Association of Suppressive Myeloid Cell Enrichment with Aggressive Oropharynx Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Biospecimens
2.2. Clinical Data Collection
2.3. Sample Processing
2.4. Data Analysis
3. Results
3.1. Demographics and Clinical Parameters
3.2. GeneExpression
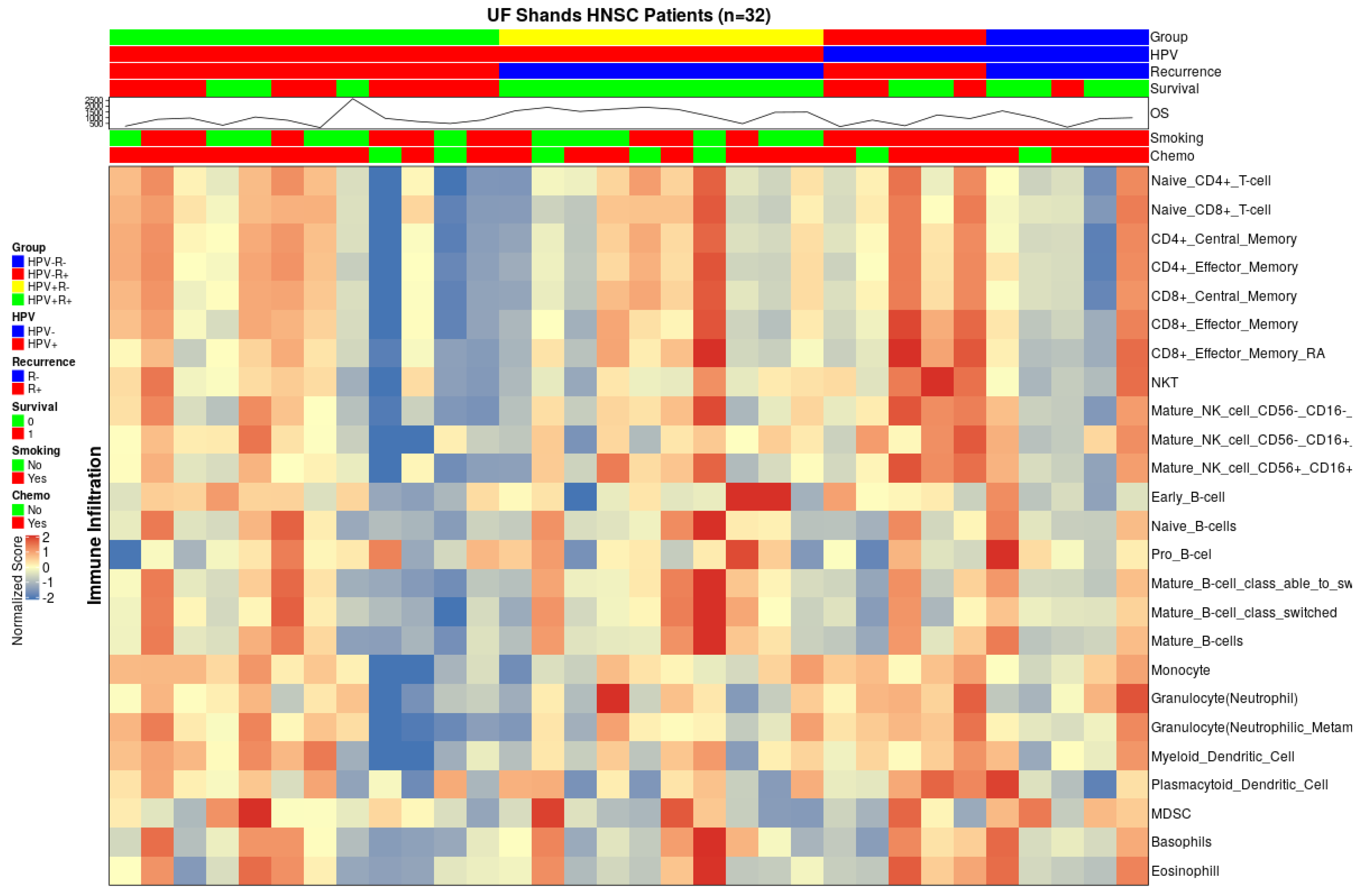
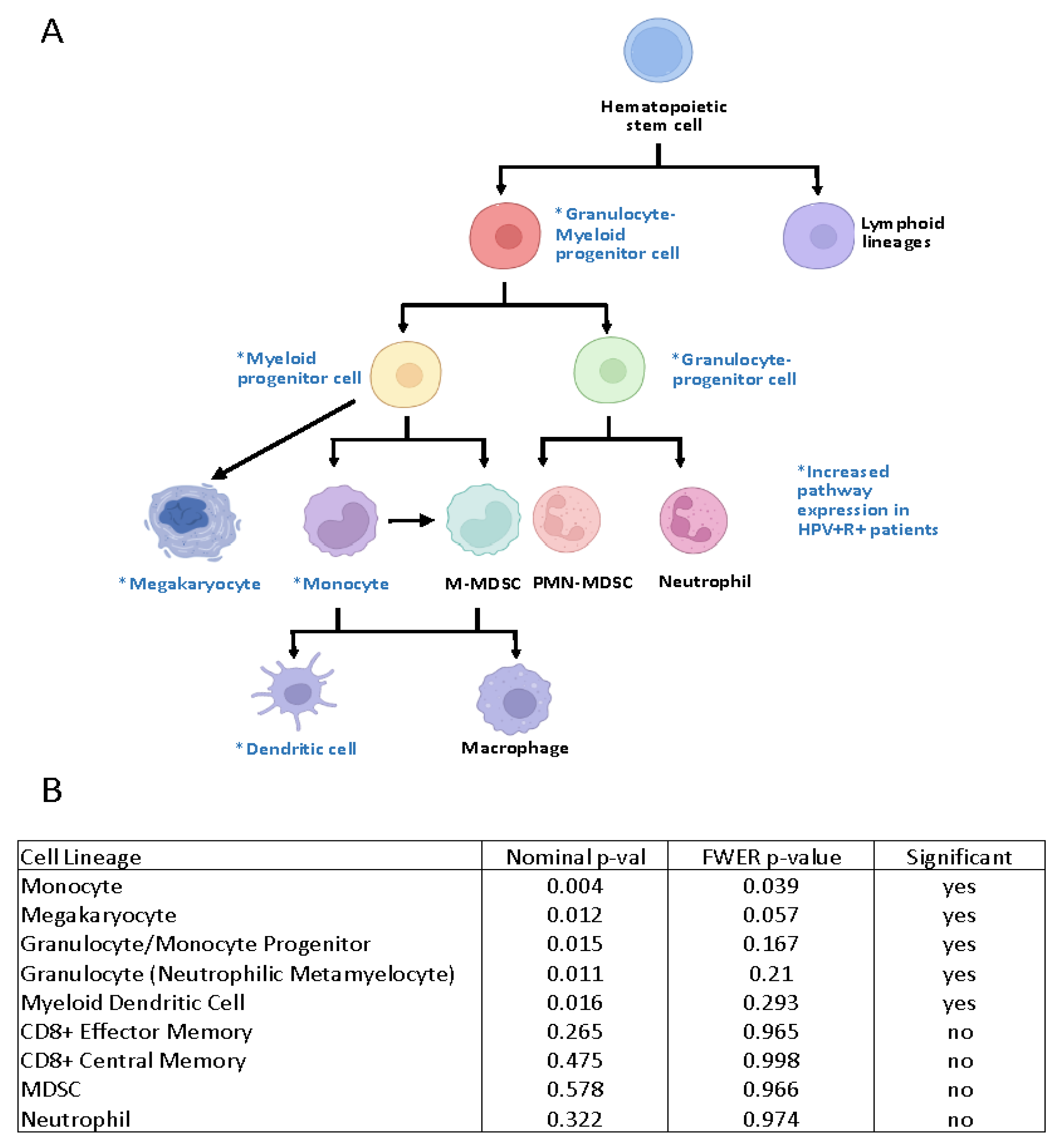
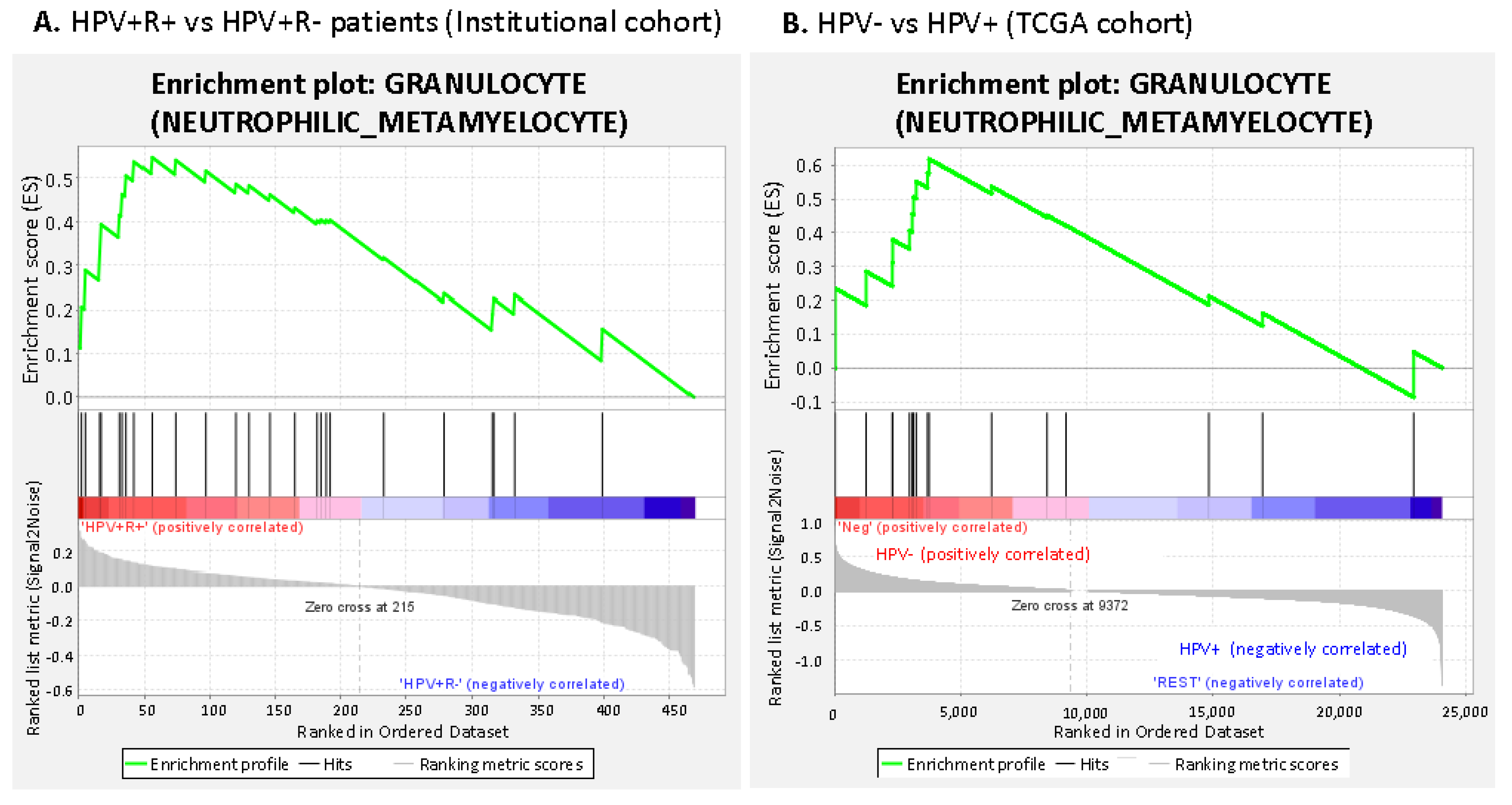
3.2.1. Suppressive Myeloid Cell Signatures Are Enriched in Aggressive OPSCC
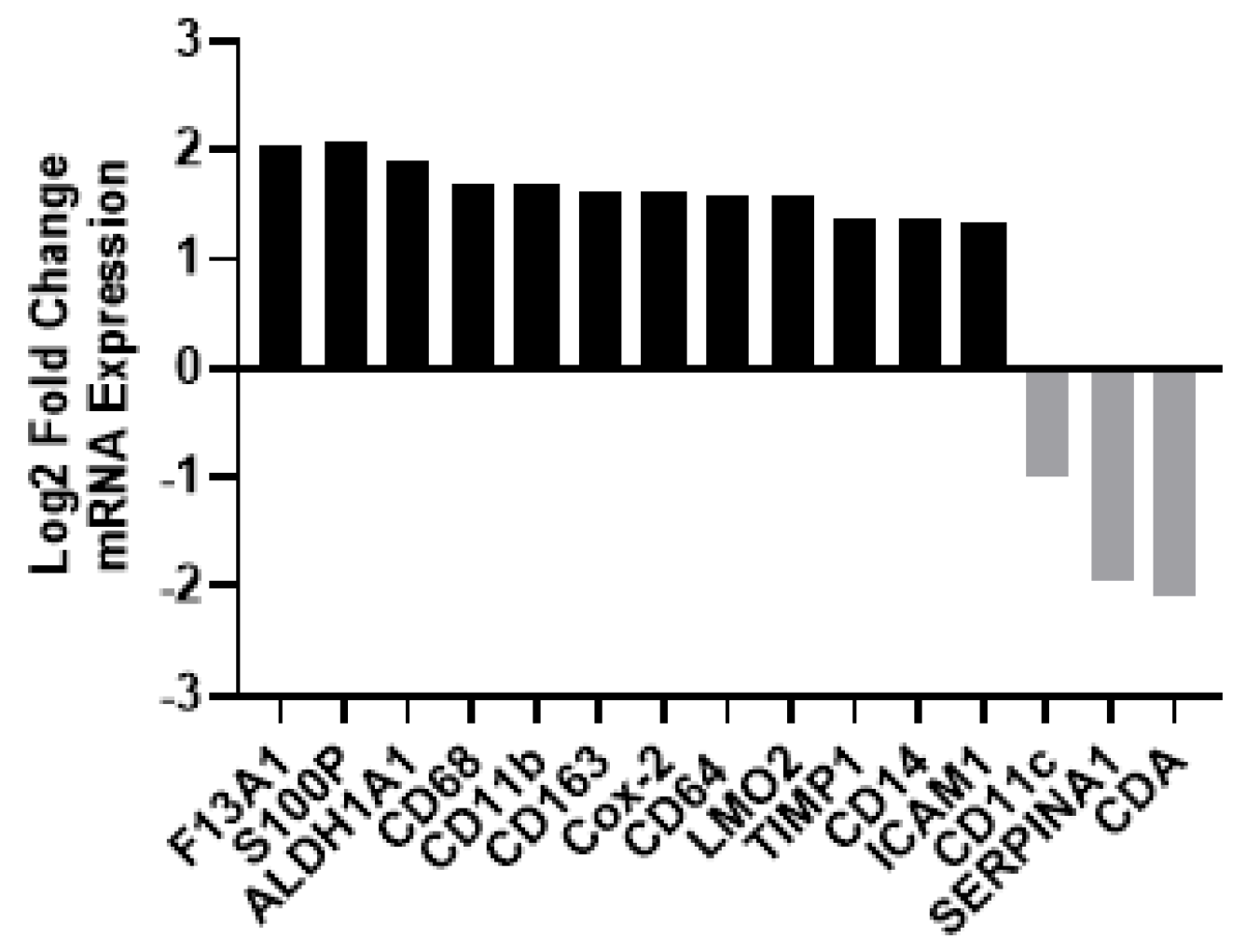
3.2.2. Gene Expression Profiling Demonstrated Increases in M2-Like Macrophage-Associated Genes in Patients with Recurrent HPV+ Cancer
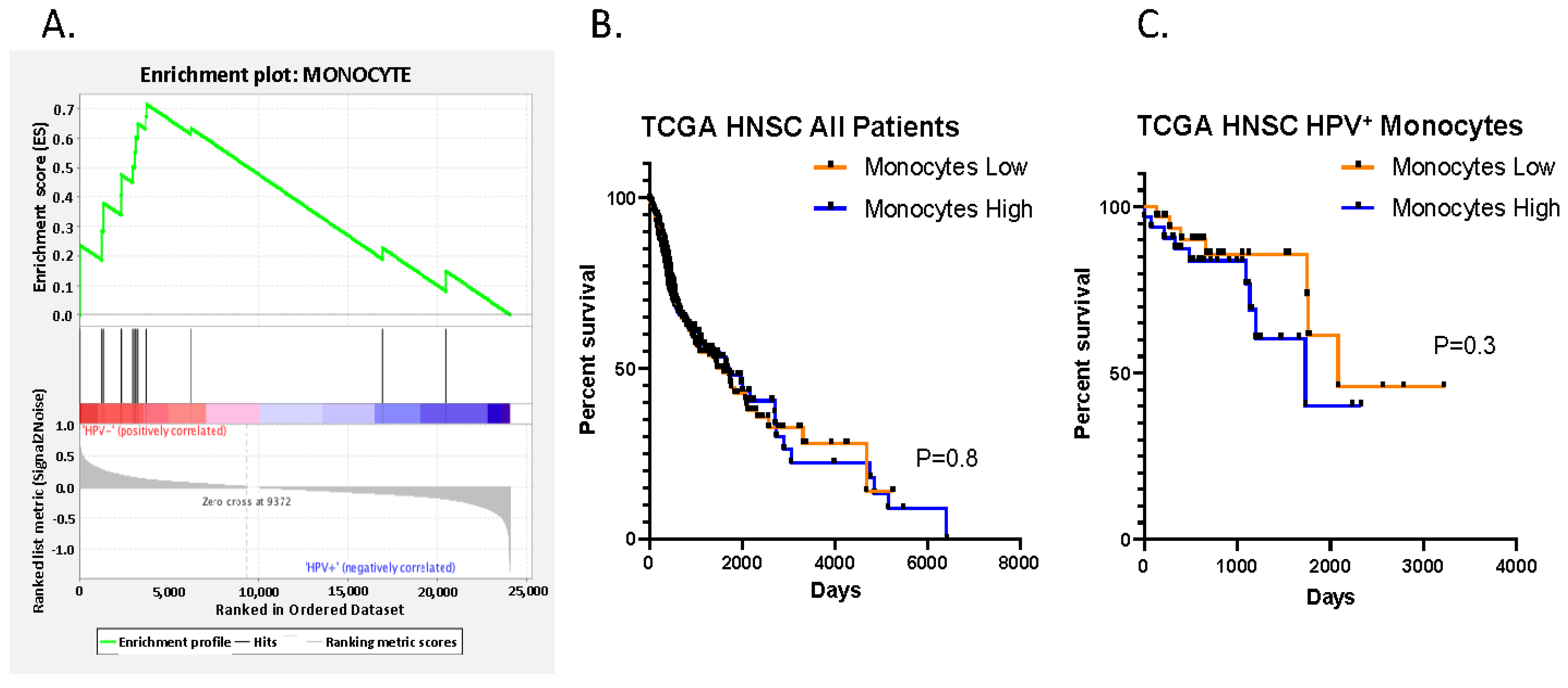
3.2.3. Increased Suppressive Monocyte/Macrophage Cell Expression Is Present in HPV- Negative vs. HPV Positive Patients in the TCGA Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.P.; Saha, S.; Yu, M.; Kimple, R.J.; Lambert, P.F. Prevalence of human papillomavirus in oropharyngeal squamous cell carcinoma in the United States across time. Chem. Res. Toxicol. 2014, 27, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Adelstein, D.J.; Ridge, J.A.; Gillison, M.L.; Chaturvedi, A.K.; D’Souza, G.; Gravitt, P.E.; Westra, W.; Psyrri, A.; Kast, W.M.; Koutsky, L.A.; et al. Head and neck squamous cell cancer and the human papillomavirus: Summary of a National Cancer Institute State of the Science Meeting, November 9–10, 2008, Washington, D.C. Head Neck 2009, 31, 1393–1422. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Zhang, Q.; Jordan, R.; Xiao, W.; Westra, W.H.; Trotti, A.; Spencer, S.; Harris, J.; Chung, C.H.; Ang, K.K. Tobacco smoking and increased risk of death and progression for patients with p16-positive and p16-negative oropharyngeal cancer. J. Clin. Oncol. 2012, 30, 2102–2111. [Google Scholar] [CrossRef]
- Lechien, J.R.; Seminerio, I.; Descamps, G.; Mat, Q.; Mouawad, F.; Hans, S.; Julieron, M.; Dequanter, D.; Vanderhaegen, T.; Journe, F.; et al. Impact of HPV Infection on the Immune System in Oropharyngeal and Non-Oropharyngeal Squamous Cell Carcinoma: A Systematic Review. Cells 2019, 8, 1061. [Google Scholar] [CrossRef]
- Ward, M.J.; Thirdborough, S.M.; Mellows, T.; Riley, C.; Harris, S.; Suchak, K.; Webb, A.; Hampton, C.; Patel, N.N.; Randall, C.J.; et al. Tumour-infiltrating lymphocytes predict for outcome in HPV-positive oropharyngeal cancer. Br. J. Cancer 2014, 110, 489–500. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef]
- Brooks, J.M.; Menezes, A.N.; Ibrahim, M.; Archer, L.; Lal, N.; Bagnall, C.J.; von Zeidler, S.V.; Valentine, H.R.; Spruce, R.J.; Batis, N.; et al. Development and Validation of a Combined Hypoxia and Immune Prognostic Classifier for Head and Neck Cancer. Clin. Cancer Res. 2019, 25, 5315–5328. [Google Scholar] [CrossRef]
- Balermpas, P.; Rodel, F.; Weiss, C.; Rodel, C.; Fokas, E. Tumor-infiltrating lymphocytes favor the response to chemoradiotherapy of head and neck cancer. Oncoimmunology 2014, 3, e27403. [Google Scholar] [CrossRef]
- Sivars, L.; Landin, D.; Grun, N.; Vlastos, A.; Marklund, L.; Nordemar, S.; Ramqvist, T.; Munck-Wikland, E.; Nasman, A.; Dalianis, T. Validation of Human Papillomavirus as a Favourable Prognostic Marker and Analysis of CD8(+) Tumour-infiltrating Lymphocytes and Other Biomarkers in Cancer of Unknown Primary in the Head and Neck Region. Anticancer Res. 2017, 37, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Seminerio, I.; Kindt, N.; Descamps, G.; Bellier, J.; Lechien, J.R.; Mat, Q.; Pottier, C.; Journé, F.; Saussez, S. High infiltration of CD68+ macrophages is associated with poor prognoses of head and neck squamous cell carcinoma patients and is influenced by human papillomavirus. Oncotarget 2018, 9, 11046–11059. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Marbaix, E.; Bouzin, C.; Hamoir, M.; Mahy, P.; Bol, V.; Grégoire, V. Immune cell infiltration in head and neck squamous cell carcinoma and patient outcome: A retrospective study. Acta Oncol. 2018, 57, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Fan, H.Y.; Tang, Y.L.; Wang, S.S.; Cao, M.X.; Wang, H.F.; Dai, L.L.; Wang, K.; Yu, X.H.; Wu, J.B.; et al. Myeloid derived suppressor cells contribute to the malignant progression of oral squamous cell carcinoma. PLoS ONE 2020, 15, e0229089. [Google Scholar] [CrossRef]
- Veglia, F.; Perego, M.; Gabrilovich, D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 2018, 19, 108–119. [Google Scholar] [CrossRef]
- Silver, N.; Dourado, J.; Hitchcock, K.; Fullerton, A.; Fredenburg, K.; Dziegielewski, P.; Danan, D.; Tighe, P.; Morris, C.; Amdur, R.; et al. Chronic opioid use in patients undergoing treatment for oropharyngeal cancer. Laryngoscope 2019, 129, 2087–2093. [Google Scholar] [CrossRef]
- Jordan, R.C.; Lingen, M.W.; Perez-Ordonez, B.; He, X.; Pickard, R.; Koluder, M.; Jiang, B.; Wakely, P.; Xiao, W.; Gillison, M.L. Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am. J. Surg. Pathol. 2012, 36, 945–954. [Google Scholar] [CrossRef]
- Hoffmann, M.; Ihloff, A.S.; Gorogh, T.; Weise, J.B.; Fazel, A.; Krams, M.; Rittgen, W.; Schwarz, E.; Kahn, T. p16(INK4a) overexpression predicts translational active human papillomavirus infection in tonsillar cancer. Int. J. Cancer 2010, 127, 1595–1602. [Google Scholar] [CrossRef]
- Friedrich, R.E.; Sperber, C.; Jäkel, T.; Röser, K.; Löning, T. Basaloid lesions of oral squamous epithelial cells and their association with HPV infection and P16 expression. Anticancer Res. 2010, 30, 1605–1612. [Google Scholar] [PubMed]
- Singhi, A.D.; Westra, W.H. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer 2010, 116, 2166–2173. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, J.; Toups, M.; Soos, T.; Arendt, C. Gene signature-based mapping of immunological systems and diseases. BMC Bioinformatics 2016, 17, 171. [Google Scholar] [CrossRef] [PubMed]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. GenePattern 2.0. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 17 November 2019).
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Bratman, S.V.; Bruce, J.P.; O’Sullivan, B.; Pugh, T.J.; Xu, W.; Yip, K.W.; Liu, F.F. Human Papillomavirus Genotype Association With Survival in Head and Neck Squamous Cell Carcinoma. JAMA Oncol. 2016, 2, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Bayik, D.; Tross, D.; Haile, L.A.; Verthelyi, D.; Klinman, D.M. Regulation of the maturation of human monocytes into immunosuppressive macrophages. Blood Adv. 2017, 1, 2510–2519. [Google Scholar] [CrossRef]
- Shabo, I.; Olsson, H.; Elkarim, R.; Sun, X.F.; Svanvik, J. Macrophage Infiltration in Tumor Stroma is Related to Tumor Cell Expression of CD163 in Colorectal Cancer. Cancer Microenviron. 2014, 7, 61–69. [Google Scholar] [CrossRef]
- Shu, Q.H.; Ge, Y.S.; Ma, H.X.; Gao, X.Q.; Pan, J.J.; Liu, D.; Xu, G.L.; Ma, J.L.; Jia, W.D. Prognostic value of polarized macrophages in patients with hepatocellular carcinoma after curative resection. J. Cell Mol. Med. 2016, 20, 1024–1035. [Google Scholar] [CrossRef]
- Mandal, R.; Şenbabaoğlu, Y.; Desrichard, A.; Havel, J.J.; Dalin, M.G.; Riaz, N.; Lee, K.-W.; Ganly, I.; Hakimi, A.A.; Chan, T.A.; et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 2016, 1, e89829. [Google Scholar] [CrossRef]
- Lin, C.H.; Chou, W.C.; Wu, Y.Y.; Lin, C.Y.; Chang, K.P.; Liao, C.T.; Ho, T.Y.; Yeh, C.M.; Liu, C.J.; Hung, S.P.; et al. Prognostic significance of dynamic changes in lymphocyte-to-monocyte ratio in patients with head and neck cancer treated with radiotherapy: Results from a large cohort study. Radiother Oncol. 2021, 154, 76–86. [Google Scholar] [CrossRef]
- Tham, T.; Olson, C.; Khaymovich, J.; Herman, S.W.; Costantino, P.D. The lymphocyte-to-monocyte ratio as a prognostic indicator in head and neck cancer: A systematic review and meta-analysis. Eur. Arch. Otorhinolaryngol. 2018, 275, 1663–1670. [Google Scholar] [CrossRef]
- Ong, H.S.; Gokavarapu, S.; Wang, L.Z.; Tian, Z.; Zhang, C.P. Low Pretreatment Lymphocyte-Monocyte Ratio and High Platelet-Lymphocyte Ratio Indicate Poor Cancer Outcome in Early Tongue Cancer. J. Oral Maxillofac. Surg. 2017, 75, 1762–1774. [Google Scholar] [CrossRef] [PubMed]
- Kano, S.; Homma, A.; Hatakeyama, H.; Mizumachi, T.; Sakashita, T.; Kakizaki, T.; Fukuda, S. Pretreatment lymphocyte-to-monocyte ratio as an independent prognostic factor for head and neck cancer. Head Neck 2017, 39, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Almatroodi, S.A.; McDonald, C.F.; Darby, I.A.; Pouniotis, D.S. Characterization of M1/M2 Tumour-Associated Macrophages (TAMs) and Th1/Th2 Cytokine Profiles in Patients with NSCLC. Cancer Microenviron. 2016, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Troiano, G.; Caponio, V.C.A.; Adipietro, I.; Tepedino, M.; Santoro, R.; Laino, L.; Lo Russo, L.; Cirillo, N.; Lo Muzio, L. Prognostic significance of CD68(+) and CD163(+) tumor associated macrophages in head and neck squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2019, 93, 66–75. [Google Scholar] [CrossRef]
- Kumar, A.T.; Knops, A.; Swendseid, B.; Martinez-Outschoom, U.; Harshyne, L.; Philp, N.; Rodeck, U.; Luginbuhl, A.; Cognetti, D.; Johnson, J.; et al. Prognostic Significance of Tumor-Associated Macrophage Content in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis. Front. Oncol. 2019, 9, 656. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulieres, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Baste, N.; Neupane, P.; Bratland, A.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]

| Patient Characteristics | |||
|---|---|---|---|
| N=37 | % (Total) | ||
| Gender | Male | 30 | 81 |
| Female | 7 | 19 | |
| HPV status | Negative | 11 (6 with recurrence) | 30 |
| Positive | 26 (14 with recurrence) | 70 | |
| Race | White | 31 | 84 |
| African American | 4 | 11 | |
| Other | 2 | 5 | |
| Smoking history | Never | 14 | 38 |
| Current or former | 23 | 62 | |
| Subsite | Tongue Base | 10 | 27 |
| Tonsil | 27 | 73 | |
| N Status | Negative | 9 | 24 |
| Positive | 28 | 76 | |
| Overall Stage | 1 | 11 | 30 |
| 2 | 6 | 16 | |
| 3 | 13 | 35 | |
| 4 | 7 | 19 | |
| Treatment | Chemoradiotherapy | 27 | 73 |
| Radiation alone | 10 | 27 |
| Risk Factor | No Recurrence N = 12 | Recurrence N = 14 | OR | Lower CI | Upper CI | p-Value | ||
|---|---|---|---|---|---|---|---|---|
| Gender | Male | 10 | 14 | 5.9 | 0.1 | 263.8 | 0.362 | |
| Female | 2 | 0 | ||||||
| Smoking Status | Never | 8 | 6 | 2.4 | 0.5 | 11.5 | 0.278 | |
| Former/current | 4 | 8 | ||||||
| Subsite | Tonsil | 11 | 9 | 0.2 | 0 | 1.6 | 0.124 | |
| Tongue | 1 | 5 | ||||||
| T Stage | 1, 2 | 9 | 3 | 0.2 | 0 | 0.9 | 0.035 | |
| 3, 4 | 3 | 11 | ||||||
| N Status | Negative | 2 | 2 | 0.2 | 0.1 | 8.4 | 1 | |
| Positive | 10 | 12 | ||||||
| Treatment | CRT | 7 | 12 | 3 | 0.5 | 18.7 | 0.245 | |
| RT | 5 | 2 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Garg, R.; Fredenburg, K.; Weidert, F.; Mendez-Gomez, H.; Amdur, R.; Lee, J.-H.; Ku, J.; Kresak, J.; Staras, S.; et al. Association of Suppressive Myeloid Cell Enrichment with Aggressive Oropharynx Squamous Cell Carcinoma. Cancers 2023, 15, 2346. https://doi.org/10.3390/cancers15082346
Yang C, Garg R, Fredenburg K, Weidert F, Mendez-Gomez H, Amdur R, Lee J-H, Ku J, Kresak J, Staras S, et al. Association of Suppressive Myeloid Cell Enrichment with Aggressive Oropharynx Squamous Cell Carcinoma. Cancers. 2023; 15(8):2346. https://doi.org/10.3390/cancers15082346
Chicago/Turabian StyleYang, Changlin, Rekha Garg, Kristanna Fredenburg, Frances Weidert, Hector Mendez-Gomez, Robert Amdur, Ji-Hyun Lee, Jamie Ku, Jesse Kresak, Stephanie Staras, and et al. 2023. "Association of Suppressive Myeloid Cell Enrichment with Aggressive Oropharynx Squamous Cell Carcinoma" Cancers 15, no. 8: 2346. https://doi.org/10.3390/cancers15082346
APA StyleYang, C., Garg, R., Fredenburg, K., Weidert, F., Mendez-Gomez, H., Amdur, R., Lee, J.-H., Ku, J., Kresak, J., Staras, S., Sikora, A. G., Wang, L., McGrail, D., Mitchell, D., Sayour, E., & Silver, N. (2023). Association of Suppressive Myeloid Cell Enrichment with Aggressive Oropharynx Squamous Cell Carcinoma. Cancers, 15(8), 2346. https://doi.org/10.3390/cancers15082346






