MALDI Imaging, a Powerful Multiplex Approach to Decipher Intratumoral Heterogeneity: Combined Hepato-Cholangiocarcinomas as Proof of Concept
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. cHCC-CCA Cases Selection
2.2. MALDI Imaging
Sample Treatment and Acquisition
2.3. Data Processing
2.4. Informed Consent and Ethical Approval
3. Results
3.1. cHCC-CCA Patients
3.2. Segmentation Analysis Based on the MALDI-MSI Data
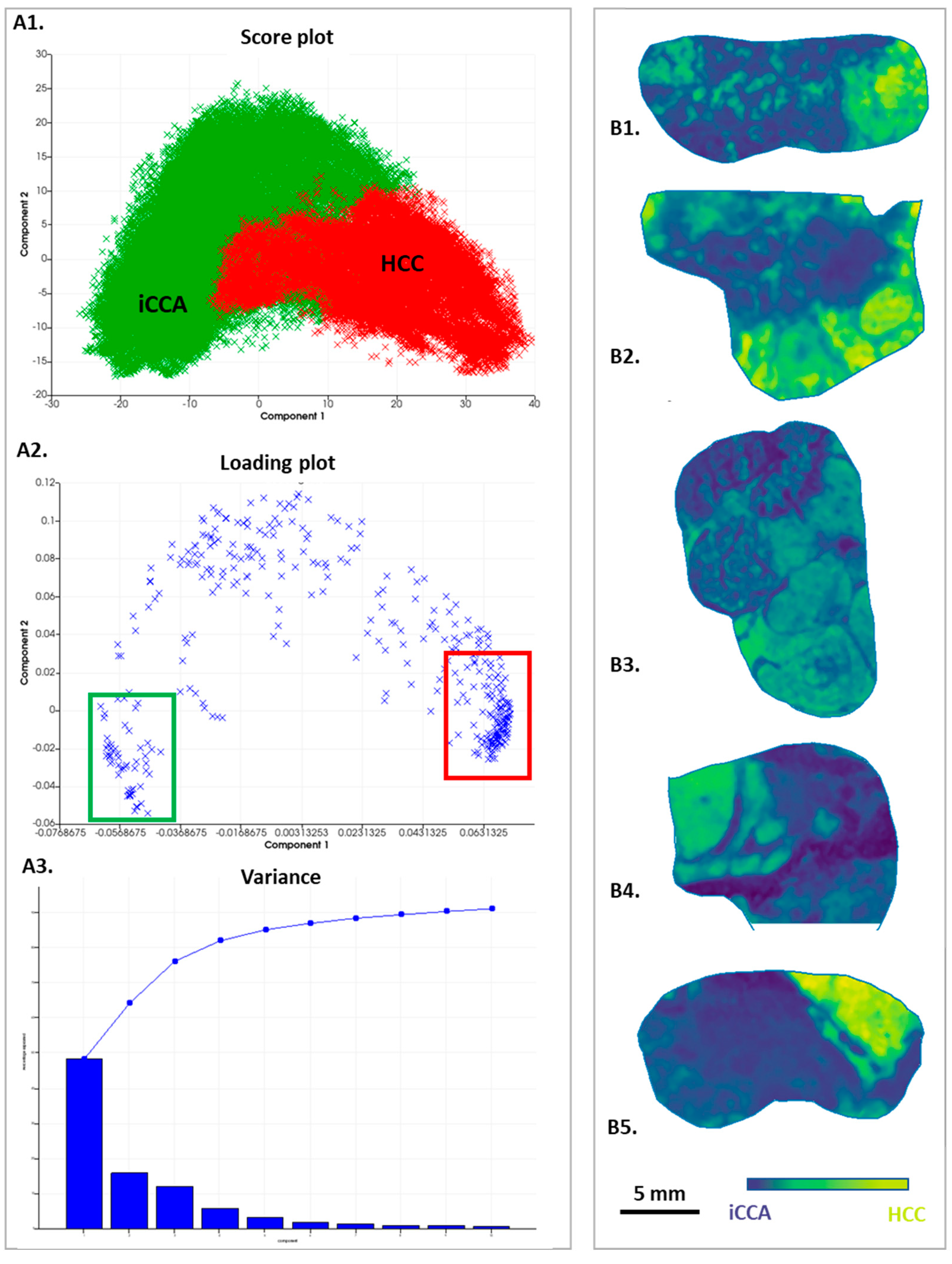
3.3. Analysis by Principal Component Analysis
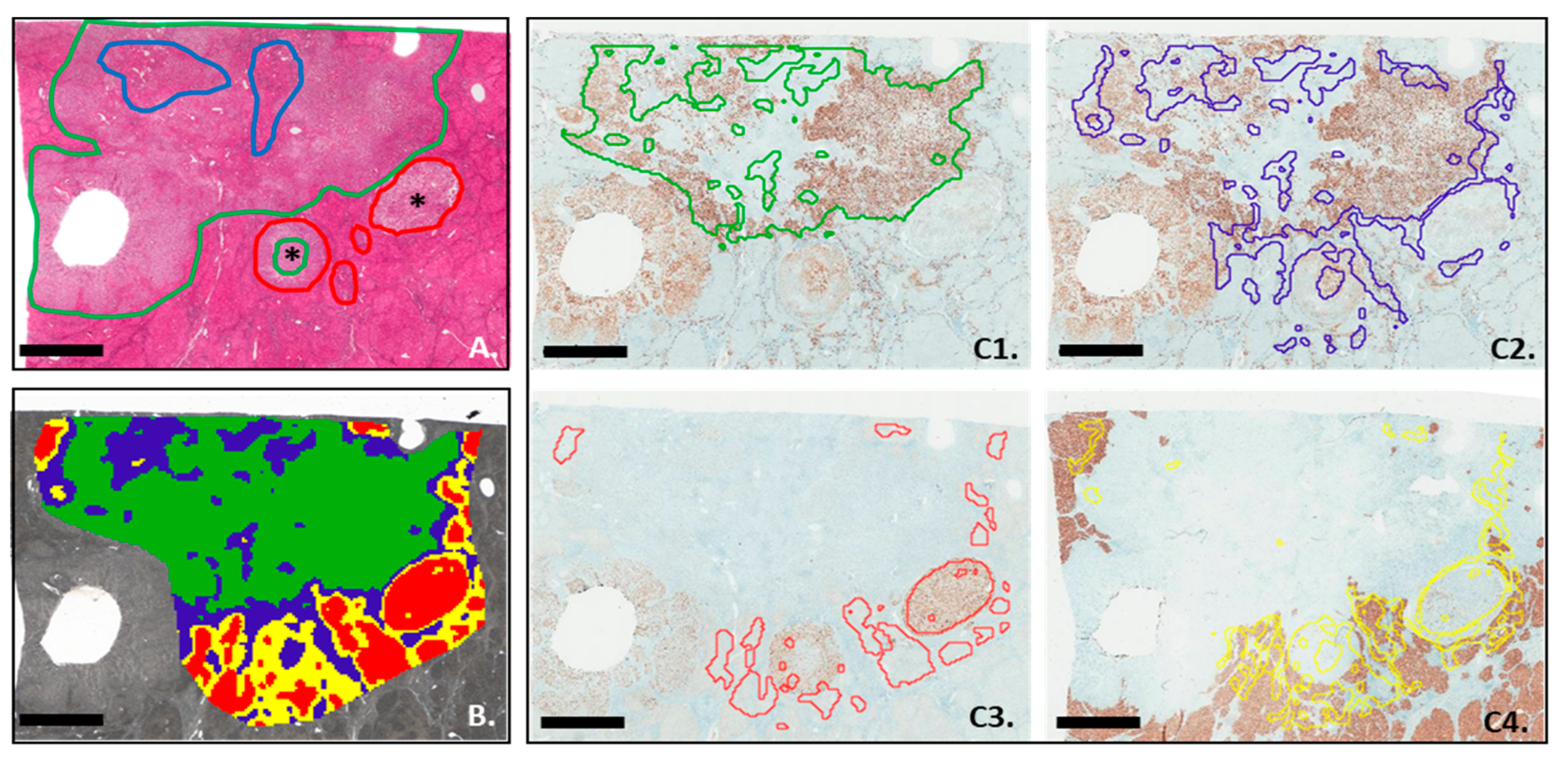
3.4. MALDI-MSI Segmentation and IHC Analysis
3.5. Comparison between In silico Digestion and IHC Analysis
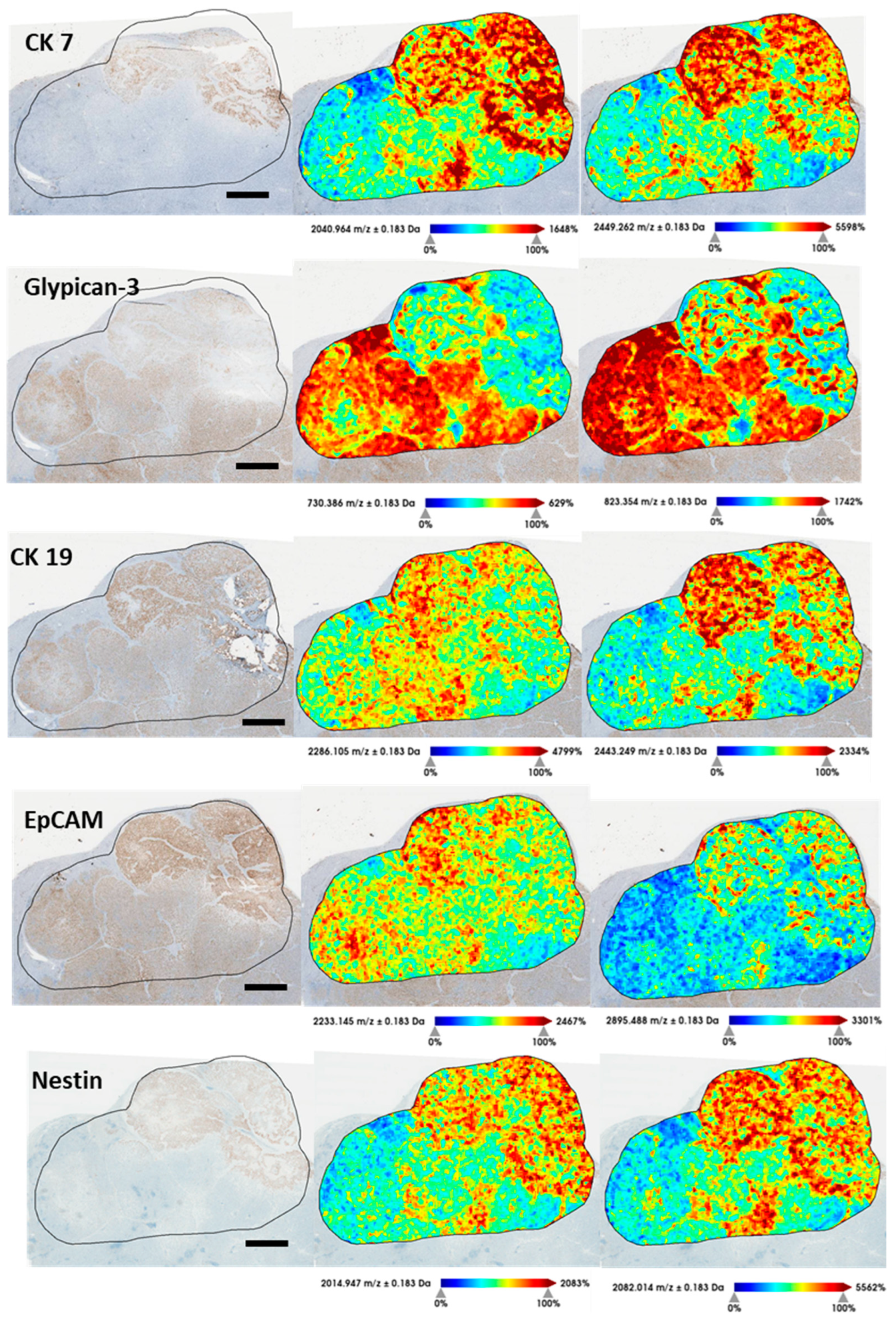
3.6. Identification of Proteins of Interest
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gigante, E.; Paradis, V.; Ronot, M.; Cauchy, F.; Soubrane, O.; Ganne-Carrié, N.; Nault, J.-C. New Insights into the Pathophysiology and Clinical Care of Rare Primary Liver Cancers. JHEP Rep. Innov. Hepatol. 2021, 3, 100174. [Google Scholar] [CrossRef] [PubMed]
- Beaufrère, A.; Calderaro, J.; Paradis, V. Combined Hepatocellular-Cholangiocarcinoma: An Update. J. Hepatol. 2021, 74, 1212–1224. [Google Scholar] [CrossRef] [PubMed]
- Sempoux, C.; Kakar, S.; Kondo, F.; Schirmacher, P. Combined Hepatocellular-Cholangiocarcinoma and Undifferentiated Primary Liver Carcinoma. WHO Classification of Tumours: Digestive System Tumours, 5th ed.; Arends, Ed.; IARC: Lyon, France, 2019. [Google Scholar]
- Brunt, E.; Aishima, S.; Clavien, P.-A.; Fowler, K.; Goodman, Z.; Gores, G.; Gouw, A.; Kagen, A.; Klimstra, D.; Komuta, M.; et al. CHCC-CCA: Consensus Terminology for Primary Liver Carcinomas with Both Hepatocytic and Cholangiocytic Differentation. Hepatology 2018, 68, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, J.; Di Tommaso, L.; Maillé, P.; Beaufrère, A.; Nguyen, C.T.; Heij, L.; Gnemmi, V.; Graham, R.P.; Charlotte, F.; Chartier, S.; et al. Nestin as a Diagnostic and Prognostic Marker for Combined Hepatocellular-Cholangiocarcinoma. J. Hepatol. 2022, 77, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Joseph, N.M.; Tsokos, C.G.; Umetsu, S.E.; Shain, A.H.; Kelley, R.K.; Onodera, C.; Bowman, S.; Talevich, E.; Ferrell, L.D.; Kakar, S.; et al. Genomic Profiling of Combined Hepatocellular-Cholangiocarcinoma Reveals Similar Genetics to Hepatocellular Carcinoma. J. Pathol. 2019, 248, 164–178. [Google Scholar] [CrossRef]
- Wang, A.; Wu, L.; Lin, J.; Han, L.; Bian, J.; Wu, Y.; Robson, S.C.; Xue, L.; Ge, Y.; Sang, X.; et al. Whole-Exome Sequencing Reveals the Origin and Evolution of Hepato-Cholangiocarcinoma. Nat. Commun. 2018, 9, 894. [Google Scholar] [CrossRef]
- Murugesan, K.; Sharaf, R.; Montesion, M.; Moore, J.A.; Pao, J.; Pavlick, D.C.; Frampton, G.M.; Upadhyay, V.A.; Alexander, B.M.; Miller, V.A.; et al. Genomic Profiling of Combined Hepatocellular Cholangiocarcinoma Reveals Genomics Similar to Either Hepatocellular Carcinoma or Cholangiocarcinoma. JCO Precis. Oncol. 2021, 5, PO.20.00397. [Google Scholar] [CrossRef]
- Caprioli, R.M. Imaging Mass Spectrometry: Molecular Microscopy for the New Age of Biology and Medicine. Proteomics 2016, 16, 1607–1612. [Google Scholar] [CrossRef]
- Le Faouder, J.; Laouirem, S.; Alexandrov, T.; Ben-Harzallah, S.; Léger, T.; Albuquerque, M.; Bedossa, P.; Paradis, V. Tumoral Heterogeneity of Hepatic Cholangiocarcinomas Revealed by MALDI Imaging Mass Spectrometry. Proteomics 2014, 14, 965–972. [Google Scholar] [CrossRef]
- Laouirem, S.; Le Faouder, J.; Alexandrov, T.; Mestivier, D.; Léger, T.; Baudin, X.; Mebarki, M.; Paradis, V.; Camadro, J.-M.; Bedossa, P. Progression from Cirrhosis to Cancer Is Associated with Early Ubiquitin Post-Translational Modifications: Identification of New Biomarkers of Cirrhosis at Risk of Malignancy. J. Pathol. 2014, 234, 452–463. [Google Scholar] [CrossRef]
- Poté, N.; Alexandrov, T.; Le Faouder, J.; Laouirem, S.; Léger, T.; Mebarki, M.; Belghiti, J.; Camadro, J.-M.; Bedossa, P.; Paradis, V. Imaging Mass Spectrometry Reveals Modified Forms of Histone H4 as New Biomarkers of Microvascular Invasion in Hepatocellular Carcinomas. Hepatology 2013, 58, 983–994. [Google Scholar] [CrossRef]
- Basu, S.S.; Agar, N.Y.R. Bringing Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging to the Clinics. Clin. Lab. Med. 2021, 41, 309–324. [Google Scholar] [CrossRef]
- Balluff, B.; Frese, C.K.; Maier, S.K.; Schöne, C.; Kuster, B.; Schmitt, M.; Aubele, M.; Höfler, H.; Deelder, A.M.; Heck, A.; et al. De Novo Discovery of Phenotypic Intratumour Heterogeneity Using Imaging Mass Spectrometry. J. Pathol. 2015, 235, 3–13. [Google Scholar] [CrossRef]
- Boskamp, T.; Lachmund, D.; Casadonte, R.; Hauberg-Lotte, L.; Kobarg, J.H.; Kriegsmann, J.; Maass, P. Using the Chemical Noise Background in MALDI Mass Spectrometry Imaging for Mass Alignment and Calibration. Anal. Chem. 2020, 92, 1301–1308. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Lindskog, I.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Hochstrasser, D.F.; Appel, R.D. Detailed Peptide Characterization Using PEPTIDEMASS—A World-Wide-Web-Accessible Tool. Electrophoresis 1997, 18, 403–408. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Schaeffer, M.; Gateau, A.; Teixeira, D.; Michel, P.-A.; Zahn-Zabal, M.; Lane, L. The NeXtProt Peptide Uniqueness Checker: A Tool for the Proteomics Community. Bioinformatics 2017, 33, 3471–3472. [Google Scholar] [CrossRef]
- Claes, B.S.R.; Krestensen, K.K.; Yagnik, G.; Grgic, A.; Kuik, C.; Lim, M.J.; Rothschild, K.J.; Vandenbosch, M.; Heeren, R.M.A. MALDI-IHC-Guided In-Depth Spatial Proteomics: Targeted and Untargeted MSI Combined. Anal. Chem. 2023, 95, 2329–2338. [Google Scholar] [CrossRef]
- Giordano, S.; Takeda, S.; Donadon, M.; Saiki, H.; Brunelli, L.; Pastorelli, R.; Cimino, M.; Soldani, C.; Franceschini, B.; Di Tommaso, L.; et al. Rapid Automated Diagnosis of Primary Hepatic Tumour by Mass Spectrometry and Artificial Intelligence. Liver Int. 2020, 40, 3117–3124. [Google Scholar] [CrossRef]
- Giordano, S.; Siciliano, A.M.; Donadon, M.; Soldani, C.; Franceschini, B.; Lleo, A.; Di Tommaso, L.; Cimino, M.; Torzilli, G.; Saiki, H.; et al. Versatile Mass Spectrometry-Based Intraoperative Diagnosis of Liver Tumor in a Multiethnic Cohort. Appl. Sci. 2022, 12, 4244. [Google Scholar] [CrossRef]
- Roth, L.; Srivastava, S.; Lindzen, M.; Sas-Chen, A.; Sheffer, M.; Lauriola, M.; Enuka, Y.; Noronha, A.; Mancini, M.; Lavi, S.; et al. SILAC Identifies LAD1 as a Filamin-Binding Regulator of Actin Dynamics in Response to EGF and a Marker of Aggressive Breast Tumors. Sci. Signal. 2018, 11, eaan0949. [Google Scholar] [CrossRef] [PubMed]
- Sohoni, S.; Ghosh, P.; Wang, T.; Kalainayakan, S.P.; Vidal, C.; Dey, S.; Konduri, P.C.; Zhang, L. Elevated Heme Synthesis and Uptake Underpin Intensified Oxidative Metabolism and Tumorigenic Functions in Non-Small Cell Lung Cancer Cells. Cancer Res. 2019, 79, 2511–2525. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, X.; Liu, Y.; Ma, Y.; Kong, P.; Bai, T.; Han, M.; Li, B. Inhibition of ALAS1 Activity Exerts Anti-Tumour Effects on Colorectal Cancer in Vitro. Saudi J. Gastroenterol. Off. J. Saudi Gastroenterol. Assoc. 2020, 26, 144–152. [Google Scholar] [CrossRef]
- Wang, Y.; Han, J.; Zhou, H.; Ai, S.; Wan, D. A Prognosis Marker Dynein Cytoplasmic 1 Heavy Chain 1 Correlates with EMT and Immune Signature in Liver Hepatocellular Carcinoma by Bioinformatics and Experimental Analysis. Dis. Markers 2022, 2022, 6304859. [Google Scholar] [CrossRef]
- Huang, H.-L.; Yao, H.-S.; Wang, Y.; Wang, W.-J.; Hu, Z.-Q.; Jin, K.-Z. Proteomic Identification of Tumor Biomarkers Associated with Primary Gallbladder Cancer. World J. Gastroenterol. WJG 2014, 20, 5511–5518. [Google Scholar] [CrossRef]
- Barry, J.A.; Ait-Belkacem, R.; Hardesty, W.M.; Benakli, L.; Andonian, C.; Licea-Perez, H.; Stauber, J.; Castellino, S. Multicenter Validation Study of Quantitative Imaging Mass Spectrometry. Anal. Chem. 2019, 91, 6266–6274. [Google Scholar] [CrossRef]
- Deininger, S.-O.; Bollwein, C.; Casadonte, R.; Wandernoth, P.; Gonçalves, J.P.L.; Kriegsmann, K.; Kriegsmann, M.; Boskamp, T.; Kriegsmann, J.; Weichert, W.; et al. Multicenter Evaluation of Tissue Classification by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Anal. Chem. 2022, 94, 8194–8201. [Google Scholar] [CrossRef]
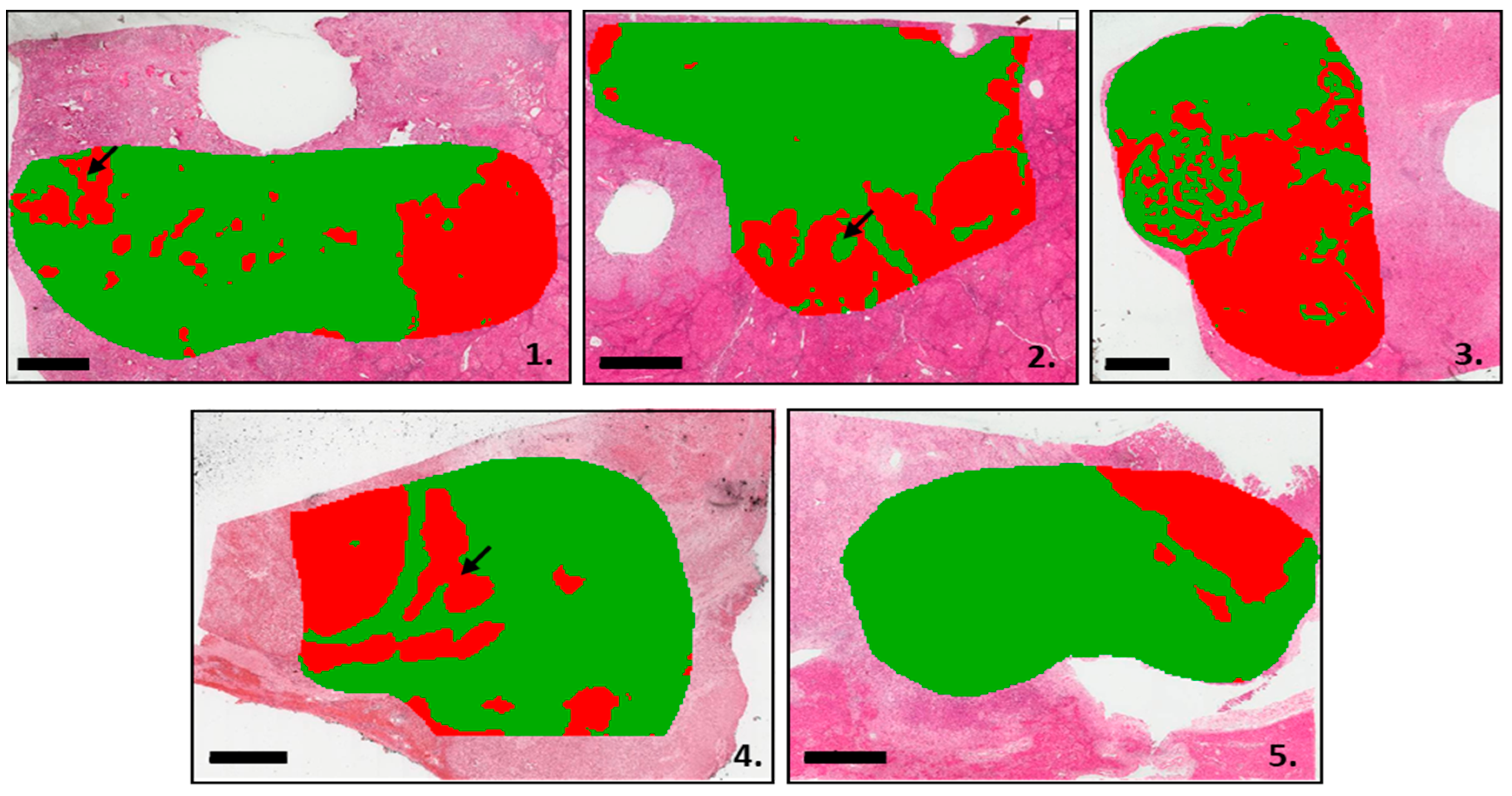

| Sex | Age (Y) | Advanced Fibrosis (≥F3) | Risk Factors | BMI | Tobacco Smoke | Satellite Nodules | Vascular Invasion | Perineural Invasion | Tumor Size (mm) |
|---|---|---|---|---|---|---|---|---|---|
| F | 65 | Yes | Alcohol, MS | 40 | Yes | Yes | Yes | Yes | 42 |
| M | 57 | Yes | Alcohol, MS | 27 | Yes | No | Yes | Yes | 25 |
| F | 29 | Yes | Genetic | 44 | Yes | No | No | No | 35 |
| M | 40 | No | HBV | 31 | Yes | No | Yes | Yes | 40 |
| M | 77 | No | MS | 24 | Yes | Yes | Yes | Yes | 70 |
| iCCA (Green) | HCC (Red) | ||||
|---|---|---|---|---|---|
| Patients | Total of Spectra | Number of Spectra | % | Number of Spectra | % |
| 1 | 20,438 | 14,602 | 71.4 | 5836 | 28.6 |
| 2 | 14,968 | 10,549 | 70.5 | 4419 | 29.5 |
| 3 | 20,650 | 9612 | 46.5 | 11038 | 53.5 |
| 4 | 16,360 | 11,584 | 70.8 | 4776 | 29.2 |
| 5 | 14,158 | 11,843 | 83.6 | 2315 | 16.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gigante, E.; Cazier, H.; Albuquerque, M.; Laouirem, S.; Beaufrère, A.; Paradis, V. MALDI Imaging, a Powerful Multiplex Approach to Decipher Intratumoral Heterogeneity: Combined Hepato-Cholangiocarcinomas as Proof of Concept. Cancers 2023, 15, 2143. https://doi.org/10.3390/cancers15072143
Gigante E, Cazier H, Albuquerque M, Laouirem S, Beaufrère A, Paradis V. MALDI Imaging, a Powerful Multiplex Approach to Decipher Intratumoral Heterogeneity: Combined Hepato-Cholangiocarcinomas as Proof of Concept. Cancers. 2023; 15(7):2143. https://doi.org/10.3390/cancers15072143
Chicago/Turabian StyleGigante, Elia, Hélène Cazier, Miguel Albuquerque, Samira Laouirem, Aurélie Beaufrère, and Valérie Paradis. 2023. "MALDI Imaging, a Powerful Multiplex Approach to Decipher Intratumoral Heterogeneity: Combined Hepato-Cholangiocarcinomas as Proof of Concept" Cancers 15, no. 7: 2143. https://doi.org/10.3390/cancers15072143
APA StyleGigante, E., Cazier, H., Albuquerque, M., Laouirem, S., Beaufrère, A., & Paradis, V. (2023). MALDI Imaging, a Powerful Multiplex Approach to Decipher Intratumoral Heterogeneity: Combined Hepato-Cholangiocarcinomas as Proof of Concept. Cancers, 15(7), 2143. https://doi.org/10.3390/cancers15072143






