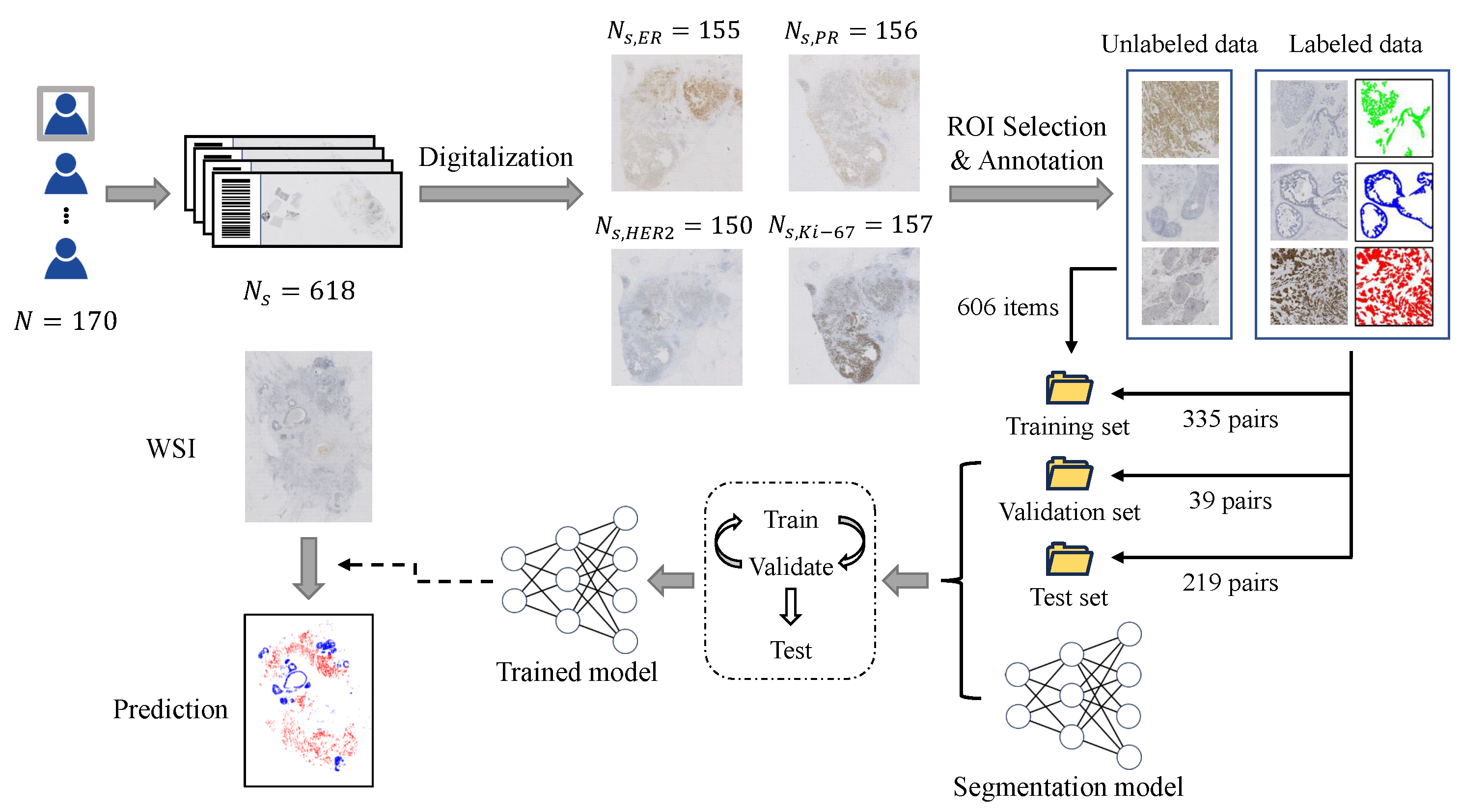
2.1. Dataset Construction
2.2. Training Framework
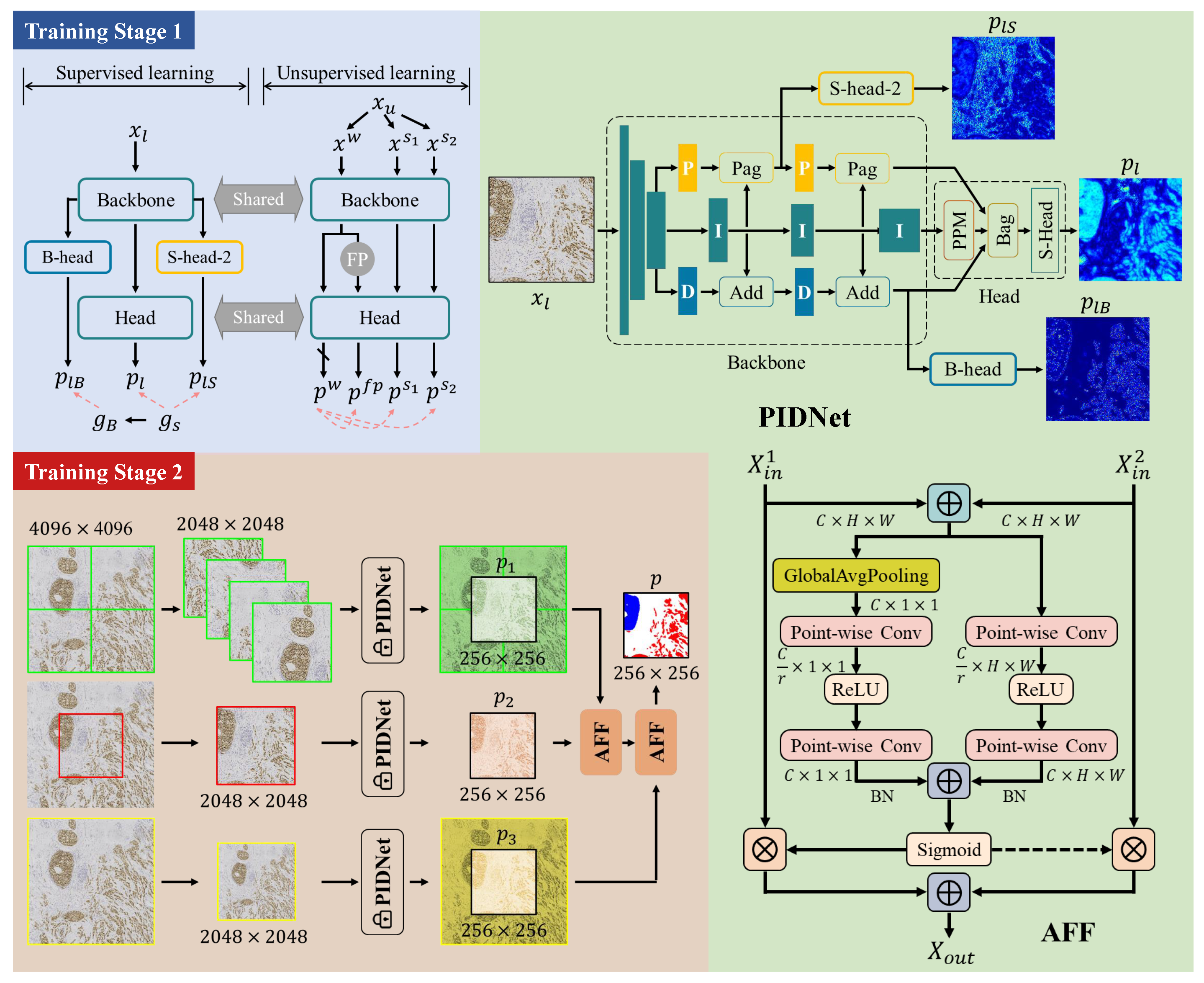
2.2.1. Initial Segmentation Model
2.2.2. Training Stage 1: Semi-Supervised Learning
2.2.3. Training Stage 2: Training of Multi-Scale Fusion Modules
- Cropped in the four corners according to the size of ;
- Cropped in the center according to the size of ;
- Scaled to the size of .
3. Results
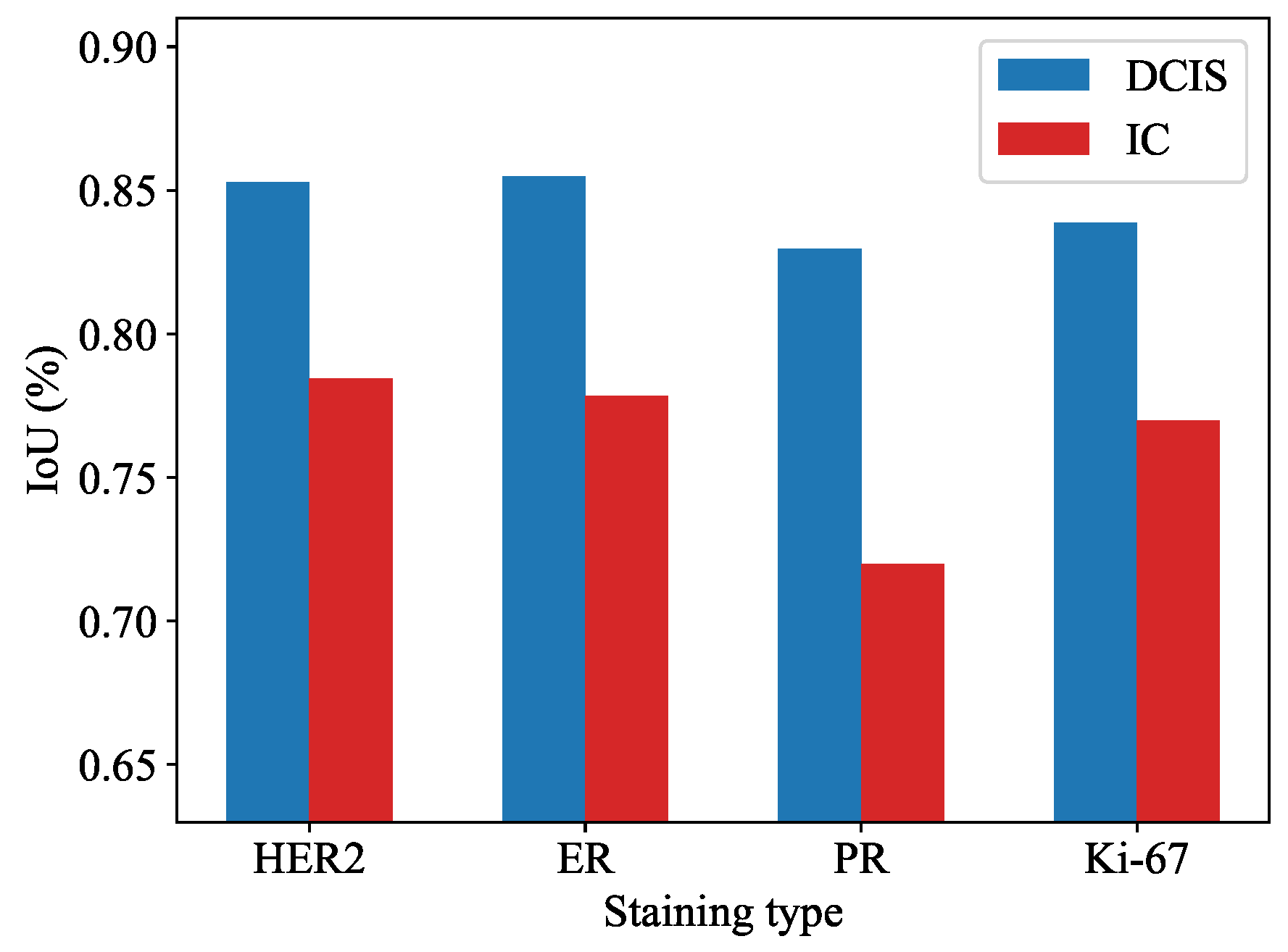
3.1. Quantitatively Experiments for Segmentation Task
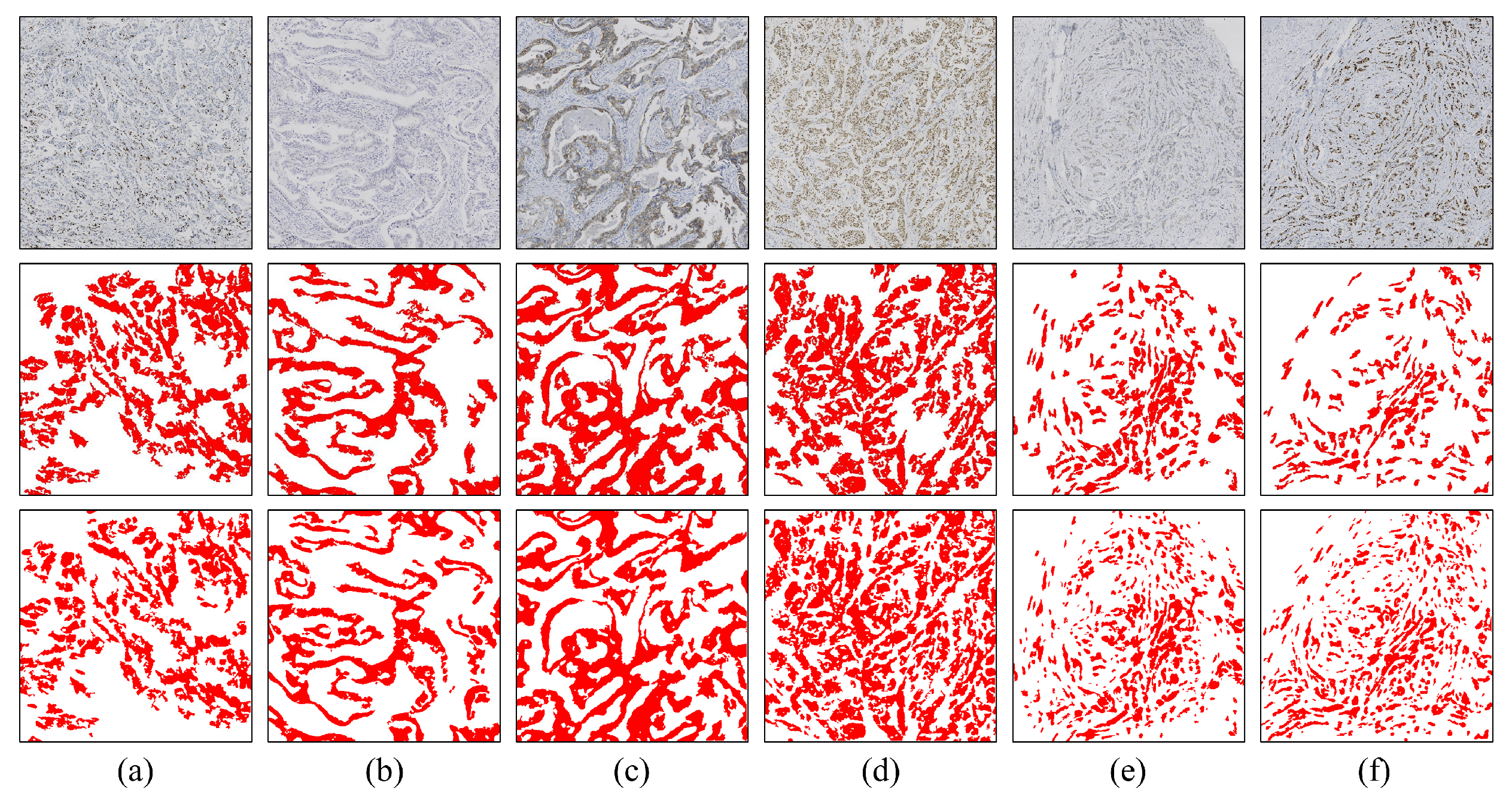
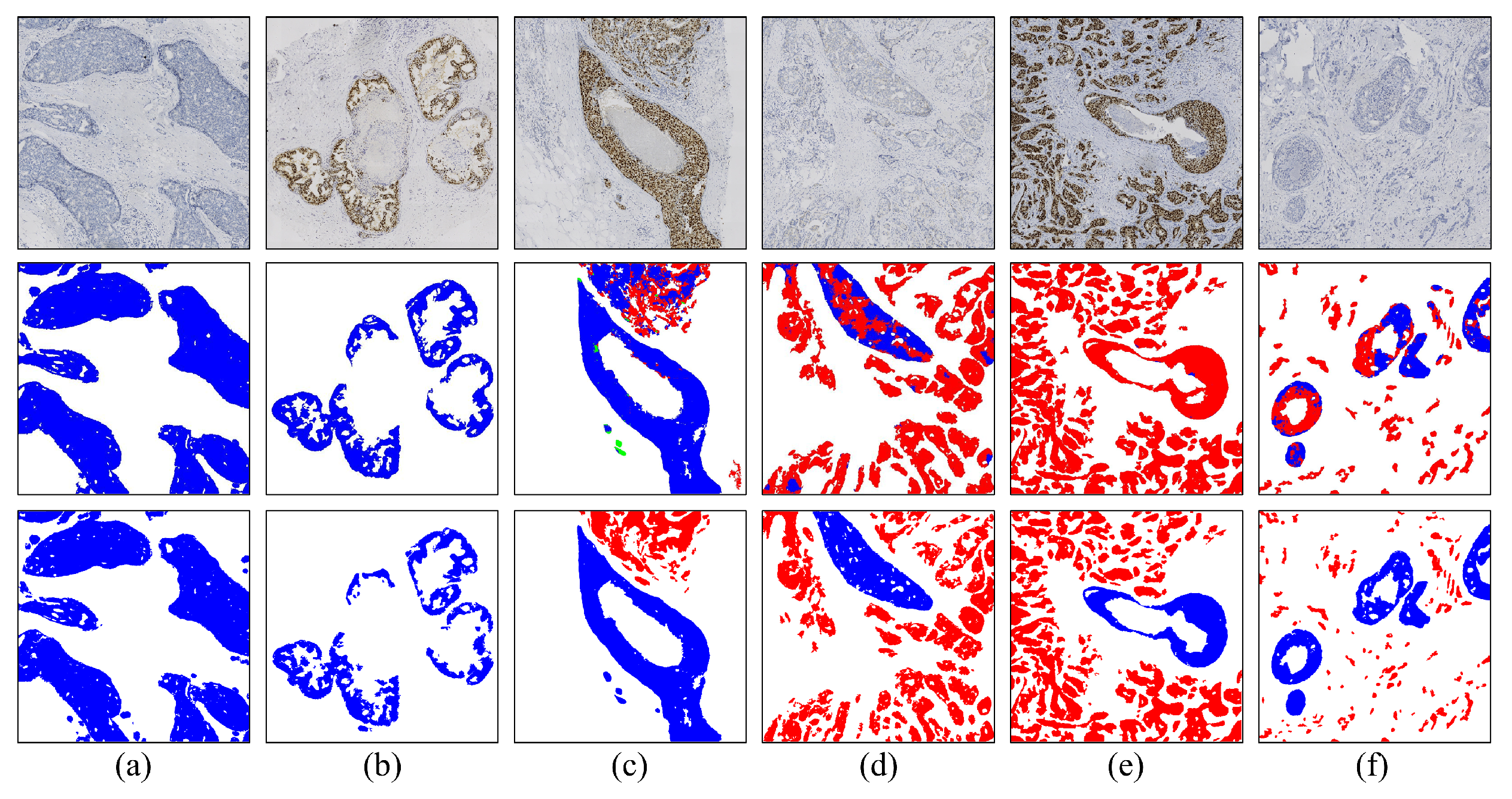
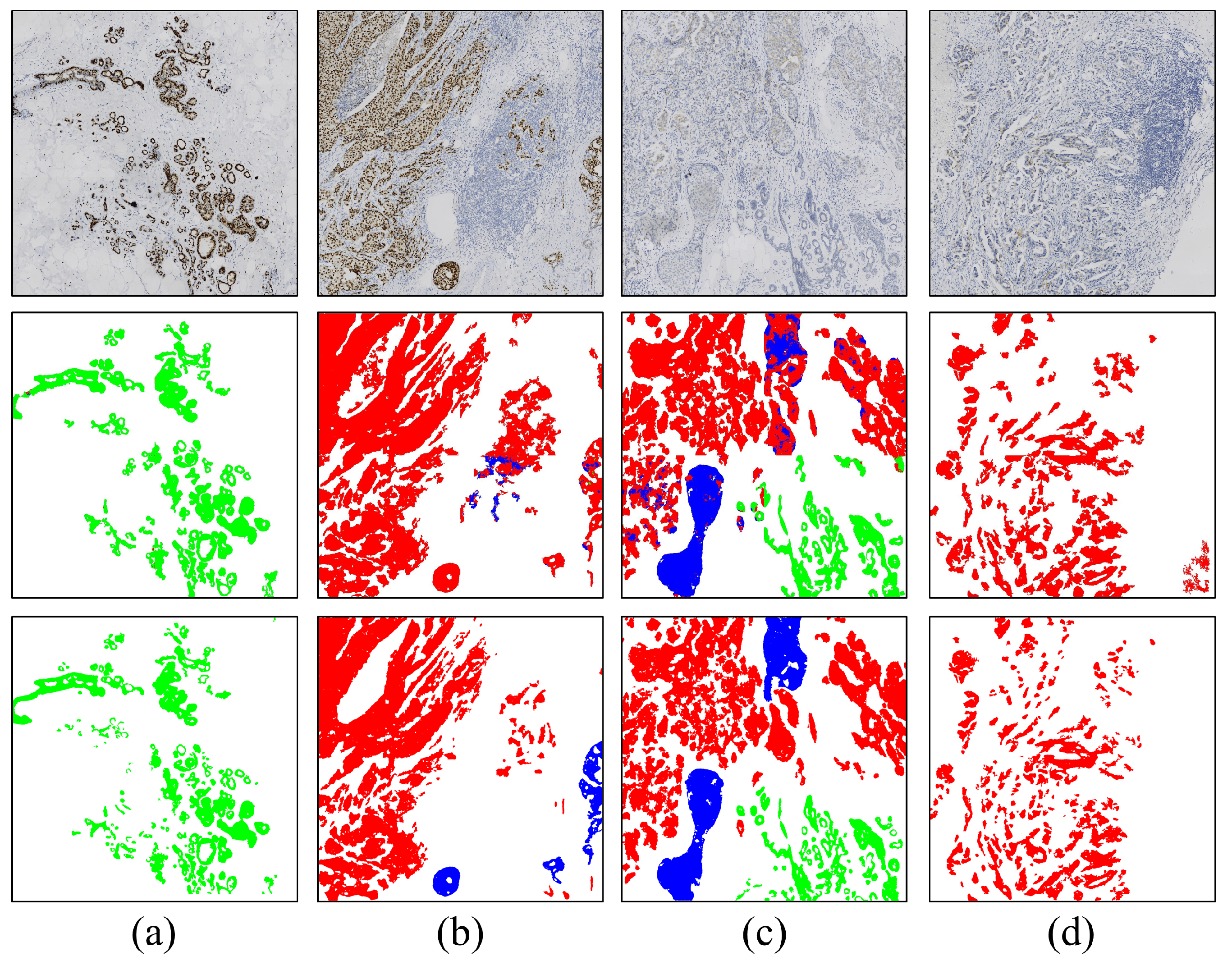
3.2. Visual Analysis
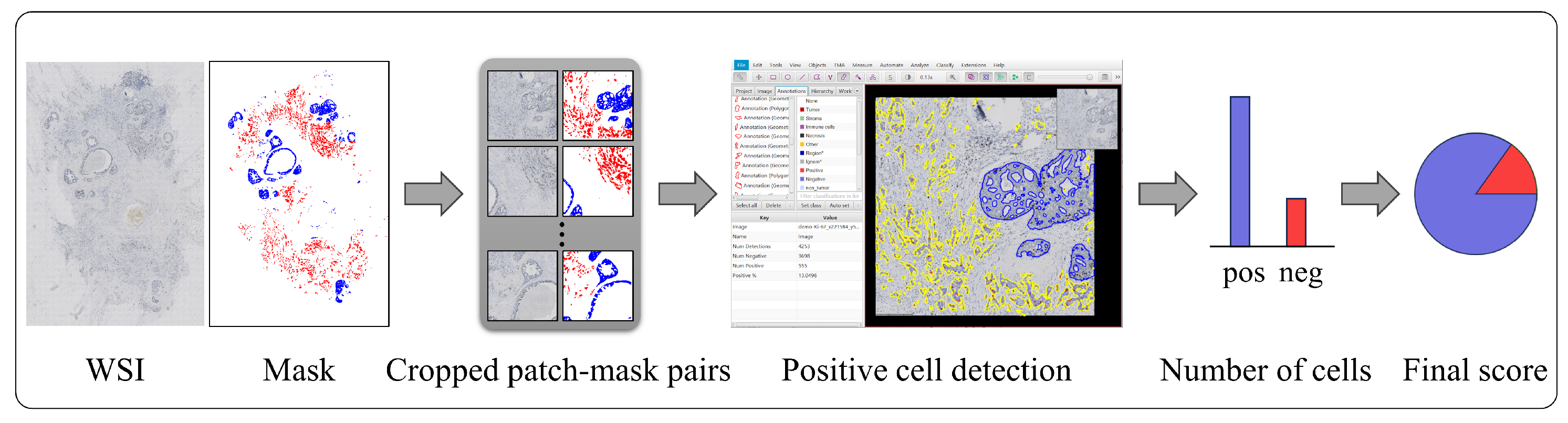
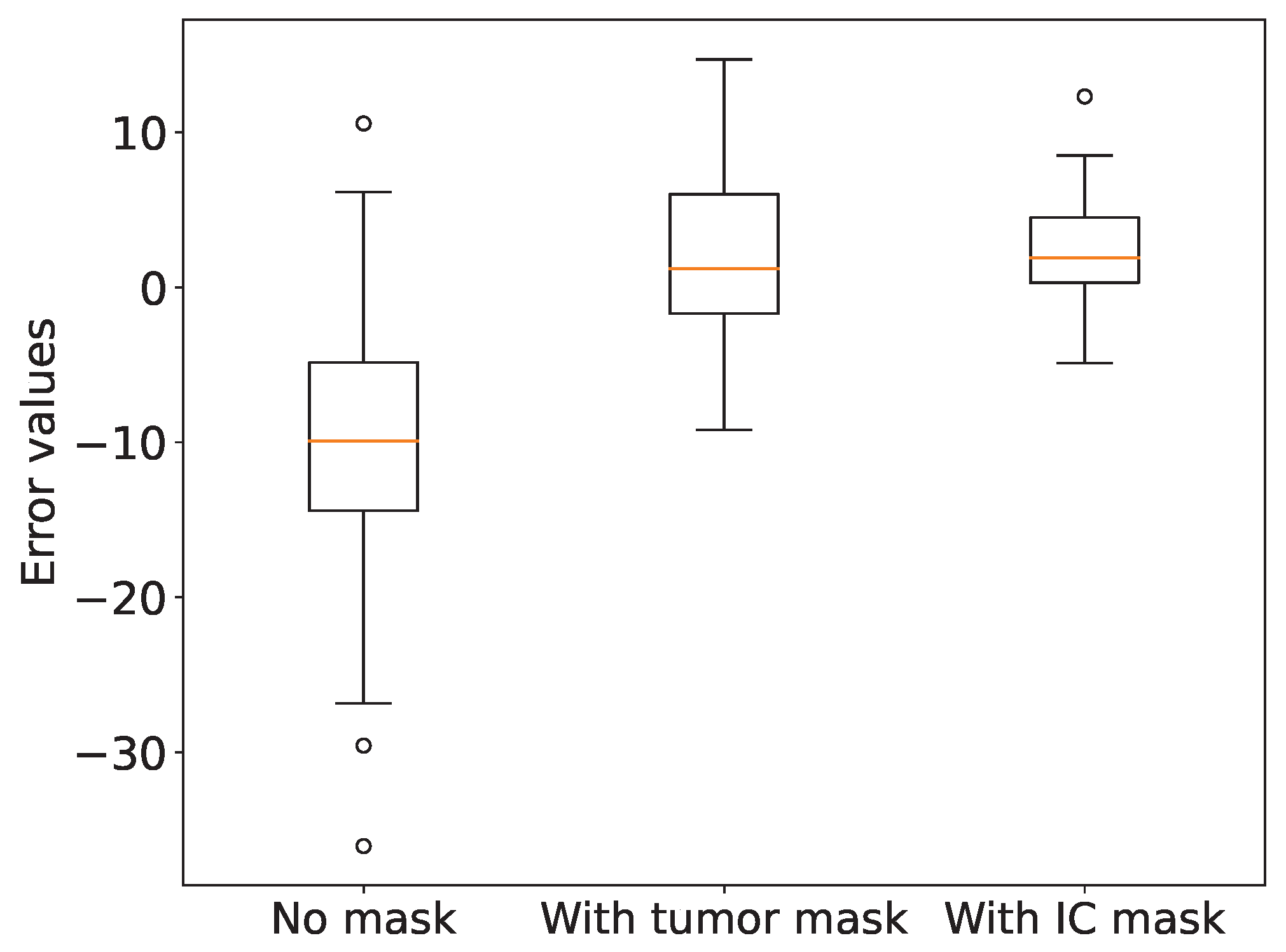
3.3. Role of Invasive Carcinoma Mask in Ki-67 Quantification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADB | auxiliary derivative branch |
| ADH | atypical ductal hyperplasia |
| AFF | attentional feature fusion |
| DCIS | ductal carcinoma in situ |
| ER | estrogen receptor |
| HER2 | human epidermal growth factor receptor 2 |
| IBC-NST | invasive breast carcinoma of no special type |
| IC | invasive carcinoma |
| IHC | immunohistochemistry |
| PR | progesterone receptor |
| ROI | region of Interest |
| SGD | stochastic gradient descent |
| UDH | usual ductal hyperplasia |
| WSI | whole-slide image |
References
- Chhikara, B.S.; Parang, K. Global Cancer Statistics 2022: The trends projection analysis. Chem. Biol. Lett. 2023, 10, 451. [Google Scholar]
- WHO. WHO Classification of Tumors–Breast Tumors, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2019. [Google Scholar]
- Zhang, L.; Huang, Y.; Feng, Z.; Wang, X.; Li, H.; Song, F.; Liu, L.; Li, J.; Zheng, H.; Wang, P.; et al. Comparison of breast cancer risk factors among molecular subtypes: A case-only study. Cancer Med. 2019, 8, 1882–1892. [Google Scholar] [CrossRef]
- Zaha, D.C. Significance of immunohistochemistry in breast cancer. World J. Clin. Oncol. 2014, 5, 382. [Google Scholar] [CrossRef] [PubMed]
- Dabbs, D.J. Diagnostic Immunohistochemistry E-Book: Theranostic and Genomic Applications; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Mathew, T.; Niyas, S.; Johnpaul, C.; Kini, J.R.; Rajan, J. A novel deep classifier framework for automated molecular subtyping of breast carcinoma using immunohistochemistry image analysis. Biomed. Signal Process. Control. 2022, 76, 103657. [Google Scholar] [CrossRef]
- López, C.; Lejeune, M.; Salvadó, M.T.; Escrivà, P.; Bosch, R.; Pons, L.E.; Álvaro, T.; Roig, J.; Cugat, X.; Baucells, J.; et al. Automated quantification of nuclear immunohistochemical markers with different complexity. Histochem. Cell Biol. 2008, 129, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Qaiser, T.; Mukherjee, A.; Reddy Pb, C.; Munugoti, S.D.; Tallam, V.; Pitkäaho, T.; Lehtimäki, T.; Naughton, T.; Berseth, M.; Pedraza, A.; et al. Her 2 challenge contest: A detailed assessment of automated her 2 scoring algorithms in whole slide images of breast cancer tissues. Histopathology 2018, 72, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Gavrielides, M.A.; Gallas, B.D.; Lenz, P.; Badano, A.; Hewitt, S.M. Observer variability in the interpretation of HER2/neu immunohistochemical expression with unaided and computer-aided digital microscopy. Arch. Pathol. Lab. Med. 2011, 135, 233–242. [Google Scholar] [CrossRef]
- Chung, Y.R.; Jang, M.H.; Park, S.Y.; Gong, G.; Jung, W.H. Interobserver variability of Ki-67 measurement in breast cancer. J. Pathol. Transl. Med. 2016, 50, 129–137. [Google Scholar] [CrossRef]
- Leung, S.C.; Nielsen, T.O.; Zabaglo, L.A.; Arun, I.; Badve, S.S.; Bane, A.L.; Bartlett, J.M.; Borgquist, S.; Chang, M.C.; Dodson, A.; et al. Analytical validation of a standardised scoring protocol for Ki67 immunohistochemistry on breast cancer excision whole sections: An international multicentre collaboration. Histopathology 2019, 75, 225–235. [Google Scholar] [CrossRef]
- Cai, L.; Yan, K.; Bu, H.; Yue, M.; Dong, P.; Wang, X.; Li, L.; Tian, K.; Shen, H.; Zhang, J.; et al. Improving Ki67 assessment concordance by the use of an artificial intelligence-empowered microscope: A multi-institutional ring study. Histopathology 2021, 79, 544–555. [Google Scholar] [CrossRef]
- Fisher, N.C.; Loughrey, M.B.; Coleman, H.G.; Gelbard, M.D.; Bankhead, P.; Dunne, P.D. Development of a semi-automated method for tumour budding assessment in colorectal cancer and comparison with manual methods. Histopathology 2022, 80, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Hondelink, L.M.; Hüyük, M.; Postmus, P.E.; Smit, V.T.; Blom, S.; von der Thüsen, J.H.; Cohen, D. Development and validation of a supervised deep learning algorithm for automated whole-slide programmed death-ligand 1 tumour proportion score assessment in non-small cell lung cancer. Histopathology 2022, 80, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Ba, W.; Wang, S.; Shang, M.; Zhang, Z.; Wu, H.; Yu, C.; Xing, R.; Wang, W.; Wang, L.; Liu, C.; et al. Assessment of deep learning assistance for the pathological diagnosis of gastric cancer. Mod. Pathol. 2022, 35, 1262–1268. [Google Scholar] [CrossRef]
- Song, Z.; Zou, S.; Zhou, W.; Huang, Y.; Shao, L.; Yuan, J.; Gou, X.; Jin, W.; Wang, Z.; Chen, X.; et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat. Commun. 2020, 11, 4294. [Google Scholar] [CrossRef] [PubMed]
- Van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.; Zhao, Z.; Chen, X.F.; Sauer, J.; Saraf, S.A.; Jialdasani, R.; Taghipour, K.; Sathe, A.; Khor, L.Y.; Lim, K.H.; et al. A promising deep learning-assistive algorithm for histopathological screening of colorectal cancer. Sci. Rep. 2022, 12, 2222. [Google Scholar] [CrossRef] [PubMed]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef] [PubMed]
- Geread, R.S.; Sivanandarajah, A.; Brouwer, E.R.; Wood, G.A.; Androutsos, D.; Faragalla, H.; Khademi, A. Pinet—An automated proliferation index calculator framework for Ki67 breast cancer images. Cancers 2020, 13, 11. [Google Scholar] [CrossRef]
- Feng, M.; Deng, Y.; Yang, L.; Jing, Q.; Zhang, Z.; Xu, L.; Wei, X.; Zhou, Y.; Wu, D.; Xiang, F.; et al. Automated quantitative analysis of Ki-67 staining and HE images recognition and registration based on whole tissue sections in breast carcinoma. Diagn. Pathol. 2020, 15, 65. [Google Scholar] [CrossRef]
- Huang, Z.; Ding, Y.; Song, G.; Wang, L.; Geng, R.; He, H.; Du, S.; Liu, X.; Tian, Y.; Liang, Y.; et al. Bcdata: A large-scale dataset and benchmark for cell detection and counting. In Proceedings of the Medical Image Computing and Computer Assisted Intervention 23rd International Conference (MICCAI 2020), Lima, Peru, 4–8 October 2020; Proceedings, Part V 23. Springer: Berlin/Heidelberg, Germany, 2020; pp. 289–298. [Google Scholar]
- Negahbani, F.; Sabzi, R.; Pakniyat Jahromi, B.; Firouzabadi, D.; Movahedi, F.; Kohandel Shirazi, M.; Majidi, S.; Dehghanian, A. PathoNet introduced as a deep neural network backend for evaluation of Ki-67 and tumor-infiltrating lymphocytes in breast cancer. Sci. Rep. 2021, 11, 8489. [Google Scholar] [CrossRef]
- Valkonen, M.; Isola, J.; Ylinen, O.; Muhonen, V.; Saxlin, A.; Tolonen, T.; Nykter, M.; Ruusuvuori, P. Cytokeratin-supervised deep learning for automatic recognition of epithelial cells in breast cancers stained for ER, PR, and Ki-67. IEEE Trans. Med. Imaging 2019, 39, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Qaiser, T.; Rajpoot, N.M. Learning where to see: A novel attention model for automated immunohistochemical scoring. IEEE Trans. Med. Imaging 2019, 38, 2620–2631. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Hou, W.; Wu, K.; Bai, Y.; Long, M.; Diao, X.; Jia, L.; Niu, D.; Li, X. Using Whole Slide Gray Value Map to Predict HER2 Expression and FISH Status in Breast Cancer. Cancers 2022, 14, 6233. [Google Scholar] [CrossRef] [PubMed]
- Priego-Torres, B.M.; Lobato-Delgado, B.; Atienza-Cuevas, L.; Sanchez-Morillo, D. Deep learning-based instance segmentation for the precise automated quantification of digital breast cancer immunohistochemistry images. Expert Syst. Appl. 2022, 193, 116471. [Google Scholar] [CrossRef]
- Huang, J.; Mei, L.; Long, M.; Liu, Y.; Sun, W.; Li, X.; Shen, H.; Zhou, F.; Ruan, X.; Wang, D.; et al. Bm-net: Cnn-based mobilenet-v3 and bilinear structure for breast cancer detection in whole slide images. Bioengineering 2022, 9, 261. [Google Scholar] [CrossRef]
- Van Rijthoven, M.; Balkenhol, M.; Siliņa, K.; Van Der Laak, J.; Ciompi, F. HookNet: Multi-resolution convolutional neural networks for semantic segmentation in histopathology whole-slide images. Med. Image Anal. 2021, 68, 101890. [Google Scholar] [CrossRef]
- Ni, H.; Liu, H.; Wang, K.; Wang, X.; Zhou, X.; Qian, Y. WSI-Net: Branch-based and hierarchy-aware network for segmentation and classification of breast histopathological whole-slide images. In Proceedings of the Machine Learning in Medical Imaging: 10th International Workshop, Held in Conjunction with MICCAI 2019 (MLMI 2019), Shenzhen, China, 13 October 2019; Proceedings 10. Springer: Berlin/Heidelberg, Germany, 2019; pp. 36–44. [Google Scholar]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Xu, J.; Xiong, Z.; Bhattacharyya, S.P. PIDNet: A Real-Time Semantic Segmentation Network Inspired by PID Controllers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 19529–19539. [Google Scholar]
- Takikawa, T.; Acuna, D.; Jampani, V.; Fidler, S. Gated-scnn: Gated shape cnns for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 5229–5238. [Google Scholar]
- Yang, L.; Qi, L.; Feng, L.; Zhang, W.; Shi, Y. Revisiting weak-to-strong consistency in semi-supervised semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7236–7246. [Google Scholar]
- Dai, Y.; Gieseke, F.; Oehmcke, S.; Wu, Y.; Barnard, K. Attentional feature fusion. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2021; pp. 3560–3569. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention: 18th International Conference (MICCAI 2015), Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Huynh, C.; Tran, A.T.; Luu, K.; Hoai, M. Progressive semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 16755–16764. [Google Scholar]
- Li, Q.; Yang, W.; Liu, W.; Yu, Y.; He, S. From contexts to locality: Ultra-high resolution image segmentation via locality-aware contextual correlation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 7252–7261. [Google Scholar]

| Method | IoU (DCIS, %) | IoU (IC, %) | Average |
|---|---|---|---|
| ResNet50 [36] + Unet [37] | |||
| Mit-b5 [38] + Unet | |||
| MagNet [39] | |||
| FCtL [40] | |||
| PIDNet | |||
| Proposed |
| Method | IoU (DCIS, %) | IoU (IC, %) | Average |
|---|---|---|---|
| PIDNet | |||
| PIDNet + Unimatch | |||
| PIDNet + AFF | |||
| Proposed |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhen, T.; Fu, Y.; Wang, Y.; He, Y.; Han, A.; Shi, H. AI-Powered Segmentation of Invasive Carcinoma Regions in Breast Cancer Immunohistochemical Whole-Slide Images. Cancers 2024, 16, 167. https://doi.org/10.3390/cancers16010167
Liu Y, Zhen T, Fu Y, Wang Y, He Y, Han A, Shi H. AI-Powered Segmentation of Invasive Carcinoma Regions in Breast Cancer Immunohistochemical Whole-Slide Images. Cancers. 2024; 16(1):167. https://doi.org/10.3390/cancers16010167
Chicago/Turabian StyleLiu, Yiqing, Tiantian Zhen, Yuqiu Fu, Yizhi Wang, Yonghong He, Anjia Han, and Huijuan Shi. 2024. "AI-Powered Segmentation of Invasive Carcinoma Regions in Breast Cancer Immunohistochemical Whole-Slide Images" Cancers 16, no. 1: 167. https://doi.org/10.3390/cancers16010167
APA StyleLiu, Y., Zhen, T., Fu, Y., Wang, Y., He, Y., Han, A., & Shi, H. (2024). AI-Powered Segmentation of Invasive Carcinoma Regions in Breast Cancer Immunohistochemical Whole-Slide Images. Cancers, 16(1), 167. https://doi.org/10.3390/cancers16010167







