Prediction of Deep Myometrial Infiltration, Clinical Risk Category, Histological Type, and Lymphovascular Space Invasion in Women with Endometrial Cancer Based on Clinical and T2-Weighted MRI Radiomic Features
Abstract
Simple Summary
Abstract
1. Introduction
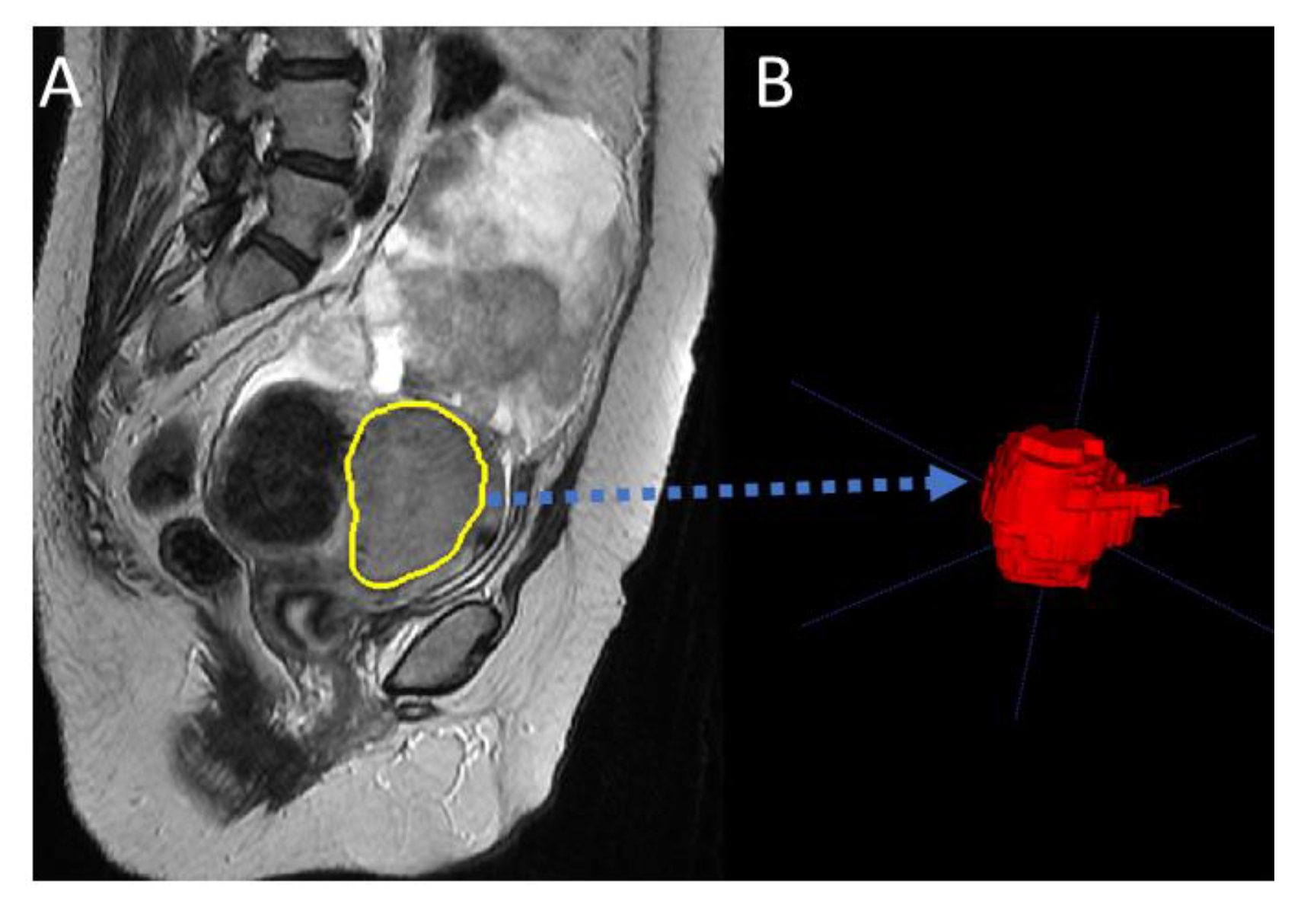
2. Materials and Methods
2.1. Training and Testing Datasets
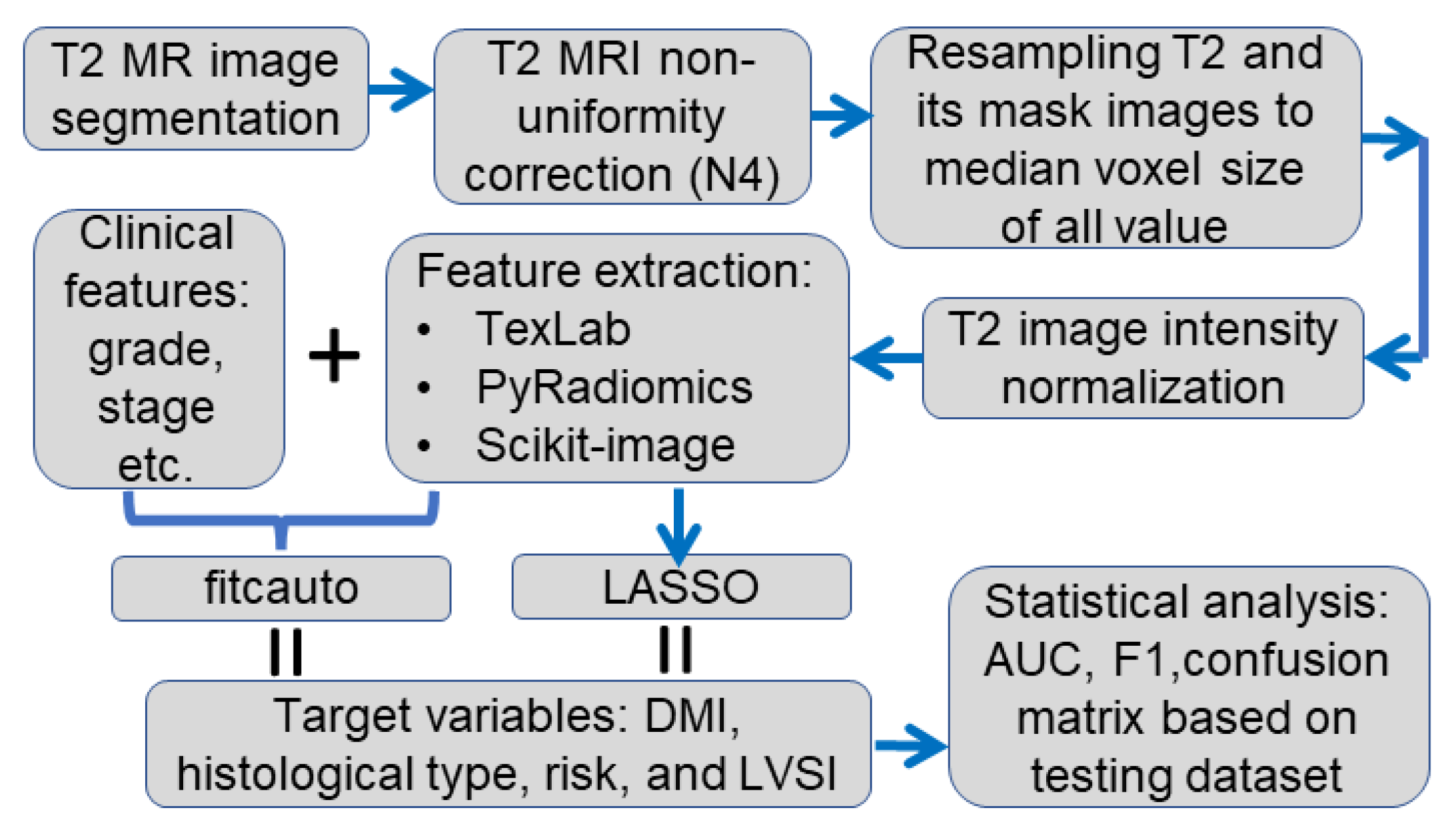
2.2. Data Analysis Pipeline
2.3. Patient DMI
2.4. Cohort Treatment and Clinical Risk Classification
2.5. Statistical Analysis
3. Results
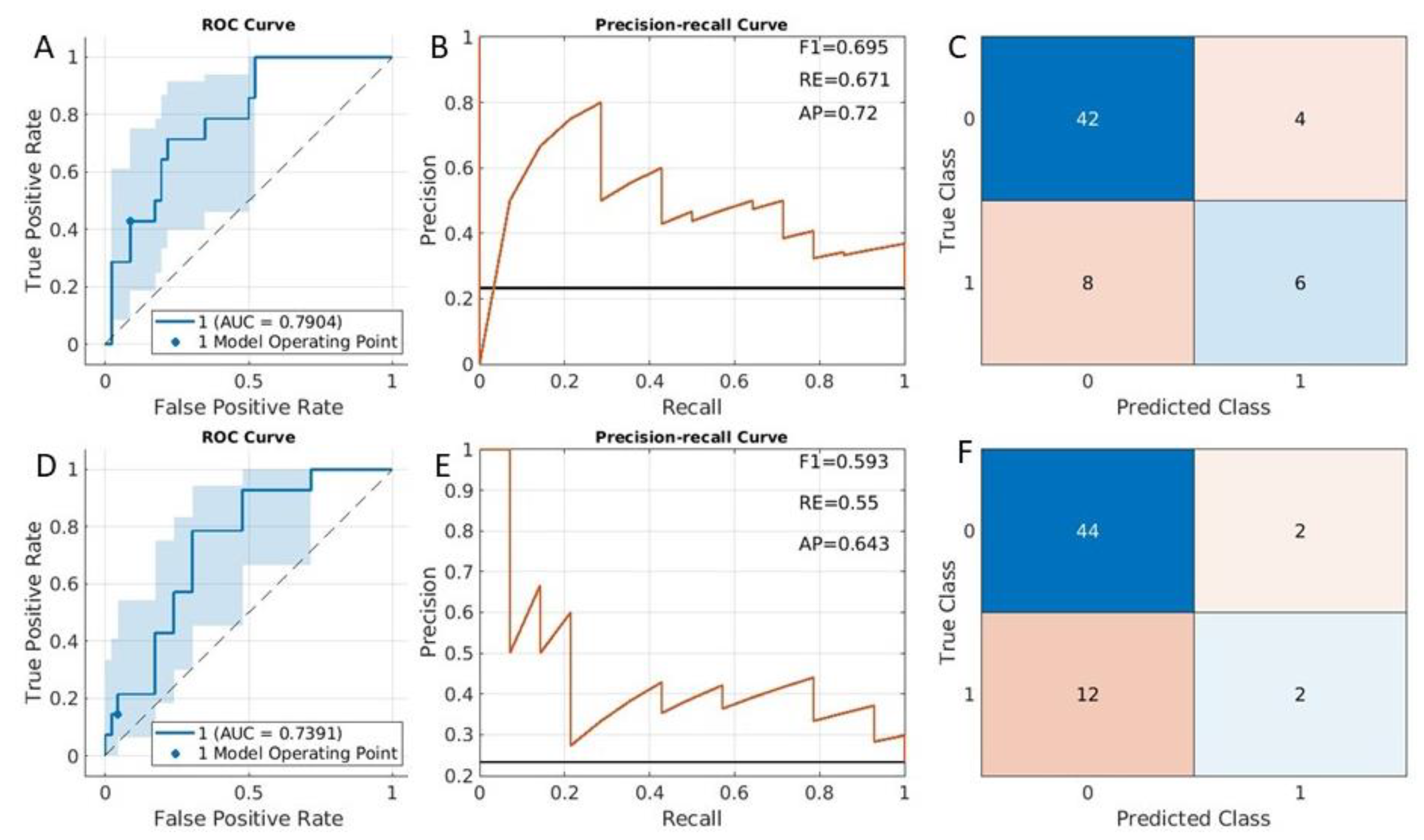
3.1. DMI Prediction Results
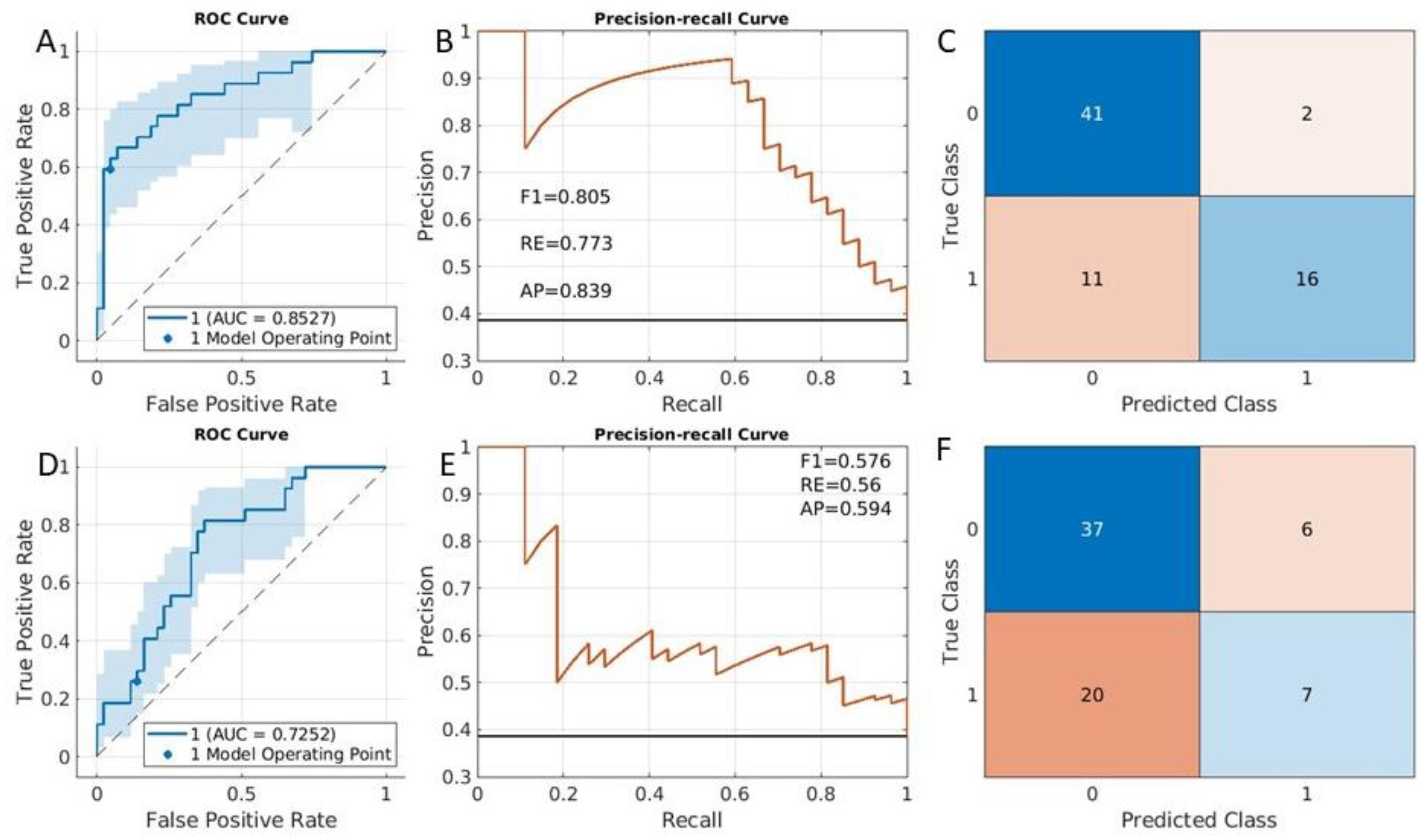
3.2. Clinical Risk Classification
3.3. Histological Type Classification
3.4. LVSI Classification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koskas, M.; Amant, F.; Mirza, M.R.; Creutzberg, C.L. Cancer of the corpus uteri: 2021 update. Int. J. Gynecol. Obstet. 2021, 155, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Nougaret, S.; Horta, M.; Sala, E.; Lakhman, Y.; Thomassin-Naggara, I.; Kido, A.; Masselli, G.; Bharwani, N.; Sadowski, E.; Ertmer, A.; et al. Endometrial Cancer MRI staging: Updated Guidelines of the European Society of Urogenital Radiology. Eur. Radiol. 2019, 29, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, R.; Gui, B.; Maresca, G.; Fanfani, F.; Bonomo, L. Endometrial cancer: Magnetic resonance imaging. Abdom. Imag. 2005, 30, 626–636. [Google Scholar] [CrossRef]
- Bouche, C.; Gomes David, M.; Salleron, J.; Rauch, P.; Leufflen, L.; Buhler, J.; Marchal, F. Evaluation of Pre-Therapeutic Assessment in Endometrial Cancer Staging. Diagnostics 2020, 10, 1045. [Google Scholar] [CrossRef]
- Soneji, N.D.; Bharwani, N.; Ferri, A.; Stewart, V.; Rockall, A. Pre-operative MRI staging of endometrial cancer in a multicentre cancer network: Can we match single centre study results? Eur. Radiol. 2018, 28, 4725–4734. [Google Scholar] [CrossRef]
- Fasmer, K.E.; Hodneland, E.; Dybvik, J.A.; Wagner-Larsen, K.; Trovik, J.; Salvesen, Ø.; Krakstad, C.; Haldorsen, I.H. Whole-volume tumor MRI radiomics for prognostic modeling in endometrial cancer. J. Magn. Reson. Imag. 2021, 53, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Beddy, P.; Moyle, P.; Kataoka, M.; Yamamoto, A.K.; Joubert, I.; Lomas, D.; Crawford, R.; Sala, E. Evaluation of Depth of Myometrial Invasion and Overall Staging in Endometrial Cancer: Comparison of Diffusion-weighted and Dynamic Contrast-enhanced MR Imaging. Radiology 2012, 262, 530–537. [Google Scholar] [CrossRef]
- Lefebvre, T.L.; Ueno, Y.; Dohan, A.; Chatterjee, A.; Vallières, M.; Winter-Reinhold, E.; Saif, S.; Levesque, I.R.; Zeng, X.Z.; Forghani, R.; et al. Development and Validation of Multiparametric MRI–based Radiomics Models for Preoperative Risk Stratification of Endometrial Cancer. Radiology 2022, 305, 375–386. [Google Scholar] [CrossRef]
- Miccò, M.; Gui, B.; Russo, L.; Boldrini, L.; Lenkowicz, J.; Cicogna, S.; Cosentino, F.; Restaino, G.; Avesani, G.; Panico, C.; et al. Preoperative Tumor Texture Analysis on MRI for High-Risk Disease Prediction in Endometrial Cancer: A Hypothesis-Generating Study. J. Pers. Med. 2022, 12, 1854. [Google Scholar] [CrossRef]
- Stanzione, A.; Cuocolo, R.; Del Grosso, R.; Nardiello, A.; Romeo, V.; Travaglino, A.; Raffone, A.; Bifulco, G.; Zullo, F.; Insabato, L.; et al. Deep Myometrial Infiltration of Endometrial Cancer on MRI: A Radiomics-Powered Machine Learning Pilot Study. Acad. Radiol. 2021, 28, 737–744. [Google Scholar] [CrossRef]
- Jónsdóttir, B.; Marcickiewicz, J.; Borgfeldt, C.; Bjurberg, M.; Dahm-Kähler, P.; Flöter-Rådestad, A.; Hellman, K.; Holmberg, E.; Kjølhede, P.; Rosenberg, P.; et al. Preoperative and intraoperative assessment of myometrial invasion in endometrial cancer—A Swedish Gynecologic Cancer Group (SweGCG) study. Acta Obstet. Gynecol. Scand. 2021, 100, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.; Amin, T.; Thanatsis, N.; Naftalin, J.; Jurkovic, D. A prospective comparison of the diagnostic accuracies of ultrasound and magnetic resonance imaging in preoperative staging of endometrial cancer. J. Gynecol. Oncol. 2022, 33, e22. [Google Scholar] [CrossRef]
- Dickson Michelson, E.L.; Kram, J.J.F.; Heslin, K.; Baugh, D.; Bamra, V.; Hu, J.; Shukla, A.; Kamelle, S.A. Can Magnetic Resonance Imaging Predict Pathologic Findings for Endometrioid Endometrial Cancer? J. Patient Cent. Res. Rev. 2020, 7, 206–212. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Barr, C.E.; Crosbie, E.J. Current and Emerging Prognostic Biomarkers in Endometrial Cancer. Front. Oncol. 2022, 12, 890908. [Google Scholar] [CrossRef]
- Manganaro, L.; Nicolino, G.M.; Dolciami, M.; Martorana, F.; Stathis, A.; Colombo, I.; Rizzo, S. Radiomics in cervical and endometrial cancer. Br. J. Radiol. 2021, 94, 20201314. [Google Scholar] [CrossRef]
- Pecorelli, S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int. J. Gynaecol. Obstet. 2009, 150, 103–104. [Google Scholar] [CrossRef]
- Mendiola, M.; Pellinen, T.; Ramon-Patino, J.L.; Berjon, A.; Bruck, O.; Heredia-Soto, V.; Turkki, R.; Escudero, J.; Hemmes, A.; Garcia de la Calle, L.E.; et al. Prognostic implications of tumor-infiltrating T cells in early-stage endometrial cancer. Mod. Pathol. 2022, 35, 256–265. [Google Scholar] [CrossRef]
- Mainenti, P.P.; Stanzione, A.; Cuocolo, R.; del Grosso, R.; Danzi, R.; Romeo, V.; Raffone, A.; Di Spiezio Sardo, A.; Giordano, E.; Travaglino, A.; et al. MRI radiomics: A machine learning approach for the risk stratification of endometrial cancer patients. Eur. J. Radiol. 2022, 149, 110226. [Google Scholar] [CrossRef]
- Ghosh, A.; Singh, T.; Singla, V.; Bagga, R.; Srinivasan, R.; Khandelwal, N. DTI histogram parameters correlate with the extent of myoinvasion and tumor type in endometrial carcinoma: A preliminary analysis. Acta Radiol. 2020, 61, 675–684. [Google Scholar] [CrossRef]
- Celli, V.; Guerreri, M.; Pernazza, A.; Cuccu, I.; Palaia, I.; Tomao, F.; Di Donato, V.; Pricolo, P.; Ercolani, G.; Ciulla, S.; et al. MRI- and Histologic-Molecular-Based Radio-Genomics Nomogram for Preoperative Assessment of Risk Classes in Endometrial Cancer. Cancers 2022, 14, 5881. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, D.; Kim, H.S.; Park, S.Y.; Kim, J.Y.; Cho, S.J.; Shin, J.H.; Kim, J.H. Quality of science and reporting of radiomics in oncologic studies: Room for improvement according to radiomics quality score and TRIPOD statement. Eur. Radiol. 2020, 30, 523–536. [Google Scholar] [CrossRef]
- Li, X.; Marcus, D.; Russell, J.; Aboagye, E.O.; Ellis, L.B.; Sheeka, A.; Park, W.-H.E.; Bharwani, N.; Ghaem-Maghami, S.; Rockall, A.G. An Integrated Clinical-MR Radiomics Model to Estimate Survival Time in Patients With Endometrial Cancer. J. Magn. Reson. Imag. 2022. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imag. 2010, 29, 1310–1320. [Google Scholar] [CrossRef] [PubMed]
- Colombo, N.; Creutzberg, C.; Amant, F.; Bosse, T.; González-Martín, A.; Ledermann, J.; Marth, C.; Nout, R.; Querleu, D.; Mirza, M.R.; et al. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: Diagnosis, treatment and follow-up. Ann. Oncol. 2016, 27, 16–41. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Powers, D.M. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. preprint arXiv 2010, 16061, 2020. [Google Scholar]
- Abu-Rustum, N.R.; Zhou, Q.; Gomez, J.D.; Alektiar, K.M.; Hensley, M.L.; Soslow, R.A.; Levine, D.A.; Chi, D.S.; Barakat, R.R.; Iasonos, A. A nomogram for predicting overall survival of women with endometrial cancer following primary therapy: Toward improving individualized cancer care. Gynecol. Oncol. 2010, 116, 399–403. [Google Scholar] [CrossRef]
- Watkins, C.; Huang, X.; Latimer, N.; Tang, Y.; Wright, E.J. Adjusting overall survival for treatment switches: Commonly used methods and practical application. Pharm. Stat. 2013, 12, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Gottwald, L.; Pluta, P.; Piekarski, J.; Spych, M.; Hendzel, K.; Topczewska-Tylinska, K.; Nejc, D.; Bibik, R.; Korczyński, J.; Ciałkowska-Rysz, A. Long-term survival of endometrioid endometrial cancer patients. Arch. Med. Sci. AMS 2010, 6, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Stalberg, K.; Bjurberg, M.; Borgfeldt, C.; Carlson, J.; Dahm-Kahler, P.; Floter-Radestad, A.; Hellman, K.; Hjerpe, E.; Holmberg, E.; Kjolhede, P.; et al. Lymphovascular space invasion as a predictive factor for lymph node metastases and survival in endometrioid endometrial cancer–A Swedish Gynecologic Cancer Group (SweGCG) study. Acta Oncol. 2019, 58, 1628–1633. [Google Scholar] [CrossRef]
- Ortoft, G.; Lausten-Thomsen, L.; Hogdall, C.; Hansen, E.S.; Dueholm, M. Lymph-vascular space invasion (LVSI) as a strong and independent predictor for non-locoregional recurrences in endometrial cancer: A Danish Gynecological Cancer Group Study. J. Gynecol. Oncol. 2019, 30, e84. [Google Scholar] [CrossRef]
- Bereby-Kahane, M.; Dautry, R.; Matzner-Lober, E.; Cornelis, F.; Sebbag-Sfez, D.; Place, V.; Mezzadri, M.; Soyer, P.; Dohan, A. Prediction of tumor grade and lymphovascular space invasion in endometrial adenocarcinoma with MR imaging-based radiomic analysis. Diagn. Interv. Imag. 2020, 101, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef]


| Target Variable | Training (n) | Training (%) | Testing (n) | Testing (%) |
|---|---|---|---|---|
| DMI | ||||
| no DMI | 199 | 68% | 46 | 76% |
| DMI | 93 | 32% | 14 | 24% |
| Clinical risk | ||||
| Low | 150 | 36% | 41 | 50% |
| High | 263 | 64% | 40 | 50% |
| Histological type | ||||
| Endometrioid | 301 | 73% | 70 | 89% |
| Other types | 111 | 27% | 9 | 11% |
| LVSI | ||||
| Positive | 141 | 36% | 27 | 39% |
| Negative | 248 | 64% | 43 | 61% |
| Risk Classification | Criteria |
|---|---|
| Low | Stage 1A endometrioid grade 1–2 |
| Intermediate | Stage 1A endometrioid grade 3 Stage IB (Grade 1 and Grade 2) with endometrioid type |
| High | Stage IB Grade 3 endometrioid type Stage 2 endometrioid type Stage 1 or 2 non-endometrioid type. |
| Advanced | Stage 3 or 4 any type |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Dessi, M.; Marcus, D.; Russell, J.; Aboagye, E.O.; Ellis, L.B.; Sheeka, A.; Park, W.-H.E.; Bharwani, N.; Ghaem-Maghami, S.; et al. Prediction of Deep Myometrial Infiltration, Clinical Risk Category, Histological Type, and Lymphovascular Space Invasion in Women with Endometrial Cancer Based on Clinical and T2-Weighted MRI Radiomic Features. Cancers 2023, 15, 2209. https://doi.org/10.3390/cancers15082209
Li X, Dessi M, Marcus D, Russell J, Aboagye EO, Ellis LB, Sheeka A, Park W-HE, Bharwani N, Ghaem-Maghami S, et al. Prediction of Deep Myometrial Infiltration, Clinical Risk Category, Histological Type, and Lymphovascular Space Invasion in Women with Endometrial Cancer Based on Clinical and T2-Weighted MRI Radiomic Features. Cancers. 2023; 15(8):2209. https://doi.org/10.3390/cancers15082209
Chicago/Turabian StyleLi, Xingfeng, Michele Dessi, Diana Marcus, James Russell, Eric O. Aboagye, Laura Burney Ellis, Alexander Sheeka, Won-Ho Edward Park, Nishat Bharwani, Sadaf Ghaem-Maghami, and et al. 2023. "Prediction of Deep Myometrial Infiltration, Clinical Risk Category, Histological Type, and Lymphovascular Space Invasion in Women with Endometrial Cancer Based on Clinical and T2-Weighted MRI Radiomic Features" Cancers 15, no. 8: 2209. https://doi.org/10.3390/cancers15082209
APA StyleLi, X., Dessi, M., Marcus, D., Russell, J., Aboagye, E. O., Ellis, L. B., Sheeka, A., Park, W.-H. E., Bharwani, N., Ghaem-Maghami, S., & Rockall, A. G. (2023). Prediction of Deep Myometrial Infiltration, Clinical Risk Category, Histological Type, and Lymphovascular Space Invasion in Women with Endometrial Cancer Based on Clinical and T2-Weighted MRI Radiomic Features. Cancers, 15(8), 2209. https://doi.org/10.3390/cancers15082209






