Deep Learning Model Based on Contrast-Enhanced Computed Tomography Imaging to Predict Postoperative Early Recurrence after the Curative Resection of a Solitary Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
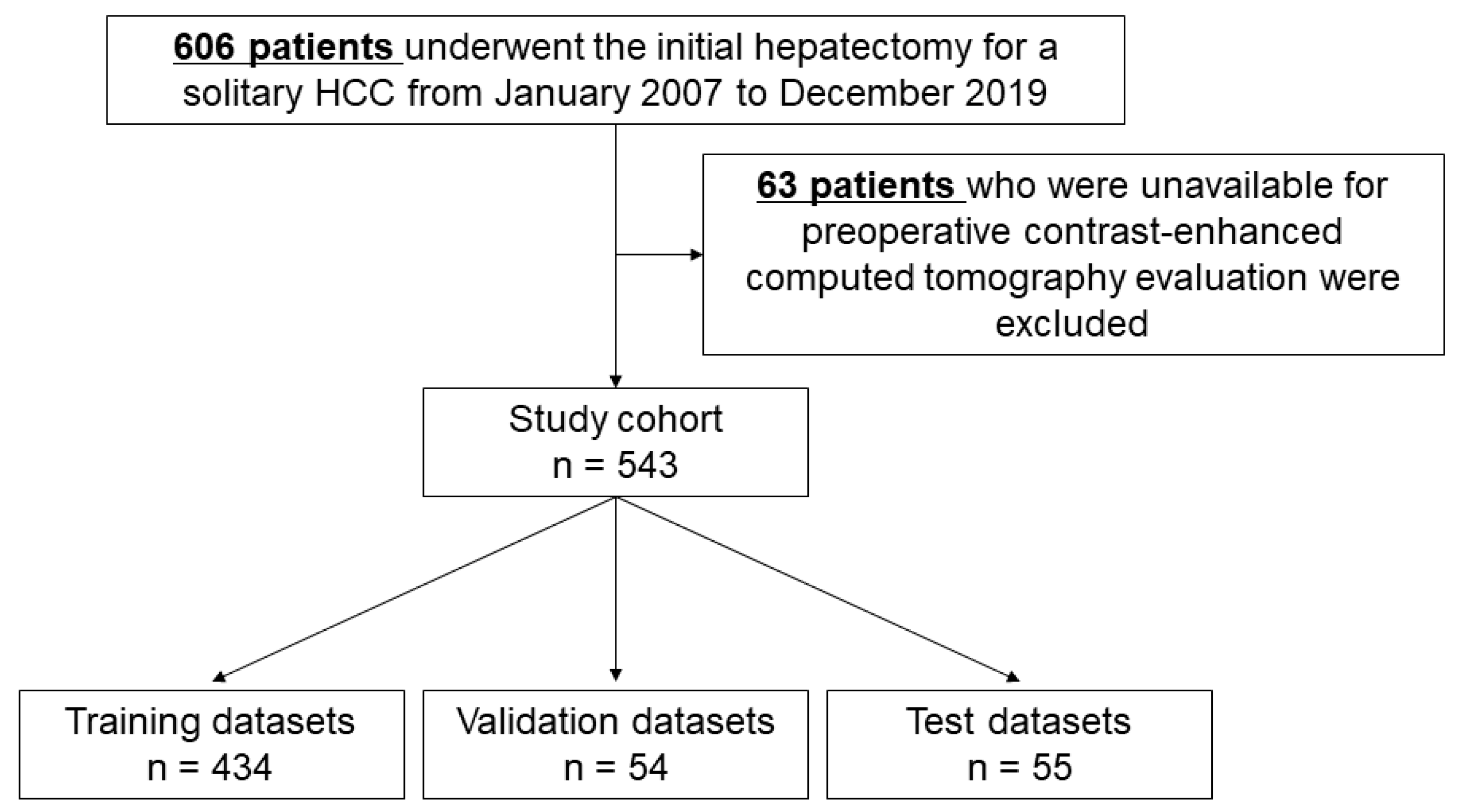
2.1. Study Cohort
2.2. Patient and Public Involvement
2.3. Postoperative Follow-Up and Recurrence Diagnosis
2.4. Data Partition
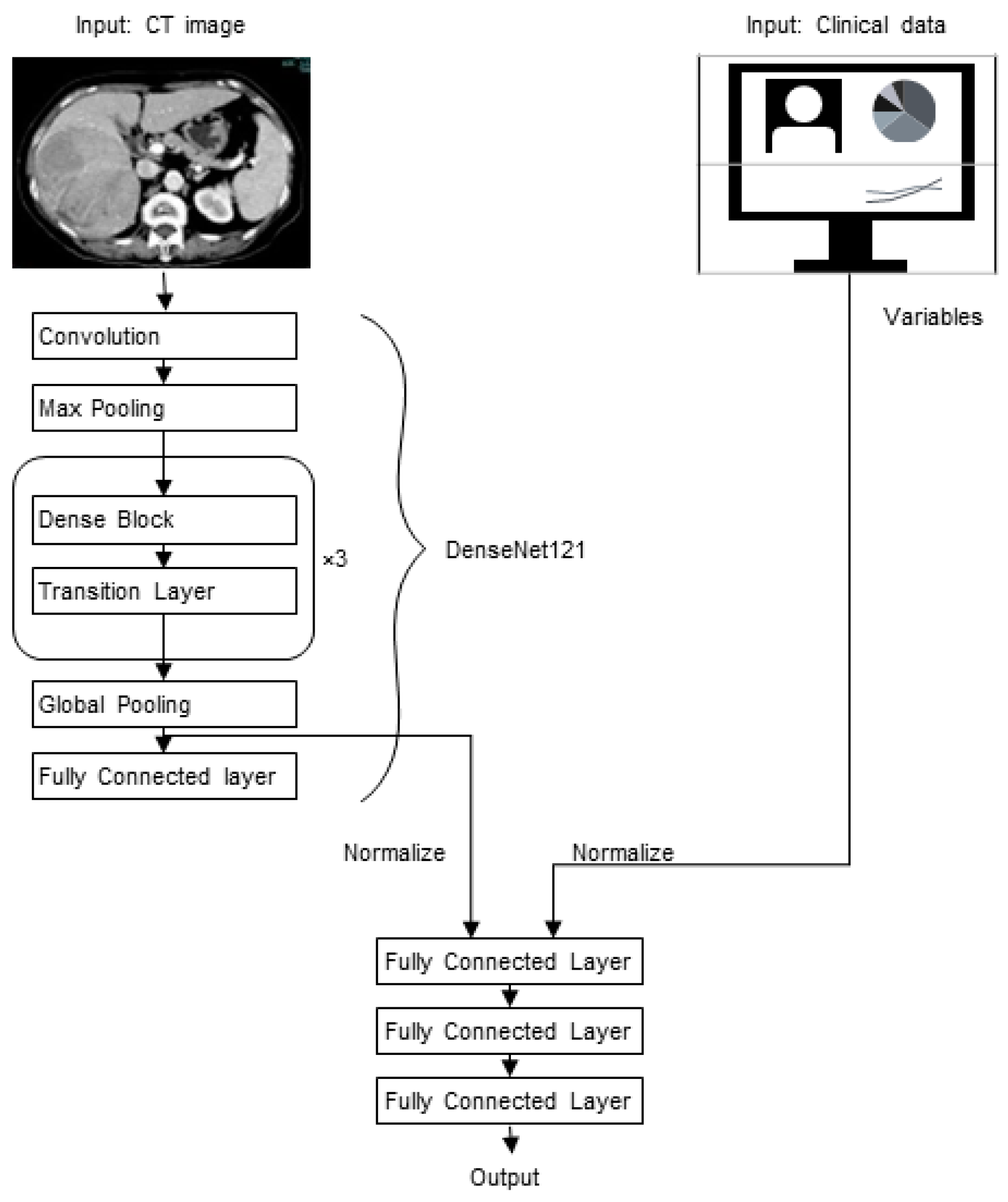
2.5. Image Processing
2.6. Model Development and Evaluation
2.7. Saliency Maps
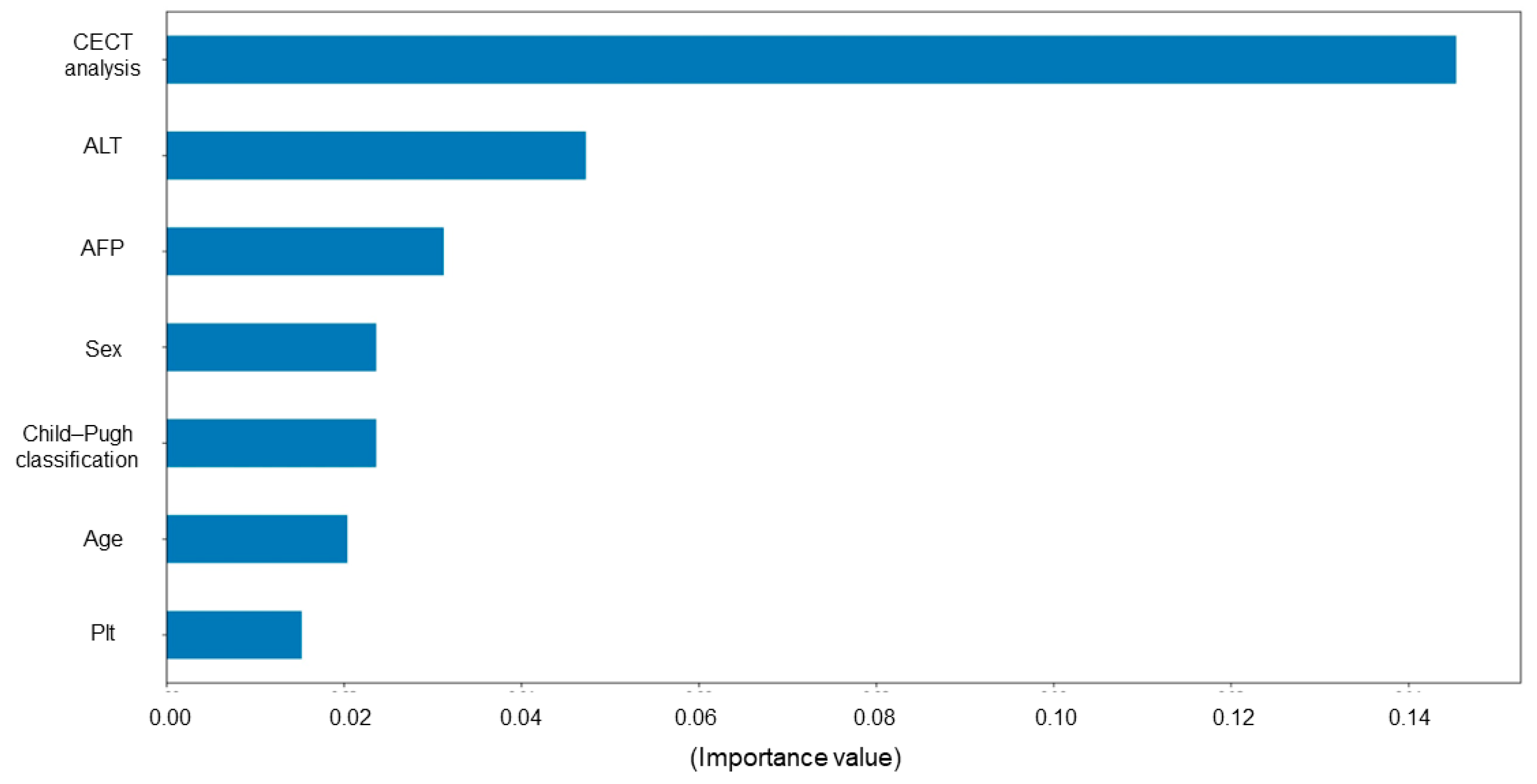
2.8. Permutation Importance
2.9. Statistical Analysis
3. Results
3.1. Patient Backgrounds and Surgical Outcomes
3.2. Patient Characteristics in Each Dataset
3.3. Model Development
3.4. Model Evaluation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thelen, A.; Benckert, C.; Tautenhahn, H.-M.; Hau, H.-M.; Bartels, M.; Linnemann, J.; Bertolini, J.; Moche, M.; Wittekind, C.; Jonas, S. Liver resection for hepatocellular carcinoma in patients without cirrhosis. Br. J. Surg. 2013, 100, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Fan, P.; Chen, S.; Tu, C.; Wan, C. Distinct Recurrence Risk Factors for Intrahepatic Metastasis and Multicenter Occurrence After Surgery in Patients with Hepatocellular Carcinoma. J. Gastrointest. Surg. 2017, 21, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Ikoma, H.; Morimura, R.; Konishi, H.; Murayama, Y.; Komatsu, S.; Shiozaki, A.; Kuriu, Y.; Kubota, T.; Nakanishi, M.; et al. Optimal duration of the early and late recurrence of hepatocellular carcinoma after hepatectomy. World J. Gastroenterol. 2015, 21, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.-M.; Kim, J.M.; Choi, G.-S.; Kwon, C.H.D.; Yi, N.-J.; Lee, K.-W.; Suh, K.S.; Joh, J.-W. Characteristics of Early Recurrence After Curative Liver Resection for Solitary Hepatocellular Carcinoma. J. Gastrointest. Surg. 2019, 23, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Kamiyama, T.; Nakanishi, K.; Yokoo, H.; Kamachi, H.; Tahara, M.; Kakisaka, T.; Tsuruga, Y.; Todo, S.; Taketomi, A. Analysis of the risk factors for early death due to disease recurrence or progression within 1 year after hepatectomy in patients with hepatocellular carcinoma. World J. Surg. Oncol. 2012, 10, 107. [Google Scholar] [CrossRef]
- Kudo, M. Targeted and immune therapies for hepatocellular carcinoma: Predictions for 2019 and beyond. World J. Gastroenterol. 2019, 25, 789–807. [Google Scholar] [CrossRef]
- Bruix, J.; Takayama, T.; Mazzaferro, V.; Chau, G.-Y.; Yang, J.; Kudo, M.; Cai, J.; Poon, R.T.; Han, K.-H.; Tak, W.Y.; et al. Adjuvant sorafenib for hepatocellular carcinoma after resection or ablation (STORM): A phase 3, randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2015, 16, 1344–1354. [Google Scholar] [CrossRef]
- Feng, Z.; Li, H.; Zhao, H.; Jiang, Y.; Liu, Q.; Chen, Q.; Wang, W.; Rong, P. Preoperative CT for Characterization of Aggressive Macrotrabecular-Massive Subtype and Vessels That Encapsulate Tumor Clusters Pattern in Hepatocellular Carcinoma. Radiology 2021, 300, 219–229. [Google Scholar] [CrossRef]
- Shinkawa, H.; Tanaka, S.; Takemura, S.; Amano, R.; Kimura, K.; Kinoshita, M.; Takahashi, K.; Matsuzaki, S.; Kubo, S. Nomograms predicting extra- and early intrahepatic recurrence after hepatic resection of hepatocellular carcinoma. Surgery 2021, 169, 922–928. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Taher, H.; Grasso, V.; Tawfik, S.; Gumbs, A. The challenges of deep learning in artificial intelligence and autonomous actions in surgery: A literature review. Artif. Intell. Surg. 2022, 2, 144–158. [Google Scholar] [CrossRef]
- Le Berre, C.; Sandborn, W.J.; Aridhi, S.; Devignes, M.-D.; Fournier, L.; Smaïl-Tabbone, M.; Danese, S.; Peyrin-Biroulet, L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology 2020, 158, 76–94.e2. [Google Scholar] [CrossRef]
- Wang, M.; Fu, F.; Zheng, B.; Bai, Y.; Wu, Q.; Wu, J.; Sun, L.; Liu, Q.; Liu, M.; Yang, Y.; et al. Development of an AI system for accurately diagnose hepatocellular carcinoma from computed tomography imaging data. Br. J. Cancer 2021, 125, 1111–1121. [Google Scholar] [CrossRef]
- Feng, S.; Yu, X.; Liang, W.; Li, X.; Zhong, W.; Hu, W.; Zhang, H.; Feng, Z.; Song, M.; Zhang, J.; et al. Development of a Deep Learning Model to Assist with Diagnosis of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 762733. [Google Scholar] [CrossRef]
- Gao, R.; Zhao, S.; Aishanjiang, K.; Cai, H.; Wei, T.; Zhang, Y.; Liu, Z.; Zhou, J.; Han, B.; Wang, J.; et al. Deep learning for differential diagnosis of malignant hepatic tumors based on multi-phase contrast-enhanced CT and clinical data. J. Hematol. Oncol. 2021, 14, 154. [Google Scholar] [CrossRef]
- Wei, J.; Jiang, H.; Zeng, M.; Wang, M.; Niu, M.; Gu, D.; Chong, H.; Zhang, Y.; Fu, F.; Zhou, M.; et al. Prediction of Microvascular Invasion in Hepatocellular Carcinoma via Deep Learning: A Multi-Center and Prospective Validation Study. Cancers 2021, 13, 2368. [Google Scholar] [CrossRef]
- Gao, W.; Wang, W.; Song, D.; Yang, C.; Zhu, K.; Zeng, M.; Rao, S.-X.; Wang, M. A predictive model integrating deep and radiomics features based on gadobenate dimeglumine-enhanced MRI for postoperative early recurrence of hepatocellular carcinoma. Radiol. Med. 2022, 127, 259–271. [Google Scholar] [CrossRef]
- Hollett, M.D.; Jeffrey, R.B.; Nino-Murcia, M.; Jorgensen, M.J.; Harris, D.P. Dual-phase helical CT of the liver: Value of arterial phase scans in the detection of small (< or = 1.5 cm) malignant hepatic neoplasms. AJR Am. J. Roentgenol. 1995, 164, 879–884. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32, 8026–8037. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ueda, D.; Yamamoto, A.; Takashima, T.; Onoda, N.; Noda, S.; Kashiwagi, S.; Morisaki, T.; Honjo, T.; Shimazaki, A.; Miki, Y. Training, Validation, and Test of Deep Learning Models for Classification of Receptor Expressions in Breast Cancers from Mammograms. JCO Precis. Oncol. 2021, 5, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Bektaş, M.; Zonderhuis, B.M.; Marquering, H.A.; Pereira, J.C.; Burchell, G.L.; van der Peet, D.L. Artificial intelligence in hepatopancreaticobiliary surgery: A systematic review. Artif. Intell. Surg. 2022, 2, 132–143. [Google Scholar] [CrossRef]
- Wang, C.J.; Hamm, C.A.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Weinreb, J.C.; Duncan, J.S.; Chapiro, J.; et al. Deep learning for liver tumor diagnosis part II: Convolutional neural network interpretation using radiologic imaging features. Eur. Radiol. 2019, 29, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, N.; Matsui, O.; Kobayashi, S.; Kitao, A.; Kozaka, K.; Inoue, D.; Yoshida, K.; Minami, T.; Koda, W.; Gabata, T. Current status of imaging biomarkers predicting the biological nature of hepatocellular carcinoma. Jpn. J. Radiol. 2019, 37, 191–208. [Google Scholar] [CrossRef]
- Ronot, M.; Purcell, Y.; Vilgrain, V. Hepatocellular Carcinoma: Current Imaging Modalities for Diagnosis and Prognosis. Dig. Dis. Sci. 2019, 64, 934–950. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Y.; Zhou, C.; Ma, X. Deep learning radiomics based on contrast enhanced computed tomography predicts microvascular invasion and survival outcome in early stage hepatocellular carcinoma. Eur. J. Surg. Oncol. 2022, 48, 1068–1077. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Z.; Rong, Y.; Zhou, B.; Bai, Y.; Wei, W.; Wang, M.; Guo, Y.; Tian, J. Deep learning provides a new computed tomography-based prognostic biomarker for recurrence prediction in high-grade serous ovarian cancer. Radiother. Oncol. 2019, 132, 171–177. [Google Scholar] [CrossRef]
- Elyan, E.; Vuttipittayamongkol, P.; Johnston, P.; Martin, K.; McPherson, K.; Moreno-García, C.F.; Jayne, C.; Sarker, M.K. Computer vision and machine learning for medical image analysis: Recent advances, challenges, and way forward. Artif. Intell. Surg. 2022, 2, 24–45. [Google Scholar] [CrossRef]
- Decker, J.M.; Sesti, J.; Turner, A.L.; Paul, S. The cassandra paradox: Looking into the crystal Ball of radiomics in thoracic surgery. Artif. Intell. Surg. 2022, 2, 57–63. [Google Scholar] [CrossRef]
- Akateh, C.; Black, S.M.; Conteh, L.; Miller, E.D.; Noonan, A.; Elliott, E.; Pawlik, T.M.; Tsung, A.; Cloyd, J.M. Neoadjuvant and adjuvant treatment strategies for hepatocellular carcinoma. World J. Gastroenterol. 2019, 25, 3704–3721. [Google Scholar] [CrossRef]
- Gumbs, A.A.; Lorenz, E.; Tsai, T.-J.; Starker, L.; Flanagan, J.; Benedetti Cacciaguerra, A.; Yu, N.J.; Bajul, M.; Chouillard, E.; Croner, R.; et al. Study: International Multicentric Minimally Invasive Liver Resection for Colorectal Liver Metastases (SIMMILR-CRLM). Cancers 2022, 14, 1379. [Google Scholar] [CrossRef]



| Variables | Value |
|---|---|
| Age, years, median (range) | 71 (19–87) |
| Sex, male/female | 401/136 |
| Comorbidities, n (%) | |
| Diabetes mellitus | 187 (34) |
| Hypertension | 287 (53) |
| Dyslipidemia | 107 (20) |
| HB surface antigen positive | 94 (17) |
| HBV-DNA < detectable levels | 39 (7.2) |
| HCV antibody positive | 279 (51) |
| HCV-SVR | 61 (11) |
| Laboratory data, median (range) | |
| Total bilirubin, mg/dL | 0.6 (0.1–2.7) |
| ALT, IU/L | 29 (6–270) |
| Albumin, g/dL | 4.0 (2.3–5.1) |
| PT, % | 94 (40–147) |
| Platelet count, ×104/μL | 15.0 (1.3–42.8) |
| AFP, ng/mL | 9.1 (1.5–283,300) |
| PIVKA-II, mAU/mL | 71 (2–3893,20) |
| Child–Pugh classification, A/B | 526/17 |
| Tumor diameter, cm, median (range) | 3 (0.7–19.5) |
| Hepatectomy procedures | |
| Partial resection | 329 (61) |
| Segmentectomy | 54 (9.9) |
| Sectionectomy | 90 (17) |
| Bisectionnectomy | 69 (13) |
| Trisectionnectomy | 1 (0.2) |
| Operative time, min, median (range) | 278 (75–776) |
| Intraoperative blood loss, g, median (range) | 280 (5–7460) |
| Postoperative complication *, n (%) | 71 (13) |
| Liver cirrhosis, n (%) | 132 (24) |
| Microvascular invasion, n (%) | 154 (28) |
| Recurrence-free survival, months, median (range) | 19 (1–170) |
| Early recurrence within 2 years, n (%) | 220 (41) |
| Intrahepatic recurrence | 195 (36) |
| Extrahepatic recurrence | 31 (5.7) |
| Observed period, months, median (range) | 46 (1–170) |
| Variables | Training Datasets (n = 434) | Validation Datasets (n = 54) | Test Datasets (n = 55) | p-Value |
|---|---|---|---|---|
| Sex, male/female | 331/103 | 37/17 | 38/17 | 0.26 |
| Age, years | 71 (31–87) | 67 (19–84) | 70 (38–82) | 0.11 |
| ALT, IU/L | 29 (8–270) | 28 (11–162) | 28 (6–126) | 0.78 |
| AFP, ng/mL | 8.9 (1.5–283,300) | 10 (2.3–109,402) | 9.4 (1.9–67,700) | 0.87 |
| PIVKA-II, mAU/mL | 64 (2–389,320) | 104 (13–57,202) | 117 (9–228,533) | 0.62 |
| Child–Pugh classification B, n (%) | 14 (3.2) | 2 (3.7) | 1 (1.8) | 0.80 |
| Platelet count, ×104/μL | 15.3 (1.3–42.8) | 14 (5.2–37.1) | 15.2 (2.2–30.3) | 0.91 |
| Tumor diameter, cm | 3 (0.9–15.0) | 3 (0.7–19.5) | 3.3 (0.9–18.2) | 0.69 |
| ≥Bisectionectomy, n (%) | 56 (13) | 6 (11) | 8 (15) | 0.87 |
| Operative time, min | 272 (93–643) | 310 (75–776) | 300 (127–563) | 0.17 |
| Intraoperative blood loss, g | 275 (5–7460) | 275 (5–3750) | 360 (5–6265) | 0.23 |
| Liver cirrhosis, n (%) | 100 (23) | 17 (32) | 15 (27) | 0.34 |
| Microvascular invasion, n (%) | 126 (29) | 17 (32) | 11 (20) | 0.33 |
| Recurrence-free survival, months | 20 (1–161) | 18 (1–113) | 17 (1–170) | 0.45 |
| Early recurrence within 2 years, n (%) | 173 (40) | 24 (44) | 23 (42) | 0.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kinoshita, M.; Ueda, D.; Matsumoto, T.; Shinkawa, H.; Yamamoto, A.; Shiba, M.; Okada, T.; Tani, N.; Tanaka, S.; Kimura, K.; et al. Deep Learning Model Based on Contrast-Enhanced Computed Tomography Imaging to Predict Postoperative Early Recurrence after the Curative Resection of a Solitary Hepatocellular Carcinoma. Cancers 2023, 15, 2140. https://doi.org/10.3390/cancers15072140
Kinoshita M, Ueda D, Matsumoto T, Shinkawa H, Yamamoto A, Shiba M, Okada T, Tani N, Tanaka S, Kimura K, et al. Deep Learning Model Based on Contrast-Enhanced Computed Tomography Imaging to Predict Postoperative Early Recurrence after the Curative Resection of a Solitary Hepatocellular Carcinoma. Cancers. 2023; 15(7):2140. https://doi.org/10.3390/cancers15072140
Chicago/Turabian StyleKinoshita, Masahiko, Daiju Ueda, Toshimasa Matsumoto, Hiroji Shinkawa, Akira Yamamoto, Masatsugu Shiba, Takuma Okada, Naoki Tani, Shogo Tanaka, Kenjiro Kimura, and et al. 2023. "Deep Learning Model Based on Contrast-Enhanced Computed Tomography Imaging to Predict Postoperative Early Recurrence after the Curative Resection of a Solitary Hepatocellular Carcinoma" Cancers 15, no. 7: 2140. https://doi.org/10.3390/cancers15072140
APA StyleKinoshita, M., Ueda, D., Matsumoto, T., Shinkawa, H., Yamamoto, A., Shiba, M., Okada, T., Tani, N., Tanaka, S., Kimura, K., Ohira, G., Nishio, K., Tauchi, J., Kubo, S., & Ishizawa, T. (2023). Deep Learning Model Based on Contrast-Enhanced Computed Tomography Imaging to Predict Postoperative Early Recurrence after the Curative Resection of a Solitary Hepatocellular Carcinoma. Cancers, 15(7), 2140. https://doi.org/10.3390/cancers15072140






