Biomarkers of Response to Low-Dose Aspirin in Familial Adenomatous Polyposis Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Study, Participant Characteristics, and Sample Collections
2.2. Assessment of Biomarkers
2.3. Animal Study
2.4. Assessment of Gene Expression by qPCR
2.5. Proteomics of FAP Colorectal Adenomas
2.5.1. Protein Extraction and Filter-Aided Sample Preparation
2.5.2. LC-MS/MS Label-Free Shotgun Proteomics
2.5.3. Raw Data Processing and Quantitative Analysis
2.6. Statistical Analysis
3. Results
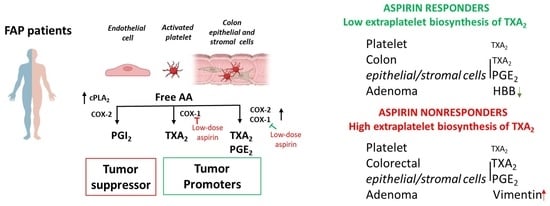
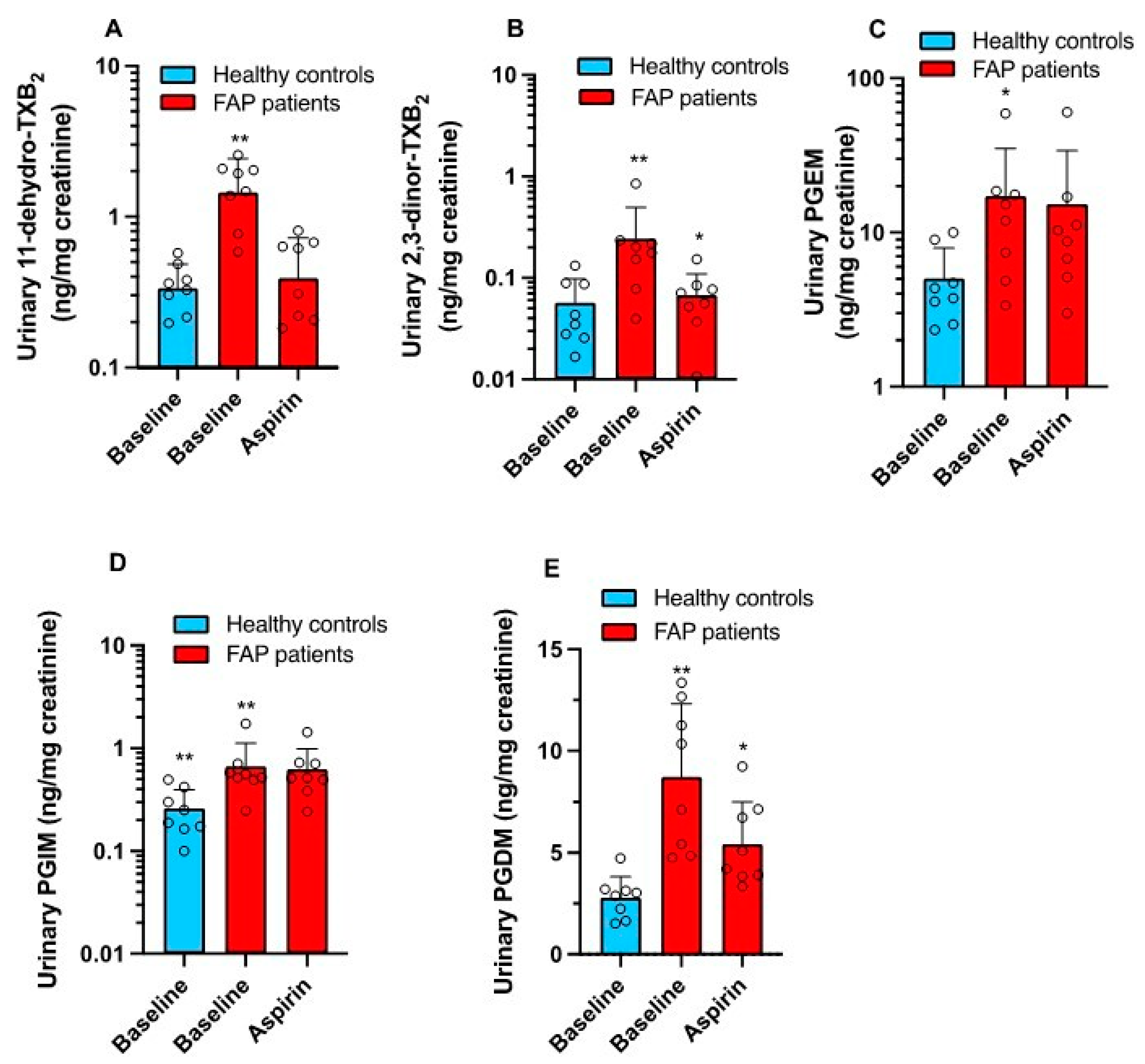
3.1. Enhanced Systemic Biosynthesis of Prostanoids in FAP
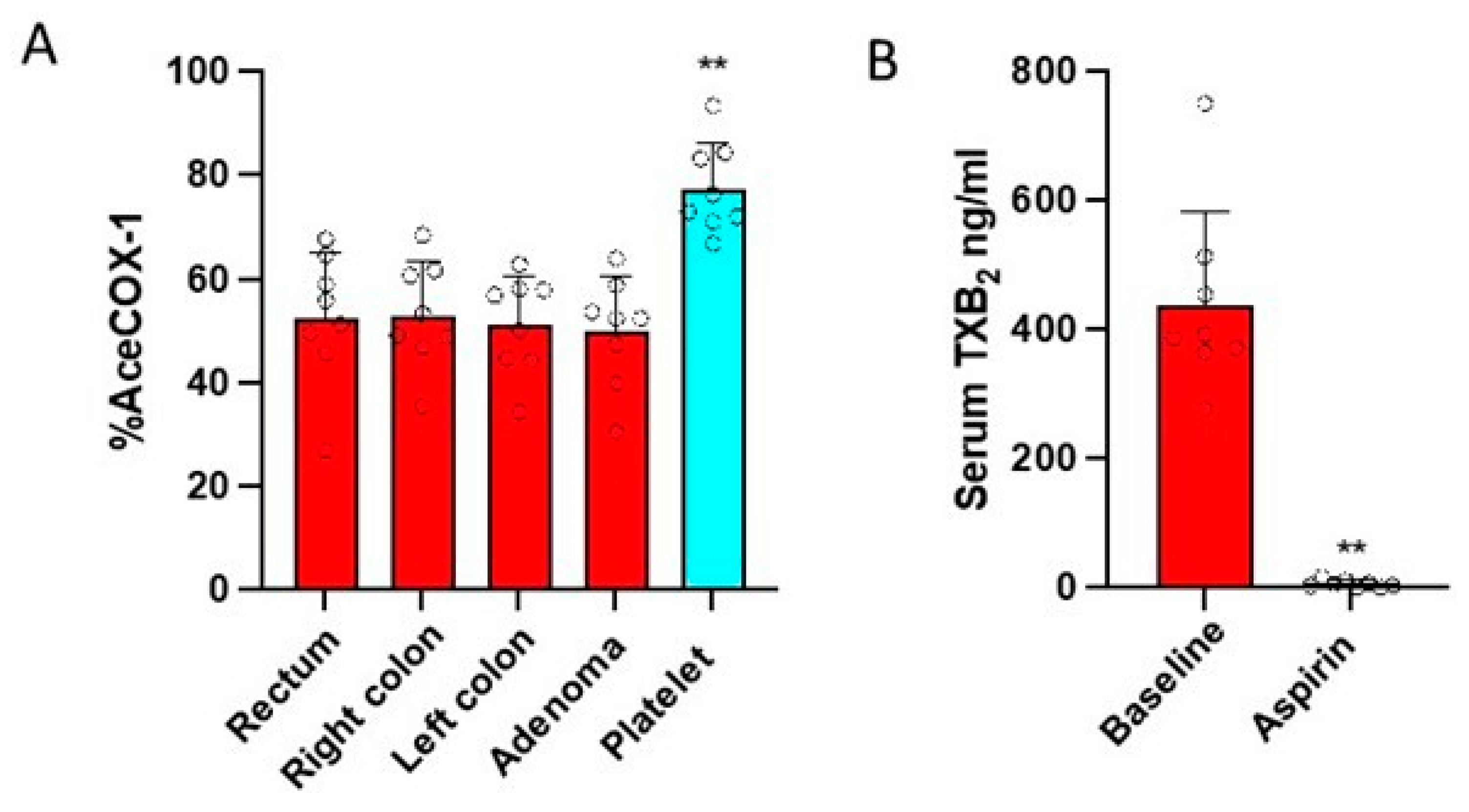
3.2. Effects of Low-Dose Aspirin on the Acetylation of COX-Isozymes
3.3. Effects of Low-Dose Aspirin on Prostanoid Biosynthesis
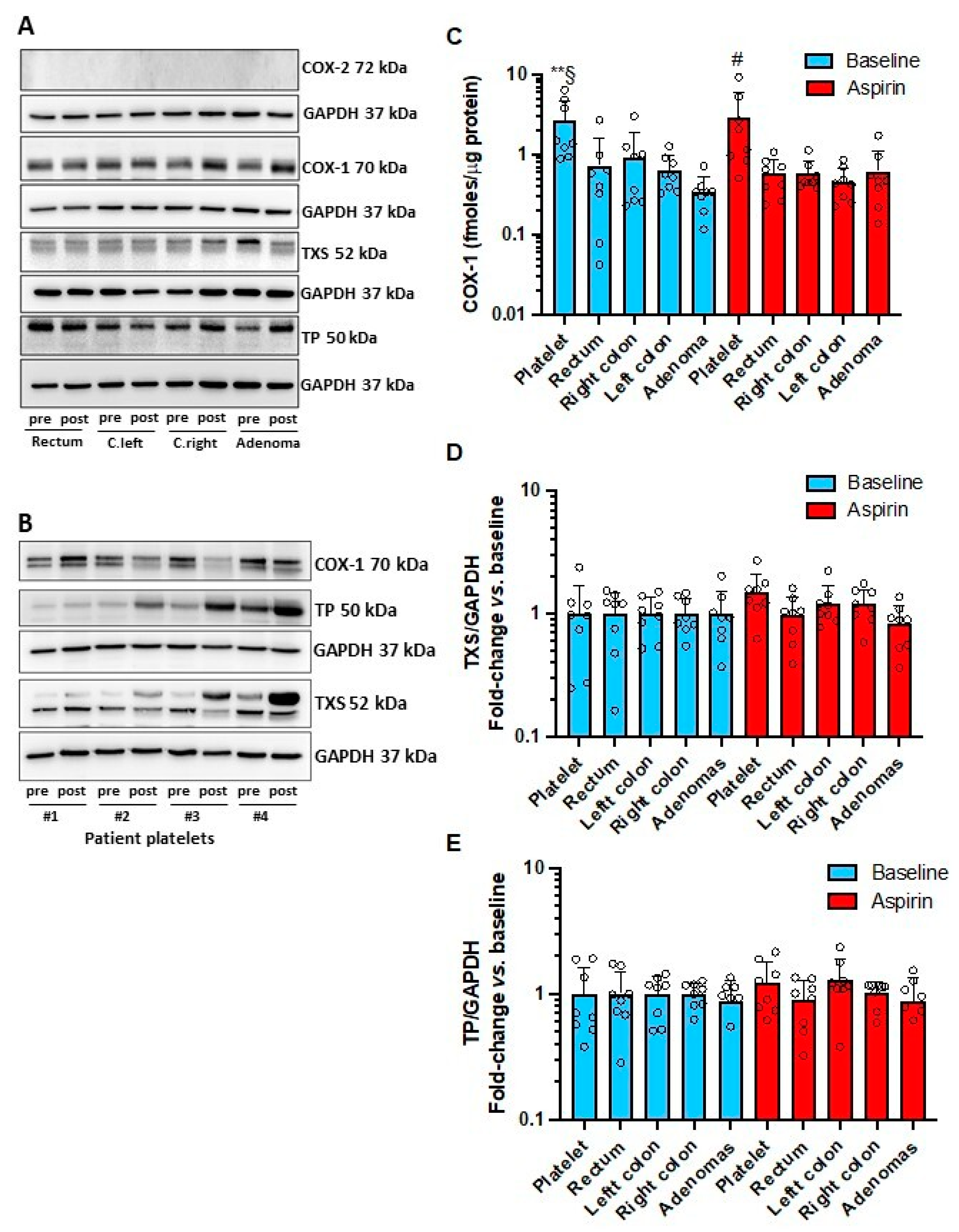
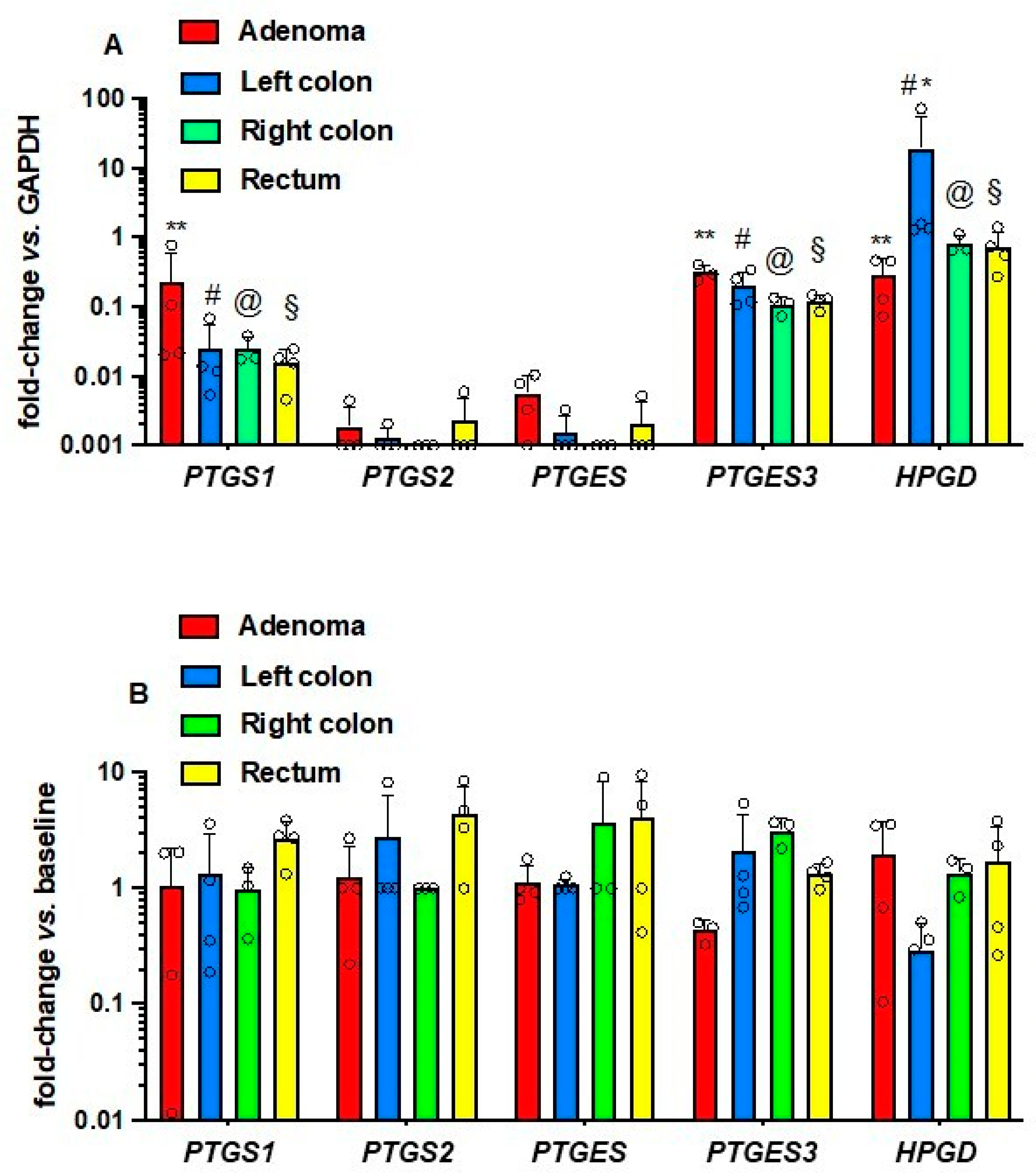
3.4. Effects of Low-Dose Aspirin on the Expression of Enzymes and Receptors of Prostanoid Pathways in Platelets and Intestinal Tissue
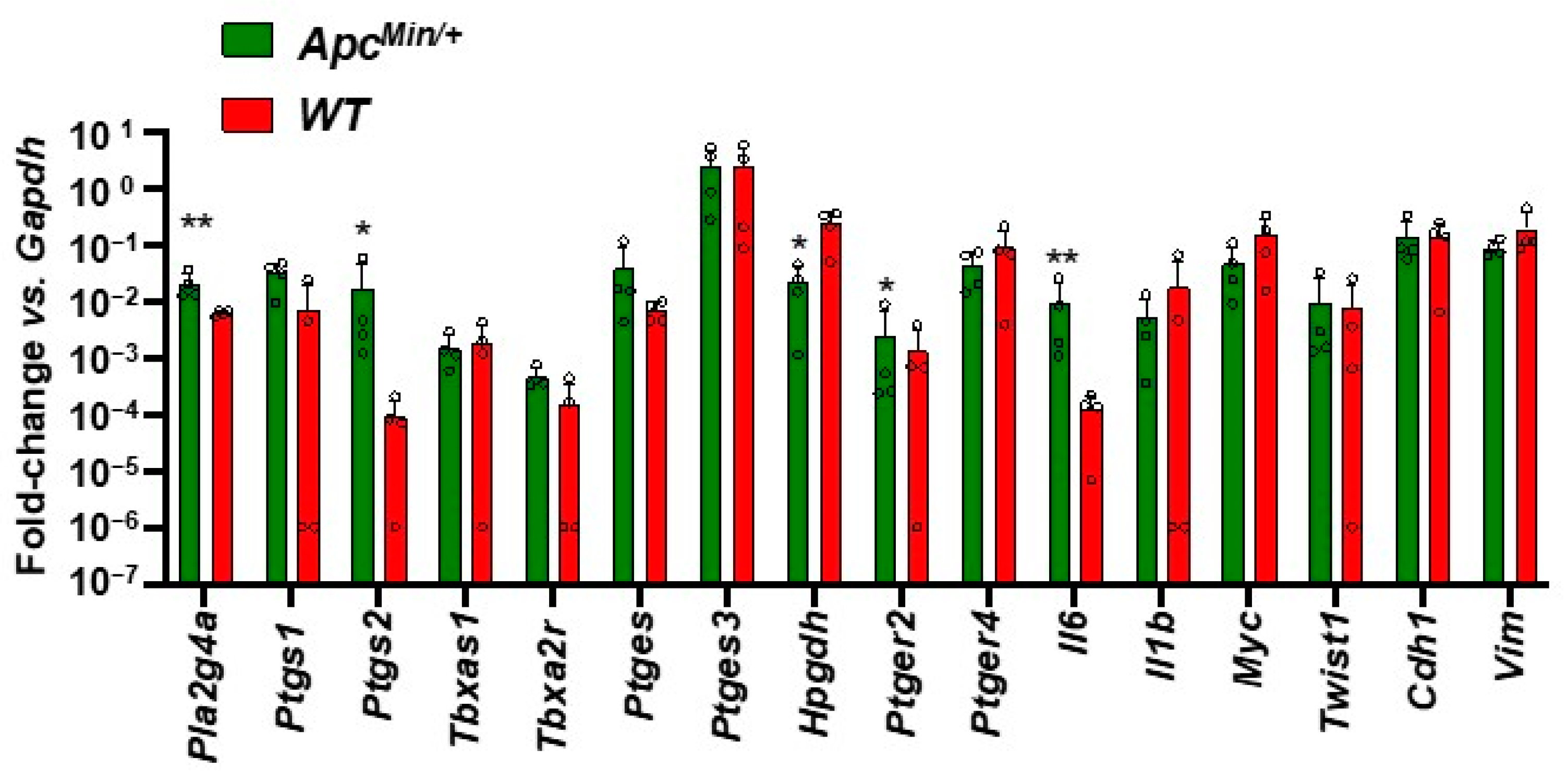
3.5. Enhanced Gene Expression of Inflammatory Pathways in the Intestinal Adenomas of ApcMin/+ vs. Normal Tissue of WT Mice
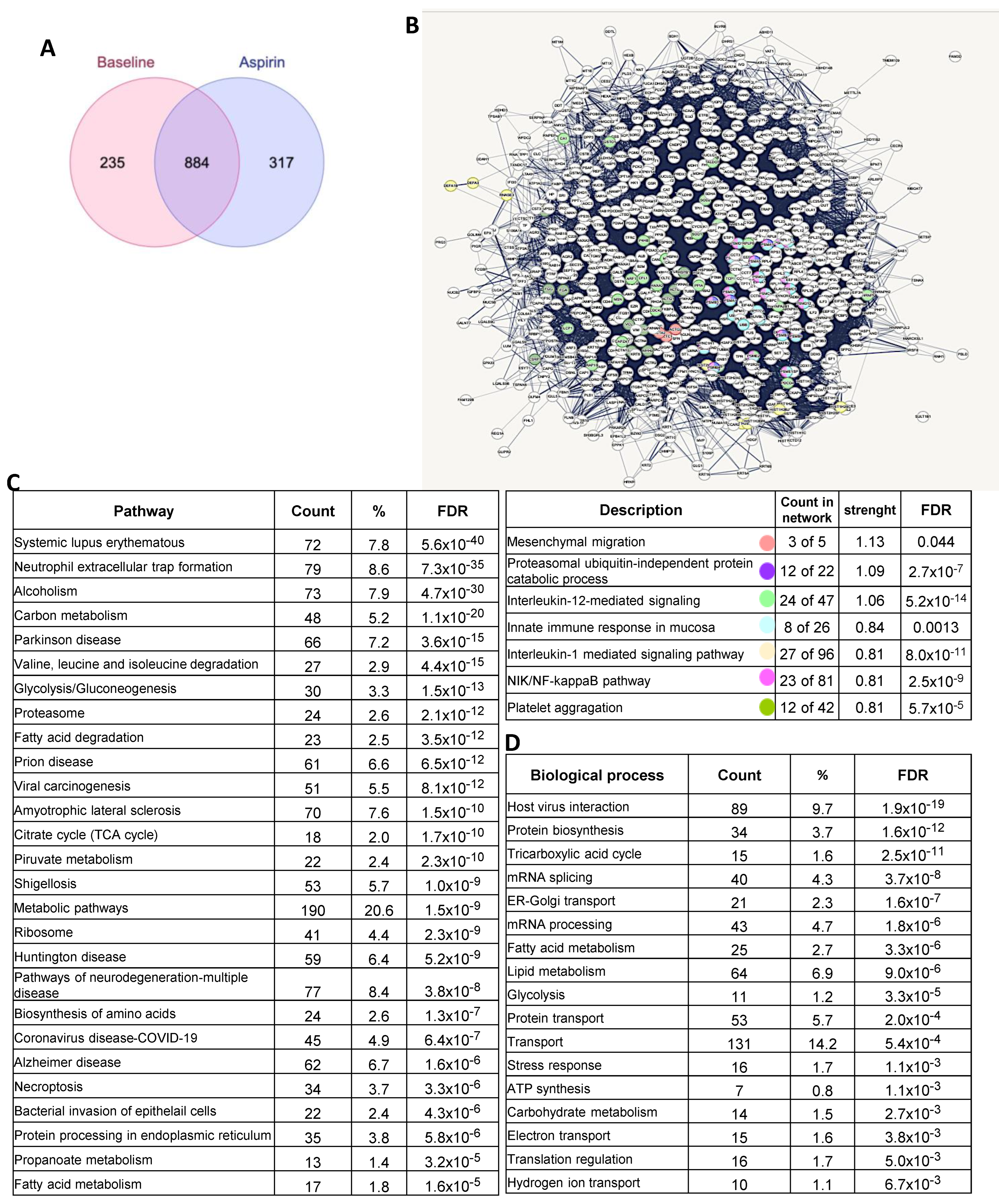
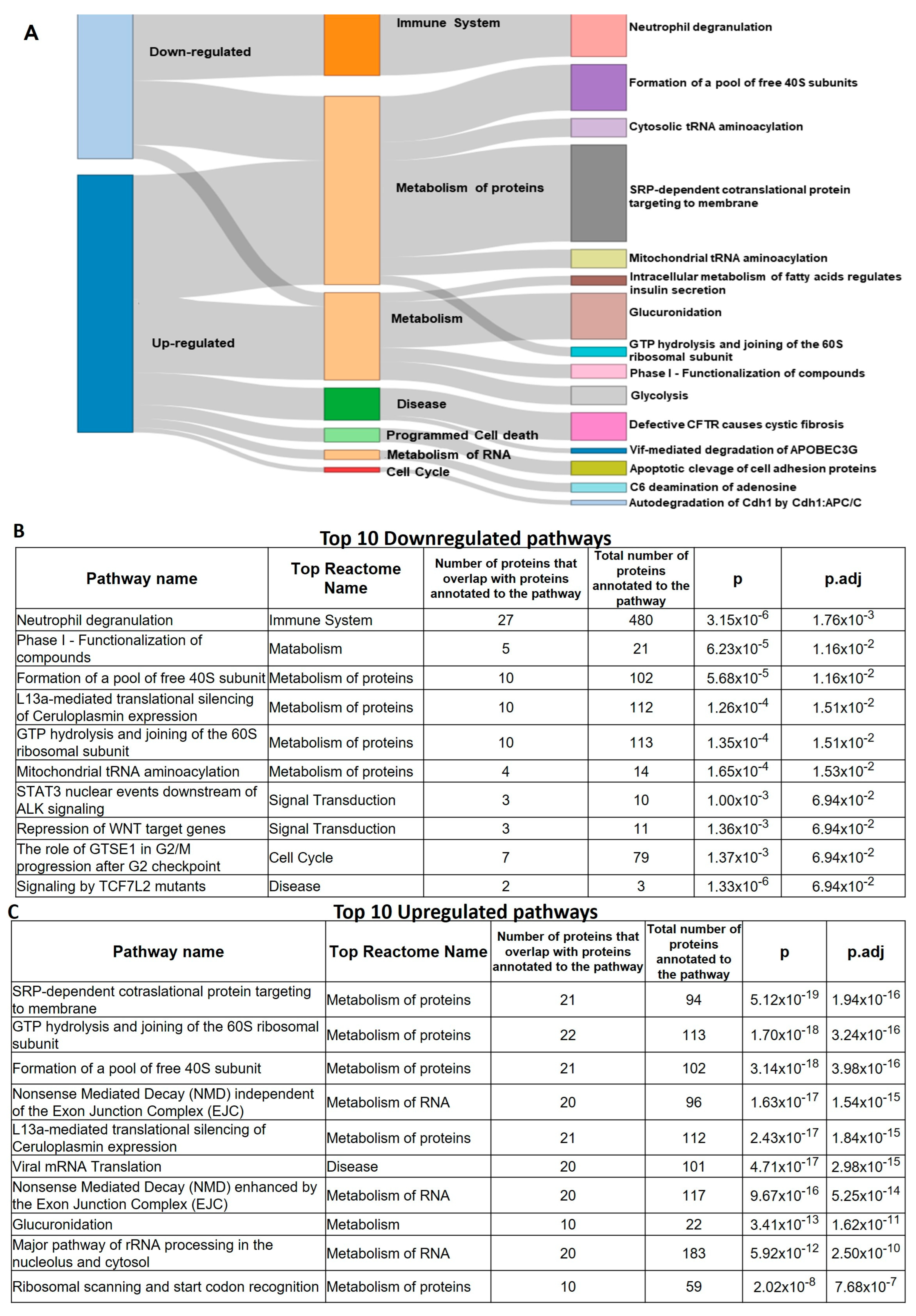
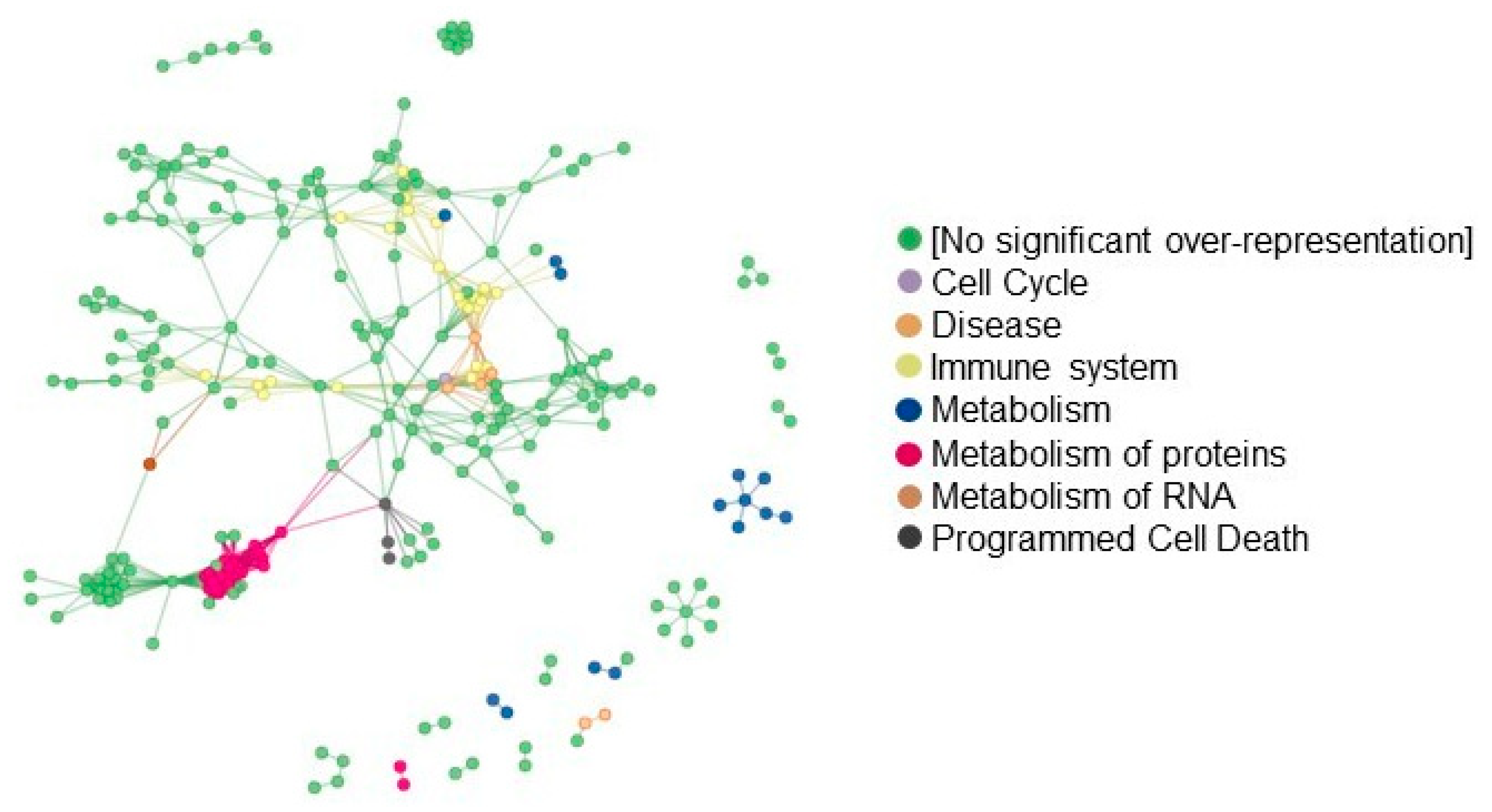
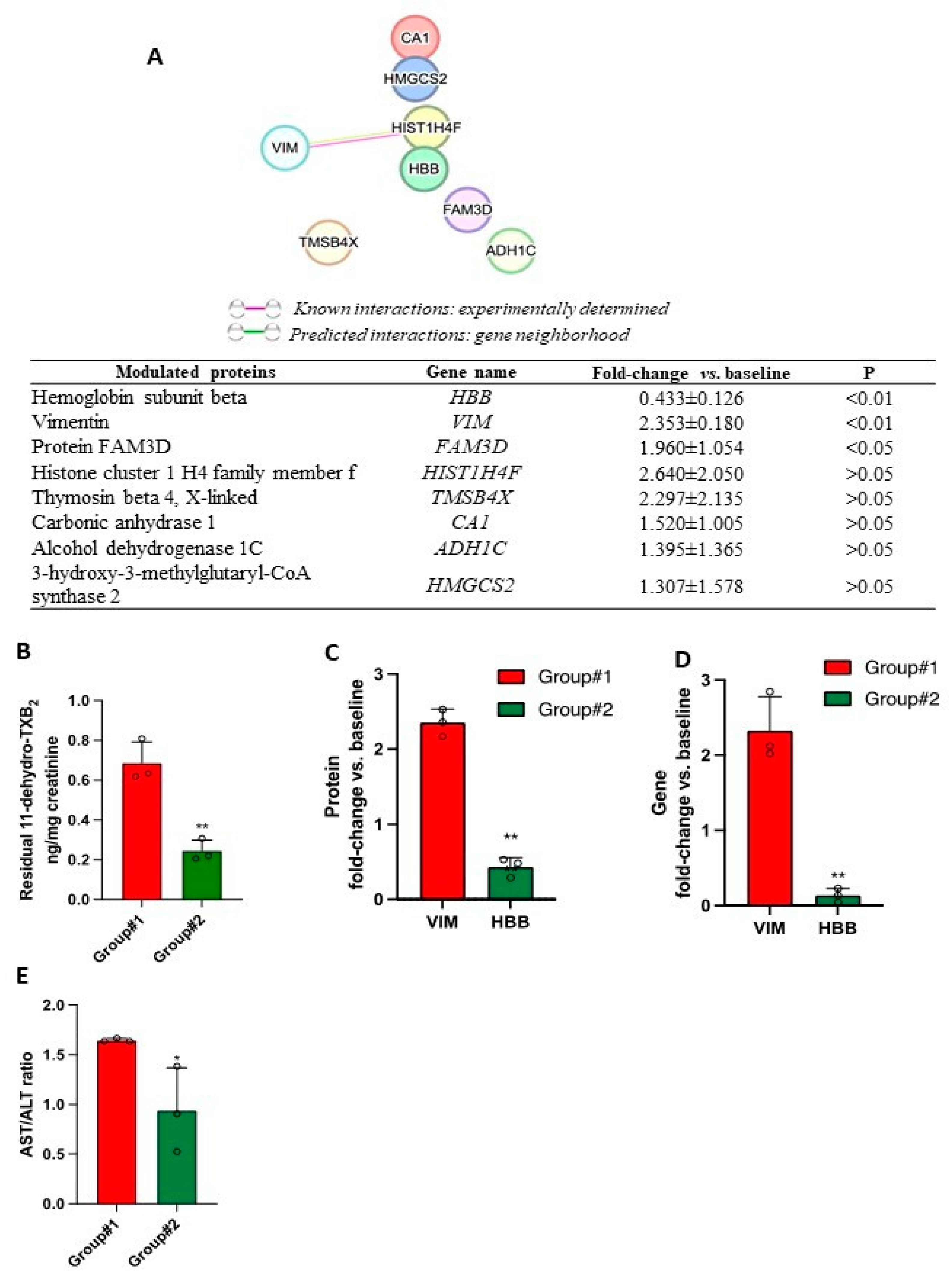
3.6. Proteomics of FAP Patients’ Adenomas and Aspirin Effects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lipton, L.; Tomlinson, I. The genetics of FAP and FAP-like syndromes. Fam. Cancer 2006, 5, 221–226. [Google Scholar] [CrossRef]
- Claes, K.; Dahan, K.; Tejpar, S.; De Paepe, A.; Bonduelle, M.; Abramowicz, M.; Verellen, C.; Franchimont, D.; Van Cutsem, E.; Kartheuser, A. The genetics of familial adenomatous polyposis (FAP) and MutYH-associated polyposis (MAP). Acta Gastroenterol. Belg. 2011, 74, 421–426. [Google Scholar]
- Half, E.; Bercovich, D.; Rozen, P. Familial adenomatous polyposis. Orphanet. J. Rare Dis. 2009, 4, 22. [Google Scholar] [CrossRef]
- Galiatsatos, P.; Foulkes, W.D. Familial adenomatous polyposis. Am. J. Gastroenterol. 2006, 101, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Giardiello, F.M. Chemoprevention in familial adenomatous polyposis. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Heiskanen, I.; Jarvinenm, H.J. Fate of the rectal stump after colectomy and ileorectal anastomosis for familial adenomatous polyposis. Int. J. Colorectal. Dis. 1997, 12, 9–13. [Google Scholar] [CrossRef]
- Burn, J.; Bishop, D.T.; Chapman, P.D.; Elliott, F.; Bertario, L.; Dunlop, M.G.; Eccles, D.; Ellis, A.; Evans, D.G.; Fodde, R.; et al. A randomized placebo-controlled prevention trial of Aspirin and/or resistant starch in young people with familial adenomatous polyposis. Cancer Prev. Res. 2011, 4, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Mutoh, M.; Sato, Y.; Doyama, H.; Tajika, M.; Tanaka, S.; Horimatsu, T.; Takeuchi, Y.; Kashida, H.; Tashiro, J.; et al. Chemoprevention with low-dose Aspirin, mesalazine, or both in patients with familial adenomatous polyposis without previous colectomy (J-FAPP Study IV): A multicentre, double-blind, randomized, two-by-two factorial design trial. Lancet Gastroenterol. Hepatol. 2021, 6, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.J.; Majerus, P.W. The mechanism of the effect of Aspirin on human platelets. Acetylation of a particulate fraction protein. J. Clin. Investig. 1975, 56, 624–632. [Google Scholar] [CrossRef]
- Lecomte, M.; Laneuville, O.; Ji, C.; DeWitt, D.L.; Smith, W.L. Acetylation of human prostaglandin endoperoxide synthase-2 (cyclooxygenase-2) by Aspirin. J. Biol. Chem. 1994, 269, 13207–13215. [Google Scholar] [CrossRef]
- Capone, M.L.; Tacconelli, S.; Sciulli, M.G.; Grana, M.; Ricciotti, E.; Minuz, P.; Di Gregorio, P.; Merciaro, G.; Patrono, C.; Patrignani, P. Clinical pharmacology of platelet, monocyte, and vascular cyclooxygenase inhibition by naproxen and low-dose Aspirin in healthy subjects. Circulation 2004, 109, 1468–1471. [Google Scholar] [CrossRef] [PubMed]
- Antithrombotic Trialists’ (ATT) Collaboration. Aspirin in the primary and secondary prevention of vascular disease: Collaborative meta-analysis of individual participant data from randomized trials. Lancet 2009, 373, 1849–1860. [Google Scholar] [CrossRef] [PubMed]
- Patrignani, P.; Tacconelli, S.; Piazuelo, E.; Di Francesco, L.; Dovizio, M.; Sostres, C.; Marcantoni, E.; Guillem-Llobat, P.; Del Boccio, P.; Zucchelli, M.; et al. Reappraisal of the clinical pharmacology of low-dose Aspirin by comparing novel direct and traditional indirect biomarkers of drug action. J. Thromb. Haemost. 2014, 12, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Patrignani, P.; Sacco, A.; Sostres, C.; Bruno, A.; Dovizio, M.; Piazuelo, E.; Di Francesco, L.; Contursi, A.; Zucchelli, M.; Schiavone, S.; et al. Low-Dose Aspirin Acetylates Cyclooxygenase-1 in Human Colorectal Mucosa: Implications for the Chemoprevention of Colorectal Cancer. Clin. Pharmacol. Ther. 2017, 102, 52–61. [Google Scholar] [CrossRef]
- Patrono, C.; Ciabattoni, G.; Pinca, E.; Pugliese, F.; Castrucci, G.; De Salvo, A.; Satta, M.A.; Peskar, B.A. Low-dose aspirin and inhibition of thromboxane B2 production in healthy subjects. Thromb. Res. 1980, 17, 317–327. [Google Scholar] [CrossRef]
- Li, X.; Fries, S.; Li, R.; Lawson, J.A.; Propert, K.J.; Diamond, S.L.; Blair, I.A.; FitzGerald, G.A.; Grosser, T. Differential impairment of aspirin-dependent platelet cyclooxygenase acetylation by nonsteroidal antiinflammatory drugs. Proc. Natl. Acad. Sci. USA 2014, 111, 16830–16835. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, S.; Contursi, A.; Falcone, L.; Mucci, M.; D’Agostino, I.; Fullone, R.; Sacco, A.; Zucchelli, M.; Bruno, A.; Ballerini, P.; et al. Characterization of cyclooxygenase-2 acetylation and prostanoid inhibition by Aspirin in cellular systems. Biochem. Pharmacol. 2020, 178, 114094. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Contursi, A.; Tacconelli, S.; Sacco, A.; Hofling, U.; Mucci, M.; Lamolinara, A.; Del Pizzo, F.; Ballerini, P.; Di Gregorio, P.; et al. The specific deletion of cyclooxygenase-1 in megakaryocytes/platelets reduces intestinal polyposis in ApcMin/+ mice. Pharmacol. Res. 2022, 185, 106506. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Tenzer, S. Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 2016, 11, 795–812. [Google Scholar] [CrossRef]
- Di Giacomo, V.; Chiavaroli, A.; Recinella, L.; Orlando, G.; Cataldi, A.; Rapino, M.; Di Valerio, V.; Ronci, M.; Leone, S.; Brunetti, L.; et al. Antioxidant and Neuroprotective Effects Induced by Cannabidiol and Cannabigerol in Rat CTX-TNA2 Astrocytes and Isolated Cortexes. Int. J. Mol. Sci. 2020, 21, 3575. [Google Scholar] [CrossRef]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteom. 2012, 11, M111.010587. [Google Scholar] [CrossRef]
- Tai, H.H.; Ensor, C.M.; Tong, M.; Zhou, H.; Yan, F. Prostaglandin catabolizing enzymes. Prostaglandins Other Lipid Mediat. 2002, 68–69, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.R.; Luongo, C.; Gould, K.A.; McNeley, M.K.; Shoemaker, A.R.; Dove, W.F. ApcMin: A mouse model for intestinal and mammary tumorigenesis. Eur. J. Cancer 1995, 31A, 1061–1064. [Google Scholar] [CrossRef] [PubMed]
- Su, L.K.; Kinzler, K.W.; Vogelstein, B.; Preisinger, A.C.; Moser, A.R.; Luongo, C.; Gould, K.A.; Dove, W.F. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 1992, 256, 668–670. [Google Scholar] [CrossRef] [PubMed]
- Leslie, C.C. Cytosolic phospholipase A2: Physiological function and role in disease. J. Lipid Res. 2015, 56, 1386–1402. [Google Scholar] [CrossRef] [PubMed]
- Dronadula, N.; Liu, Z.; Wang, C.; Cao, H.; Rao, G.N. STAT-3-dependent cytosolic phospholipase A2 expression is required for thrombin-induced vascular smooth muscle cell motility. J. Biol. Chem. 2005, 280, 3112–3120. [Google Scholar] [CrossRef] [PubMed]
- Giardiello, F.M.; Spannhake, E.W.; DuBois, R.N.; Hylind, L.M.; Robinson, C.R.; Hubbard, W.C.; Hamilton, S.R.; Yang, V.W. Prostaglandin levels in human colorectal mucosa: Effects of sulindac in patients with familial adenomatous polyposis. Dig. Dis. Sci. 1998, 43, 311–316. [Google Scholar]
- Li, H.; Liu, K.; Boardman, L.A.; Zhao, Y.; Wang, L.; Sheng, Y.; Oi, N.; Limburg, P.J.; Bode, A.M.; Dong, Z. Circulating prostaglandin biosynthesis in colorectal cancer and potential clinical significance. EBioMedicine 2015, 2, 165–171. [Google Scholar] [CrossRef]
- Sacco, A.; Bruno, A.; Contursi, A.; Dovizio, M.; Tacconelli, S.; Ricciotti, E.; Guillem-Llobat, P.; Salvatore, T.; Di Francesco, L.; Fullone, R.; et al. Platelet-Specific Deletion of Cyclooxygenase-1 Ameliorates Dextran Sulfate Sodium-Induced Colitis in Mice. J. Pharmacol. Exp. Ther. 2019, 370, 416–426. [Google Scholar] [CrossRef]
- D’Agostino, I.; Tacconelli, S.; Bruno, A.; Contursi, A.; Mucci, L.; Hu, X.; Xie, Y.; Chakraborty, R.; Jain, K.; Sacco, A.; et al. Low-dose Aspirin prevents hypertension and cardiac fibrosis when thromboxane A2 is unrestrained. Pharmacol. Res. 2021, 170, 105744. [Google Scholar] [CrossRef]
- Wang, D.; DuBois, R.N. Role of Prostanoids in Gastrointestinal Cancer. J. Clin. Investig. 2018, 128, 2732–2742. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. An inflammatory mediator, prostaglandin E2, in colorectal cancer. Cancer J. 2013, 19, 502–510. [Google Scholar] [CrossRef]
- Wang, D.; DuBois, R.N. The Role of COX-2 in Intestinal Inflammation and Colorectal Cancer. Oncogene 2010, 29, 781–788. [Google Scholar] [CrossRef]
- Wang, D.; DuBois, N. Urinary PGE-M: A promising cancer biomarker. Cancer Prev. Res. 2013, 6, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Dovizio, M.; Tacconelli, S.; Ricciotti, E.; Bruno, A.; Maier, T.J.; Anzellotti, P.; Di Francesco, L.; Sala, P.; Signoroni, S.; Bertario, L.; et al. Effects of celecoxib on prostanoid biosynthesis and circulating angiogenesis proteins in familial adenomatous polyposis. J. Pharmacol. Exp. Ther. 2012, 341, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Oshima, M.; Dinchuk, J.E.; Kargman, S.L.; Oshima, H.; Hancock, B.; Kwong, E.; Trzaskos, J.M.; Evans, J.F.; Taketo, M.M. Suppression of intestinal polyposis in Apc delta716 knockout mice by inhibition of cyclooxygenase 2 (COX-2). Cell 1996, 87, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Grosser, T.; Fries, S.; FitzGerald, G.A. Biological basis for the cardiovascular consequences of COX-2 inhibition: Therapeutic challenges and opportunities. J. Clin. Investig. 2006, 116, 4–15. [Google Scholar] [CrossRef]
- DuBois, R.N.; Awad, J.; Morrow, J.; Roberts, L.J., 2nd; Bishop, P.R. Regulation of eicosanoid production and mitogenesis in rat intestinal epithelial cells by transforming growth factor-alpha and phorbol ester. J. Clin. Investig. 1994, 93, 493–498. [Google Scholar] [CrossRef]
- Pradono, P.; Tazawa, R.; Maemondo, M.; Tanaka, M.; Usui, K.; Saijo, Y.; Hagiwara, K.; Nukiwa, T. Gene transfer of thromboxane A2 synthase and prostaglandin I2 synthase antithetically altered tumor angiogenesis and tumor growth. Cancer Res. 2002, 62, 63–66. [Google Scholar]
- Alfonso, L.; Ai, G.; Spitale, R.C.; Bhat, G.J. Molecular Targets of Aspirin and Cancer Prevention. Br. J. Cancer 2014, 111, 61–67. [Google Scholar] [CrossRef]
- Ricciotti, E.; Wangensteen, K.J.; FitzGerald, G.A. Aspirin in Hepatocellular Carcinoma. Cancer Res. 2021, 81, 3751–3761. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Miyamoto, D.T.; Wittner, B.S.; Sullivan, J.P.; Aceto, N.; Jordan, N.V.; Yu, M.; Karabacak, N.M.; Comaills, V.; Morris, R.; et al. Expression of β-globin by cancer cells promotes cell survival during blood-borne dissemination. Nat. Commun. 2017, 8, 14344. [Google Scholar] [CrossRef] [PubMed]
- Usman, S.; Waseem, N.H.; Nguyen, T.K.N.; Mohsin, S.; Jamal, A.; The, M.T.; Waseem, A. Vimentin Is at the Heart of Epithelial Mesenchymal Transition (EMT) Mediated Metastasis. Cancers 2021, 13, 4985. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Xu, E.; Liang, W.; Pei, X.; Chen, D.; Zheng, D.; Zhang, Y.; Zheng, C.; Wang, P.; She, S.; et al. Identification of FAM3D as a new endogenous chemotaxis agonist for the formyl peptide receptors. J. Cell Sci. 2016, 129, 1831–1842. [Google Scholar] [PubMed]
- Zhou, Y.; Jia, S.; Wang, C.; Chen, Z.; Chi, Y.; Li, J.; Xu, G.; Guan, Y.; Yang, J. FAM3A is a target gene of peroxisome proliferator-activated receptor gamma. Biochim. Biophys. Acta 2013, 1830, 4160–4170. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Ide, T.; Suzuki, M.; Mochizuki, T.; Kadowaki, T. Evidence for direct binding of fatty acids and eicosanoids to human peroxisome proliferators-activated receptor alpha. Biochem. Biophys. Res. Commun. 1999, 260, 609–613. [Google Scholar] [CrossRef]
- Chen, W.; Wang, W.; Zhou, L.; Zhou, J.; He, L.; Li, J.; Xu, X.; Wang, J.; Wang, L. Elevated AST/ALT ratio is associated with all-cause mortality and cancer incident. J. Clin. Lab. Anal. 2022, 36, e24356. [Google Scholar] [CrossRef]
- Scheipner, L.; Smolle, M.A.; Barth, D.; Posch, F.; Stotz, M.; Pichler, M.; StÖger, H.; Gerger, A.; Riedl, J.M. The AST/ALT Ratio Is an Independent Prognostic Marker for Disease-free Survival in Stage II and III Colorectal Carcinoma. Anticancer Res. 2021, 41, 429–436. [Google Scholar] [CrossRef]
- Xu, J.; Shi, X.; Pan, Y. The Association of Aspartate Aminotransferase/Alanine Aminotransferase Ratio with Diabetic Nephropathy in Patients with Type 2 Diabetes. Diabetes. Metab. Syndr. Obes. 2021, 14, 3831–3837. [Google Scholar] [CrossRef]
- Rade, J.J.; Barton, B.A.; Vasan, R.S.; Kronsberg, S.S.; Xanthakis, V.; Keaney, J.F., Jr.; Hamburg, N.M.; Kakouros, N.; Kickler, T.A. Association of thromboxane generation with survival in aspirin users and nonusers. J. Am. Coll. Cardiol. 2022, 80, 233–250. [Google Scholar] [CrossRef]

| FAP Patients (n = 8) | Healthy Subjects (n = 8) | |
|---|---|---|
| Age, y | 40.25 ± 8.73 | 41.00 ± 10.36 |
| Sex, % female | 75.00 | 75.00 |
| BMI, kg/m2 | 24.17 ± 3.45 | 23.03 ± 2.85 |
| Systolic blood pressure, mmHg | 110.80 ± 8.19 | 114.40 ± 8.21 |
| Diastolic blood pressure, mmHg | 69.00 ± 5.24 | 72 ± 3.85 |
| Platelets count (×103/mL) | 265 ± 97.17 | 252.40 ± 43.18 |
| Hematocrit (%) | 42.71 ± 1.88 | 41.89 ± 2.21 |
| Haemoglobin (g/dL) | 14.26 ± 0.89 | 13.77 ± 0.79 |
| Glycemia, mg/dL | 86.13 ± 10.01 | 92.00 ± 8.67 |
| Creatinine, mg/dL | 0.69 ± 0.16 | 0.80 ± 0.10 |
| Total cholesterol, mg/dL | 195.40 ± 30.80 | 196.00 ± 27.36 |
| Type of polyps | ||
| Serrated adenoma with low-grade dysplasia, n (%) | 1 (12.5) | 0 (0) |
| Tubular adenoma with low-grade dysplasia, n (%) | 3 (37.5) | 0 (0) |
| Tubulovillous adenoma with low-grade dysplasia, n (%) | 4 (50) | 0 (0) |
| Mutations | ||
| APC, n (%) | 7 (87.5) | 0 (0) |
| MUTYH, n (%) | 1 (12.5) | 0 (0) |
| Classification | ||
| Intermediate, n (%) | 5 (62.5) | 0 (0) |
| Attenuated, n (%) | 3 (37.5) | 0 (0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lanas, A.; Tacconelli, S.; Contursi, A.; Piazuelo, E.; Bruno, A.; Ronci, M.; Marcone, S.; Dovizio, M.; Sopeña, F.; Falcone, L.; et al. Biomarkers of Response to Low-Dose Aspirin in Familial Adenomatous Polyposis Patients. Cancers 2023, 15, 2457. https://doi.org/10.3390/cancers15092457
Lanas A, Tacconelli S, Contursi A, Piazuelo E, Bruno A, Ronci M, Marcone S, Dovizio M, Sopeña F, Falcone L, et al. Biomarkers of Response to Low-Dose Aspirin in Familial Adenomatous Polyposis Patients. Cancers. 2023; 15(9):2457. https://doi.org/10.3390/cancers15092457
Chicago/Turabian StyleLanas, Angel, Stefania Tacconelli, Annalisa Contursi, Elena Piazuelo, Annalisa Bruno, Maurizio Ronci, Simone Marcone, Melania Dovizio, Federico Sopeña, Lorenza Falcone, and et al. 2023. "Biomarkers of Response to Low-Dose Aspirin in Familial Adenomatous Polyposis Patients" Cancers 15, no. 9: 2457. https://doi.org/10.3390/cancers15092457
APA StyleLanas, A., Tacconelli, S., Contursi, A., Piazuelo, E., Bruno, A., Ronci, M., Marcone, S., Dovizio, M., Sopeña, F., Falcone, L., Milillo, C., Mucci, M., Ballerini, P., & Patrignani, P. (2023). Biomarkers of Response to Low-Dose Aspirin in Familial Adenomatous Polyposis Patients. Cancers, 15(9), 2457. https://doi.org/10.3390/cancers15092457