Hypoxia Changes Energy Metabolism and Growth Rate in Non-Small Cell Lung Cancer Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cultivation
2.2. Quantification of Glucose Consumption and Lactate Production
2.3. Determination of Cell Proliferation
2.4. Analysis of Cell Cycle Distribution
2.5. Gene Expression Analysis
2.6. Statistical Analysis
3. Results
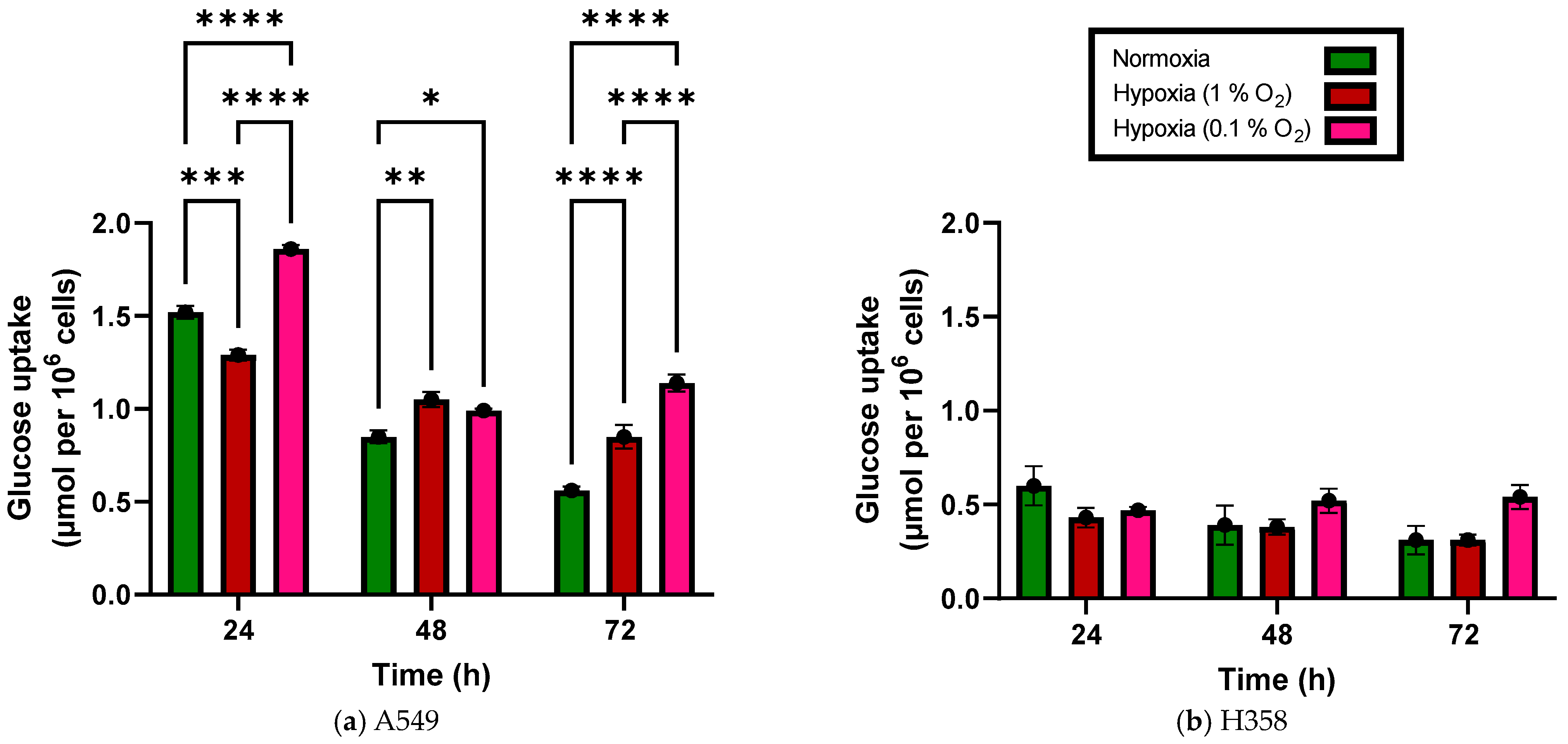
3.1. Glucose Uptake Increases under Hypoxia in NSCLC Cell Lines
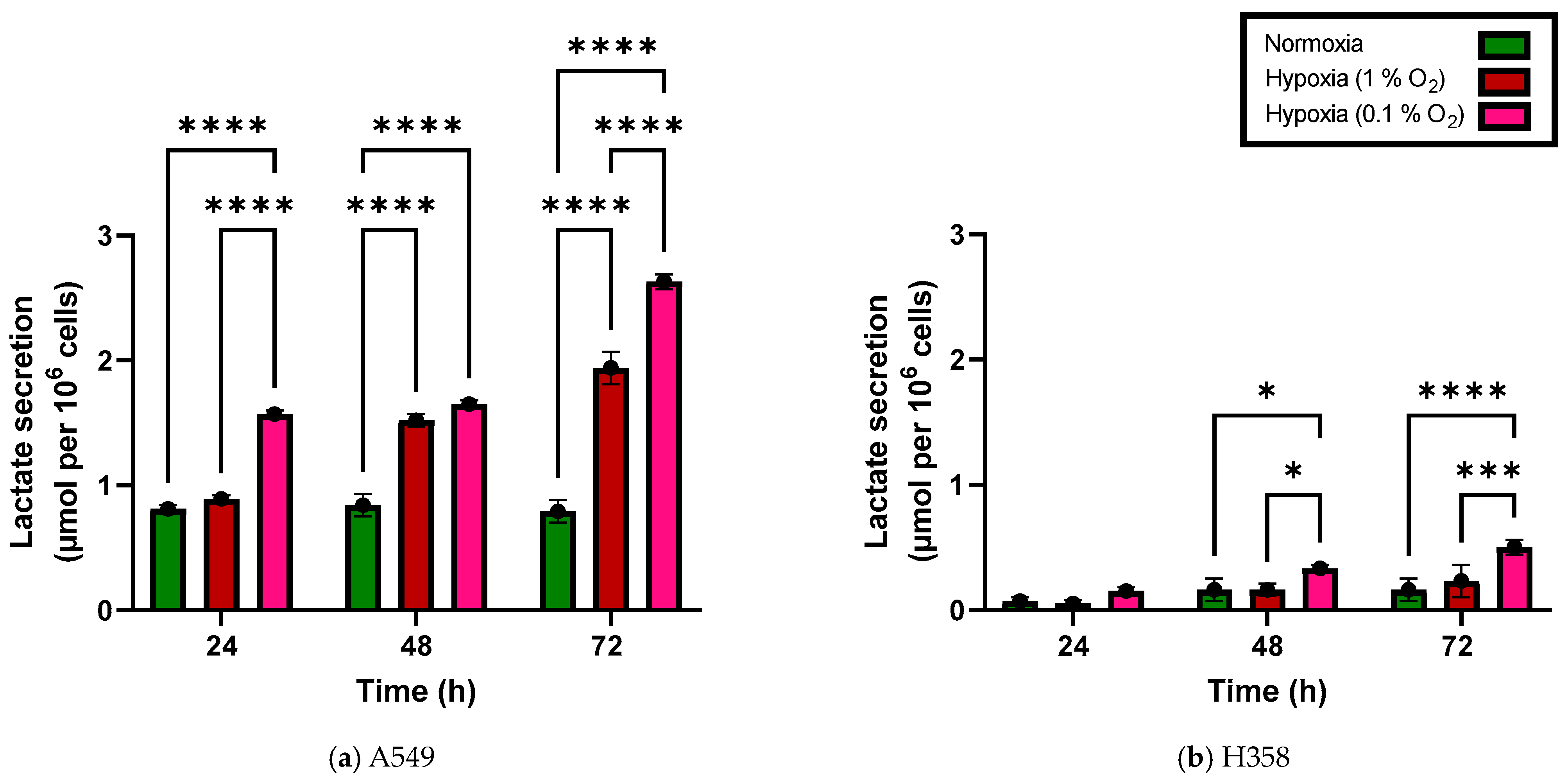
3.2. Lactate Secretion Increases under Hypoxia in NSCLC Cell Lines
3.3. NSCLC Cellular Proliferation Rate Decreases under Hypoxia
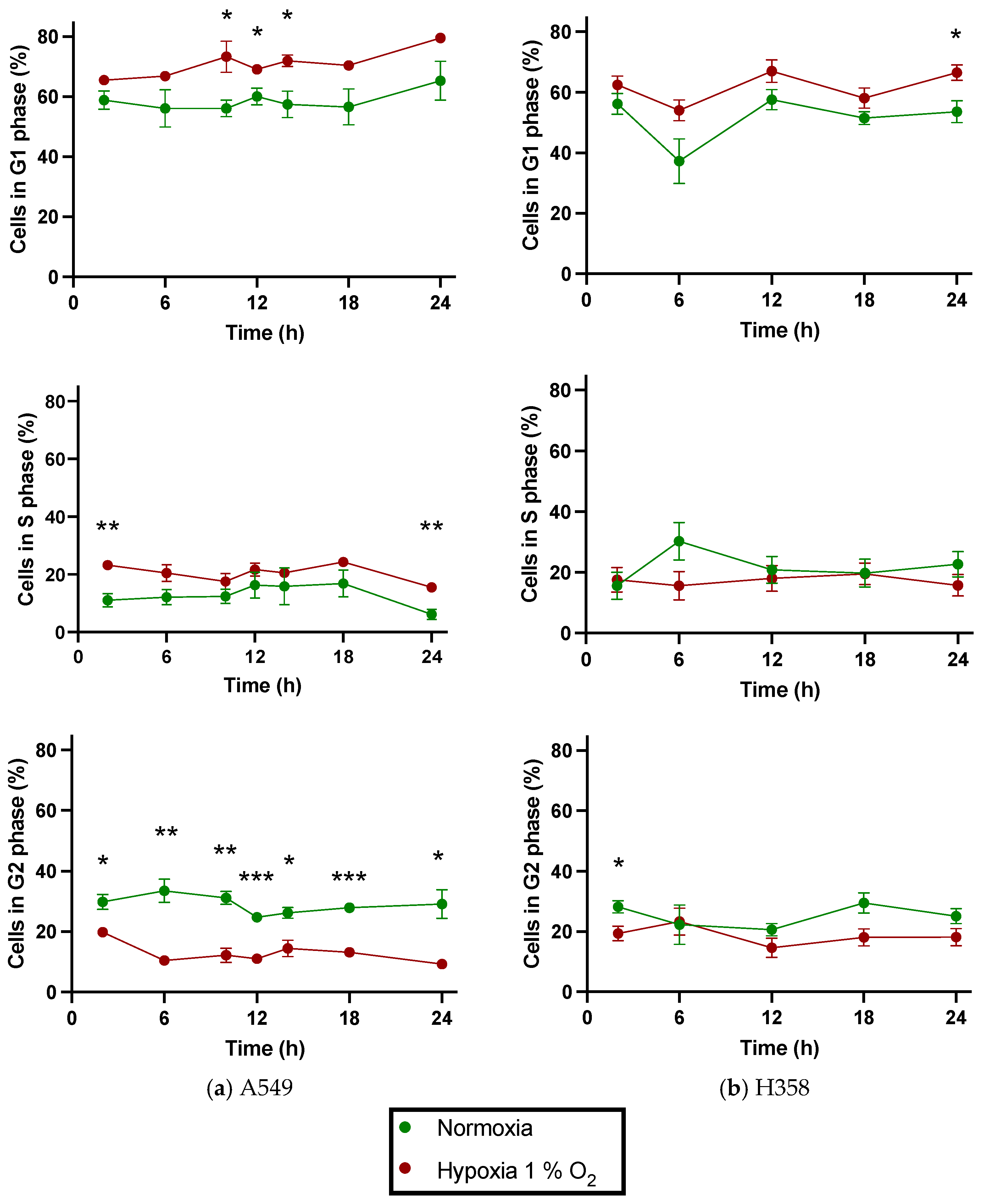
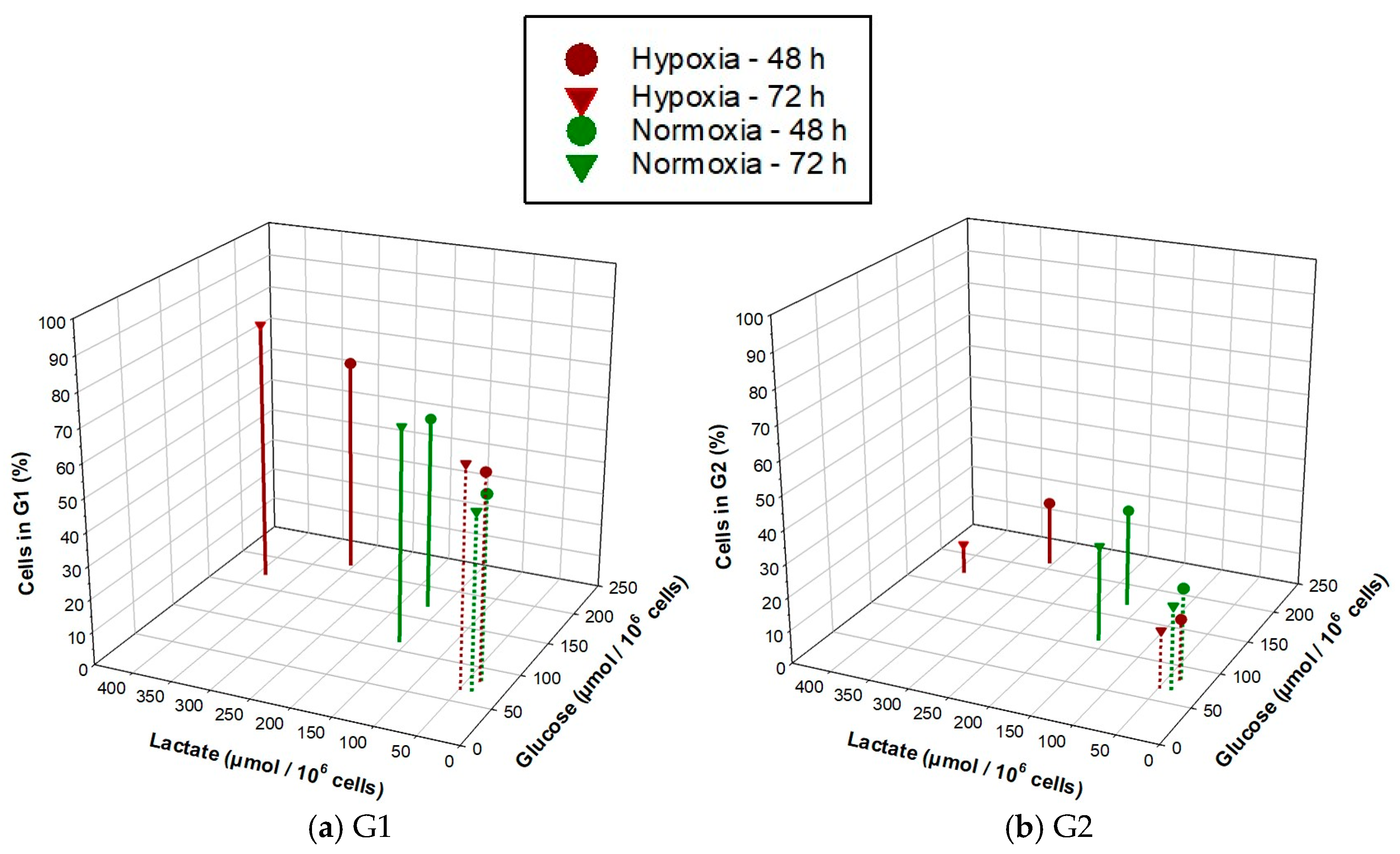
3.4. NSCLC Cell Lines Show Cell Cycle Phase Redistribution toward G1 under Hypoxia
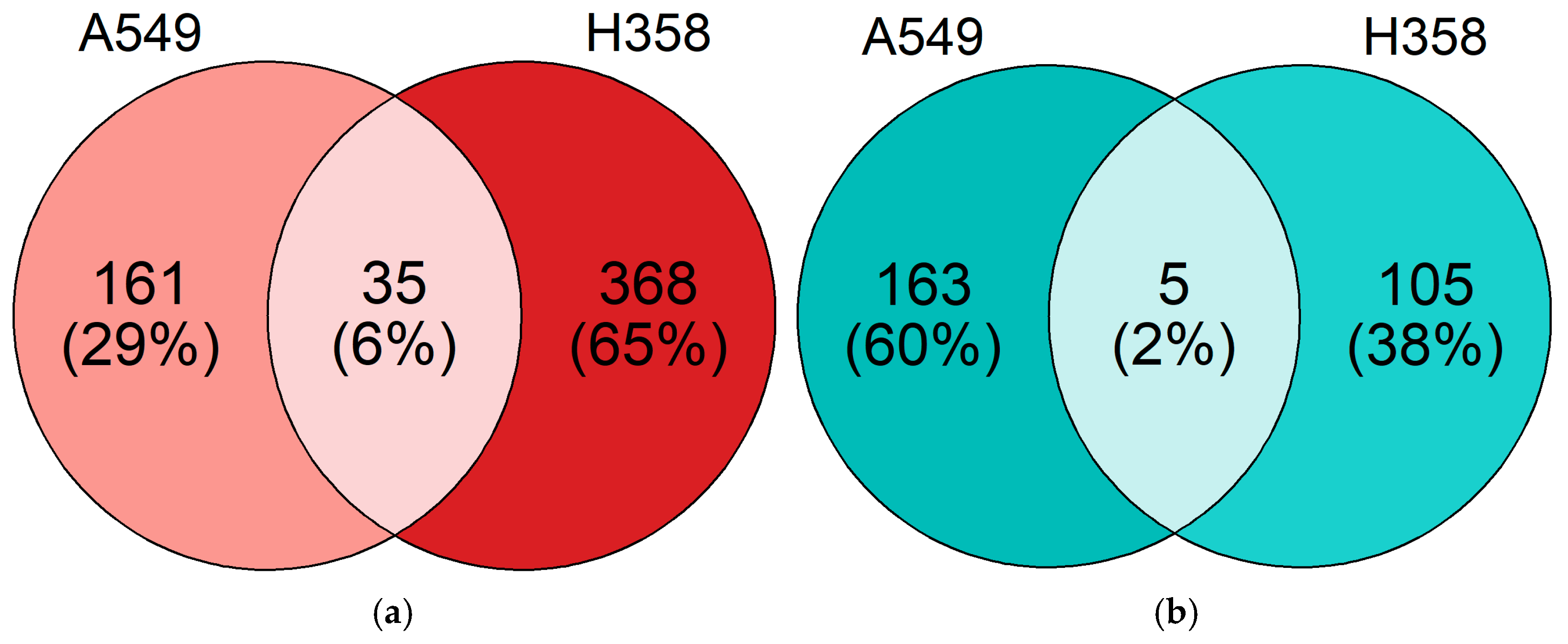
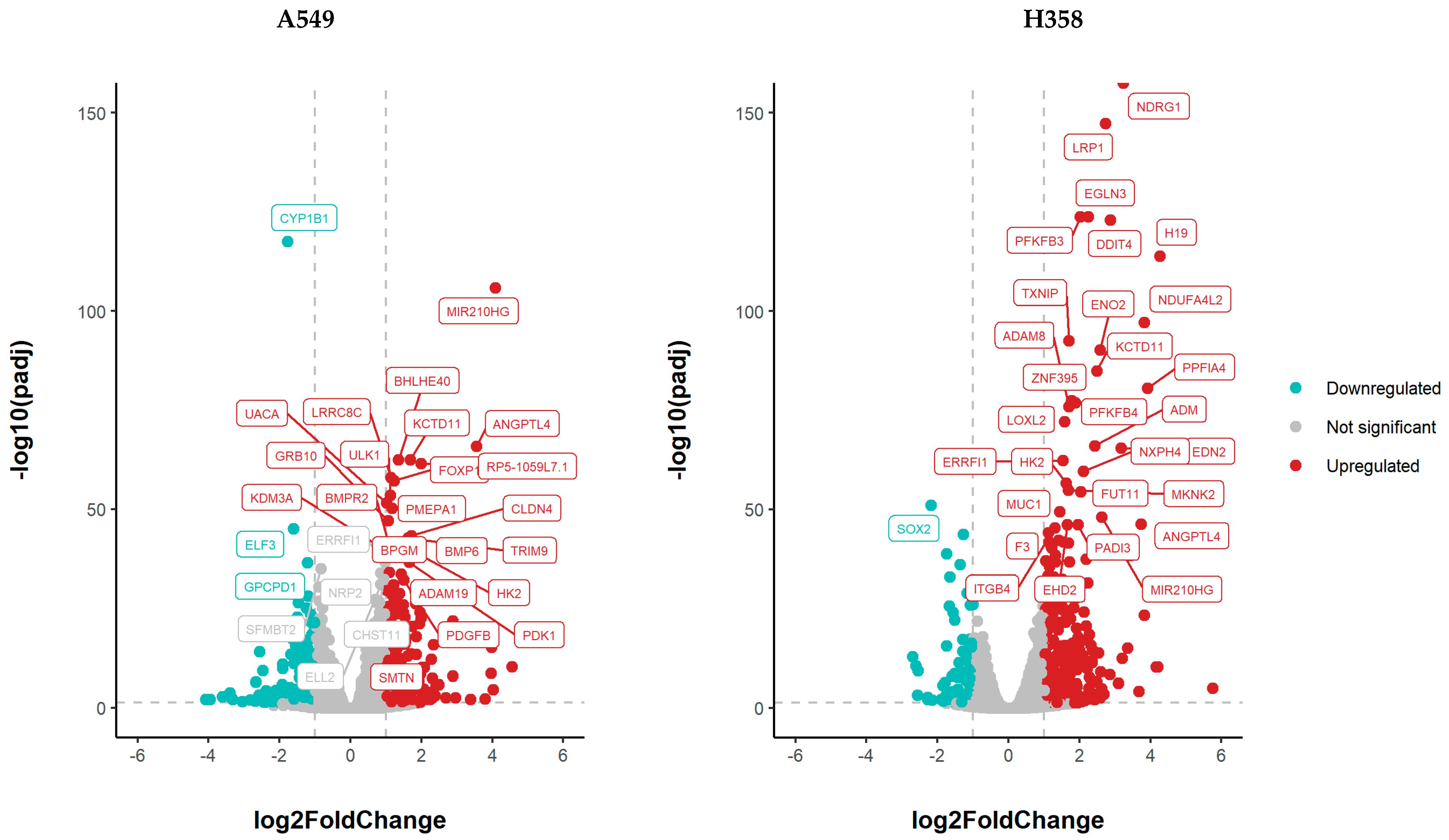
3.5. NSCLC Cell Lines Show Distinct Gene Expression Changes under Hypoxia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Cosse, J.-P.; Michiels, C. Tumour Hypoxia Affects the Responsiveness of Cancer Cells to Chemotherapy and Promotes Cancer Progression. Anti-Cancer Agents Med. Chem. Anti-Cancer Agents 2008, 8, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Span, P.N.; Bussink, J. Biology of Hypoxia. Semin. Nucl. Med. 2015, 45, 101–109. [Google Scholar] [CrossRef]
- Hockel, M.; Vaupel, P. Tumor Hypoxia: Definitions and Current Clinical, Biologic, and Molecular Aspects. JNCI J. Natl. Cancer Inst. 2001, 93, 266–276. [Google Scholar] [CrossRef]
- Eales, K.L.; Hollinshead, K.E.R.; Tennant, D.A. Hypoxia and Metabolic Adaptation of Cancer Cells. Oncogenesis 2016, 5, e190. [Google Scholar] [CrossRef]
- Kierans, S.J.; Taylor, C.T. Regulation of Glycolysis by the Hypoxia-Inducible Factor (HIF): Implications for Cellular Physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Riddle, S.R.; Ahmad, A.; Ahmad, S.; Deeb, S.S.; Malkki, M.; Schneider, B.K.; Allen, C.B.; White, C.W. Hypoxia Induces Hexokinase II Gene Expression in Human Lung Cell Line A549. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000, 278, L407–L416. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.P.; Ganapathy, V. Lactate/GPR81 Signaling and Proton Motive Force in Cancer: Role in Angiogenesis, Immune Escape, Nutrition, and Warburg Phenomenon. Pharmacol. Ther. 2020, 206, 107451. [Google Scholar] [CrossRef]
- Brown, T.P.; Bhattacharjee, P.; Ramachandran, S.; Sivaprakasam, S.; Ristic, B.; Sikder, M.O.F.; Ganapathy, V. The Lactate Receptor GPR81 Promotes Breast Cancer Growth via a Paracrine Mechanism Involving Antigen-Presenting Cells in the Tumor Microenvironment. Oncogene 2020, 39, 3292–3304. [Google Scholar] [CrossRef]
- Raychaudhuri, D.; Bhattacharya, R.; Sinha, B.P.; Liu, C.S.C.; Ghosh, A.R.; Rahaman, O.; Bandopadhyay, P.; Sarif, J.; D’Rozario, R.; Paul, S.; et al. Lactate Induces Pro-Tumor Reprogramming in Intratumoral Plasmacytoid Dendritic Cells. Front. Immunol. 2019, 10, 1878. [Google Scholar] [CrossRef]
- Wigerup, C.; Påhlman, S.; Bexell, D. Therapeutic Targeting of Hypoxia and Hypoxia-Inducible Factors in Cancer. Pharmacol. Ther. 2016, 164, 152–169. [Google Scholar] [CrossRef] [PubMed]
- Hubbi, M.E.; Semenza, G.L. Regulation of Cell Proliferation by Hypoxia-Inducible Factors. Am. J. Physiol. Cell Physiol. 2015, 309, C775–C782. [Google Scholar] [CrossRef] [PubMed]
- Lung Cancer—Non-Small Cell—Statistics. Available online: https://www.cancer.net/cancer-types/lung-cancer-non-small-cell/statistics (accessed on 23 November 2022).
- Salem, A.; Asselin, M.-C.; Reymen, B.; Jackson, A.; Lambin, P.; West, C.M.L.; O’Connor, J.P.B.; Faivre-Finn, C. Targeting Hypoxia to Improve Non–Small Cell Lung Cancer Outcome. JNCI J. Natl. Cancer Inst. 2018, 110, 14–30. [Google Scholar] [CrossRef] [PubMed]
- van Elmpt, W.; Zegers, K.; Reymen, B.; Even, A.J.G.; Dingemans, A.-M.C.; Öllers, M.; Wildberger, J.E.; Mottaghy, F.M.; Das, M.; Troost, E.G.C.; et al. Multiparametric Imaging of Patient and Tumour Heterogeneity in Non-Small-Cell Lung Cancer: Quantification of Tumour Hypoxia, Metabolism and Perfusion. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 240–248. [Google Scholar] [CrossRef]
- Schuurbiers, O.C.J.; Meijer, T.W.H.; Kaanders, J.H.A.M.; Looijen-Salamon, M.G.; de Geus-Oei, L.-F.; van der Drift, M.A.; van der Heijden, E.H.F.M.; Oyen, W.J.; Visser, E.P.; Span, P.N.; et al. Glucose Metabolism in NSCLC Is Histology-Specific and Diverges the Prognostic Potential of 18FDG-PET for Adenocarcinoma and Squamous Cell Carcinoma. J. Thorac. Oncol. 2014, 9, 1485–1493. [Google Scholar] [CrossRef]
- Hinz, T.K.; Kalkur, R.; Rabinovitch, J.; Hinkle, W.; Heasley, L.E. TP53 Null Mutations Identify Lung Cancer Cell Lines with Highest Sensitivity to the Nontaxane Microtubule Inhibitor Eribulin. Mol. Pharmacol. 2021, 100, 144–154. [Google Scholar] [CrossRef]
- Glucose-GloTM Assay Technical Manual. Available online: https://www.promega.de/resources/protocols/technical-manuals/101/glucose-glo-assay-protocol/ (accessed on 1 September 2022).
- Lactate Glo Assay Protocol. Available online: https://www.promega.de/en/resources/protocols/technical-manuals/101/lactate-glo-assay-protocol/ (accessed on 1 September 2022).
- Korzyńska, A.; Zychowicz, M. A Method of Estimation of the Cell Doubling Time on Basis of the Cell Culture Monitoring Data. Biocybern. Biomed. Eng. 2008, 28, 75–82. [Google Scholar]
- Dean, P.N.; Jett, J.H. Mathematical analysis of DNA distributions derived from flow microfluorometry. J. Cell Biol. 1974, 60, 523–527. [Google Scholar] [CrossRef]
- Schafer, Z.T.; Grassian, A.R.; Song, L.; Jiang, Z.; Gerhart-Hines, Z.; Irie, H.Y.; Gao, S.; Puigserver, P.; Brugge, J.S. Antioxidant and Oncogene Rescue of Metabolic Defects Caused by Loss of Matrix Attachment. Nature 2009, 461, 109–113. [Google Scholar] [CrossRef]
- Ma, L.; Tao, Y.; Duran, A.; Llado, V.; Galvez, A.; Barger, J.F.; Castilla, E.A.; Chen, J.; Yajima, T.; Porollo, A.; et al. Control of Nutrient Stress-Induced Metabolic Reprogramming by PKCζ in Tumorigenesis. Cell 2013, 152, 599–611. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Horn, H.F.; Vousden, K.H. Coping with Stress: Multiple Ways to Activate P53. Oncogene 2007, 26, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of P53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Levine, A.J. The Regulation of Energy Metabolism and the IGF-1/MTOR Pathways by the P53 Protein. Trends Cell Biol. 2010, 20, 427–434. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Weidberg, H.; Gonen, C.; Wilder, S.; Elazar, Z.; Oren, M. P53-Dependent Regulation of Autophagy Protein LC3 Supports Cancer Cell Survival under Prolonged Starvation. Proc. Natl. Acad. Sci. USA 2010, 107, 18511–18516. [Google Scholar] [CrossRef]
- Berkers, C.R.; Maddocks, O.D.K.; Cheung, E.C.; Mor, I.; Vousden, K.H. Metabolic Regulation by P53 Family Members. Cell Metab. 2013, 18, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Sinthupibulyakit, C.; Ittarat, W.; St. Clair, W.H.; St. Clair, D.K. P53 Protects Lung Cancer Cells against Metabolic Stress. Int. J. Oncol. 2010, 37, 1575–1581. [Google Scholar] [CrossRef] [PubMed]
- Matoba, S.; Kang, J.-G.; Patino, W.D.; Wragg, A.; Boehm, M.; Gavrilova, O.; Hurley, P.J.; Bunz, F.; Hwang, P.M. P53 Regulates Mitochondrial Respiration. Science 2006, 312, 1650–1653. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.-F.; Lin, R.-X.; Huang, J.; Zhou, Z.; Yang, J.; Guo, G.-Z.; Wang, S.-Q. Identification of Differentially Expressed Genes Contributing to Radioresistance in Lung Cancer Cells Using Microarray Analysis. Radiat. Res. 2005, 164, 27–35. [Google Scholar] [CrossRef]
- Wilkie, M.D.; Anaam, E.A.; Lau, A.S.; Rubbi, C.P.; Jones, T.M.; Boyd, M.T.; Vlatković, N. TP53 Mutations in Head and Neck Cancer Cells Determine the Warburg Phenotypic Switch Creating Metabolic Vulnerabilities and Therapeutic Opportunities for Stratified Therapies. Cancer Lett. 2020, 478, 107–121. [Google Scholar] [CrossRef]
- Martín-Bernabé, A.; Tarragó-Celada, J.; Cunin, V.; Michelland, S.; Cortés, R.; Poignant, J.; Boyault, C.; Rachidi, W.; Bourgoin-Voillard, S.; Cascante, M.; et al. Quantitative Proteomic Approach Reveals Altered Metabolic Pathways in Response to the Inhibition of Lysine Deacetylases in A549 Cells under Normoxia and Hypoxia. Int. J. Mol. Sci. 2021, 22, 3378. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef] [PubMed]
- Lampe, K.J.; Namba, R.M.; Silverman, T.R.; Bjugstad, K.B.; Mahoney, M.J. Impact of Lactic Acid on Cell Proliferation and Free Radical Induced Cell Death in Monolayer Cultures of Neural Precursor Cells. Biotechnol. Bioeng. 2009, 103, 1214–1223. [Google Scholar] [CrossRef]
- Ortmann, B.; Druker, J.; Rocha, S. Cell Cycle Progression in Response to Oxygen Levels. Cell. Mol. Life Sci. 2014, 71, 3569–3582. [Google Scholar] [CrossRef] [PubMed]
- Seim, J.; Graff, P.; Åmellem, Ø.; Landsverk, K.S.; Stokke, T.; Pettersen, E.O. Hypoxia-induced Irreversible S-phase Arrest Involves Down-regulation of Cyclin A. Cell Prolif. 2003, 36, 321–332. [Google Scholar] [CrossRef]
- Fukuchi, K.; Nakamura, K.; Ichimura, S.; Tatsumi, K.; Gomi, K. The Association of Cyclin A and Cyclin Kinase Inhibitor P21 in Response to Gamma-Irradiation Requires the CDK2 Binding Region, but Not the Cy Motif. Biochim. Biophys. Acta 2003, 1642, 163–171. [Google Scholar] [CrossRef]
- Birch, J.; Clarke, C.J.; Campbell, A.D.; Campbell, K.; Mitchell, L.; Liko, D.; Kalna, G.; Strathdee, D.; Sansom, O.J.; Neilson, M.; et al. The Initiator Methionine TRNA Drives Cell Migration and Invasion Leading to Increased Metastatic Potential in Melanoma. Biol. Open 2016, 5, 1371–1379. [Google Scholar] [CrossRef]
- Fanfone, D.; Wu, Z.; Mammi, J.; Berthenet, K.; Neves, D.; Weber, K.; Halaburkova, A.; Virard, F.; Bunel, F.; Jamard, C.; et al. Confined Migration Promotes Cancer Metastasis through Resistance to Anoikis and Increased Invasiveness. eLife 2022, 11, e73150. [Google Scholar] [CrossRef]
- Novikov, N.M.; Zolotaryova, S.Y.; Gautreau, A.M.; Denisov, E.V. Mutational Drivers of Cancer Cell Migration and Invasion. Br. J. Cancer 2021, 124, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Kang, D.; Ji, D.; Wang, X.; Zhan, W.; Fu, M.; Xin, H.-B.; Wang, J.-B. How Does Cancer Cell Metabolism Affect Tumor Migration and Invasion? Cell Adh. Migr. 2013, 7, 395–403. [Google Scholar] [CrossRef]
- Zhang, Y.; Manouchehri Doulabi, E.; Herre, M.; Cedervall, J.; Qiao, Q.; Miao, Z.; Hamidi, A.; Hellman, L.; Kamali-Moghaddam, M.; Olsson, A.-K. Platelet-Derived PDGFB Promotes Recruitment of Cancer-Associated Fibroblasts, Deposition of Extracellular Matrix and Tgfβ Signaling in the Tumor Microenvironment. Cancers 2022, 14, 1947. [Google Scholar] [CrossRef]
- Han, X.; Guo, B.; Li, Y.; Zhu, B. Tissue Factor in Tumor Microenvironment: A Systematic Review. J. Hematol. Oncol. 2014, 7, 54. [Google Scholar] [CrossRef]
- Wang, S.; Pang, L.; Liu, Z.; Meng, X. SERPINE1 Associated with Remodeling of the Tumor Microenvironment in Colon Cancer Progression: A Novel Therapeutic Target. BMC Cancer 2021, 21, 767. [Google Scholar] [CrossRef]
- Chen, S.; Li, Y.; Zhu, Y.; Fei, J.; Song, L.; Sun, G.; Guo, L.; Li, X. SERPINE1 Overexpression Promotes Malignant Progression and Poor Prognosis of Gastric Cancer. J. Oncol. 2022, 2022, 2647825. [Google Scholar] [CrossRef]
- Jiang, J.; Lu, Y.; Zhang, F.; Pan, T.; Zhang, Z.; Wan, Y.; Ren, X.; Zhang, R. Semaphorin 4B Promotes Tumor Progression and Associates with Immune Infiltrates in Lung Adenocarcinoma. BMC Cancer 2022, 22, 632. [Google Scholar] [CrossRef]
- Kim, M.; Jang, K.; Miller, P.; Picon-Ruiz, M.; Yeasky, T.M.; El-Ashry, D.; Slingerland, J.M. VEGFA Links Self-Renewal and Metastasis by Inducing Sox2 to Repress MiR-452, Driving Slug. Oncogene 2017, 36, 5199–5211. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, S.; Li, T.; Yu, L.; Zhang, Y.; Zeng, H.; Qian, X.; Bi, J.; Lin, Y. ACE2 Inhibits Breast Cancer Angiogenesis via Suppressing the VEGFa/VEGFR2/ERK Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 173. [Google Scholar] [CrossRef] [PubMed]


| Cell Line | O2 Protocol | Doubling Time (h) ± SE |
|---|---|---|
| A549 | Normoxia | 23.45 ± 0.42 |
| Hypoxia—1% O2 | 30.90 ± 2.31 | |
| Hypoxia—0.1% O2 | 53.46 ± 12.71 | |
| H358 | Normoxia | 33.28 ± 1.54 |
| Hypoxia—1% O2 | 52.34 ± 5.46 | |
| Hypoxia—0.1% O2 | 68.59 ± 5.62 |
| Cell Line | O2 Protocol | p Values at Different Time Points (h) | ||||||
|---|---|---|---|---|---|---|---|---|
| 24 | 48 | 72 | 96 | 120 | 144 | 168 | ||
| A549 | Normoxia vs. hypoxia—1% O2 | ns | ns | * | **** | **** | **** | **** |
| Normoxia vs. hypoxia—0.1% O2 | ns | ns | ** | **** | **** | **** | **** | |
| Hypoxia—1% O2 vs. hypoxia—0.1% O2 | ns | ns | ns | ns | * | * | ** | |
| H358 | Normoxia vs. hypoxia—1% O2 | ns | ns | ** | **** | **** | **** | **** |
| Normoxia vs. hypoxia—0.1% O2 | ns | ns | **** | **** | **** | **** | **** | |
| Hypoxia—1% O2 vs. hypoxia—0.1% O2 | ns | ns | * | ns | ns | ns | ns | |
| Cell Cycle Phase | O2 Protocol | Statistical Significance of Difference between Cell Lines (p Value) at Different Time Points (h) | ||||
|---|---|---|---|---|---|---|
| 2 | 6 | 12 | 18 | 24 | ||
| G1 | Normoxia | ns | ns | ns | ns | * |
| Hypoxia—1% O2 | ns | * | ns | * | ** | |
| S | Normoxia | ns | ns | ns | ns | * |
| Hypoxia—1% O2 | ns | ns | ns | ns | ns | |
| G2 | Normoxia | ns | ns | ns | ns | ns |
| Hypoxia—1% O2 | ns | ns | ns | ns | * | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nisar, H.; Sanchidrián González, P.M.; Brauny, M.; Labonté, F.M.; Schmitz, C.; Roggan, M.D.; Konda, B.; Hellweg, C.E. Hypoxia Changes Energy Metabolism and Growth Rate in Non-Small Cell Lung Cancer Cells. Cancers 2023, 15, 2472. https://doi.org/10.3390/cancers15092472
Nisar H, Sanchidrián González PM, Brauny M, Labonté FM, Schmitz C, Roggan MD, Konda B, Hellweg CE. Hypoxia Changes Energy Metabolism and Growth Rate in Non-Small Cell Lung Cancer Cells. Cancers. 2023; 15(9):2472. https://doi.org/10.3390/cancers15092472
Chicago/Turabian StyleNisar, Hasan, Paulina Mercedes Sanchidrián González, Melanie Brauny, Frederik M. Labonté, Claudia Schmitz, Marie Denise Roggan, Bikash Konda, and Christine E. Hellweg. 2023. "Hypoxia Changes Energy Metabolism and Growth Rate in Non-Small Cell Lung Cancer Cells" Cancers 15, no. 9: 2472. https://doi.org/10.3390/cancers15092472
APA StyleNisar, H., Sanchidrián González, P. M., Brauny, M., Labonté, F. M., Schmitz, C., Roggan, M. D., Konda, B., & Hellweg, C. E. (2023). Hypoxia Changes Energy Metabolism and Growth Rate in Non-Small Cell Lung Cancer Cells. Cancers, 15(9), 2472. https://doi.org/10.3390/cancers15092472










