p53 and p63 Proteoforms Derived from Alternative Splicing Possess Differential Seroreactivity in Colorectal Cancer with Distinct Diagnostic Ability from the Canonical Proteins
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasma of Patients and Controls
2.2. In Silico Modeling of the Proteins
2.3. Gateway Plasmid Construction, Gene Cloning, DNA Preparation and Protein Expression
2.4. SDS-PAGE and Western Blot Analysis
2.5. Seroreactivity luminescence and Biosensing Assays
2.6. Statistical Analysis
2.7. Data Availability
3. Results
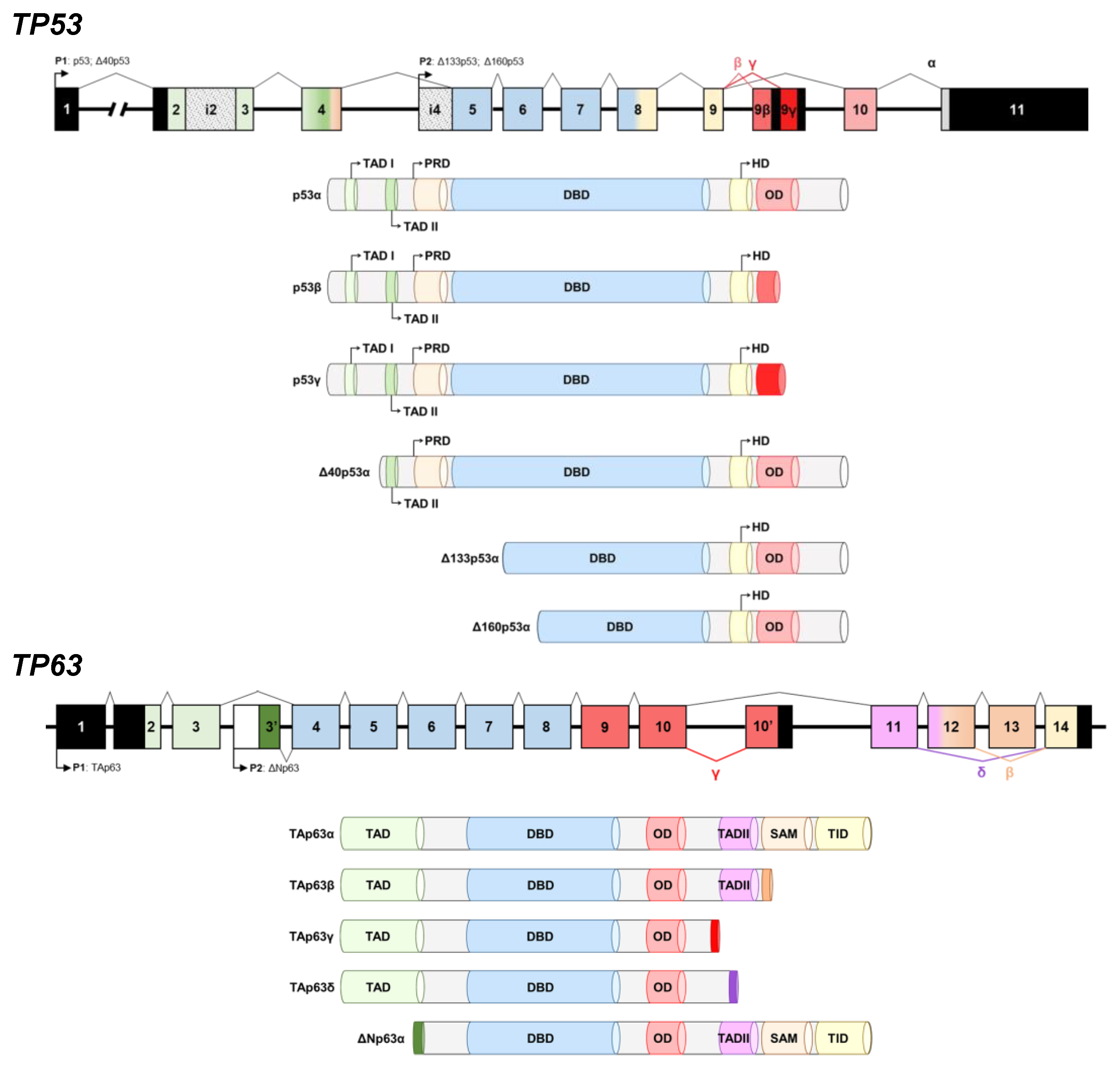
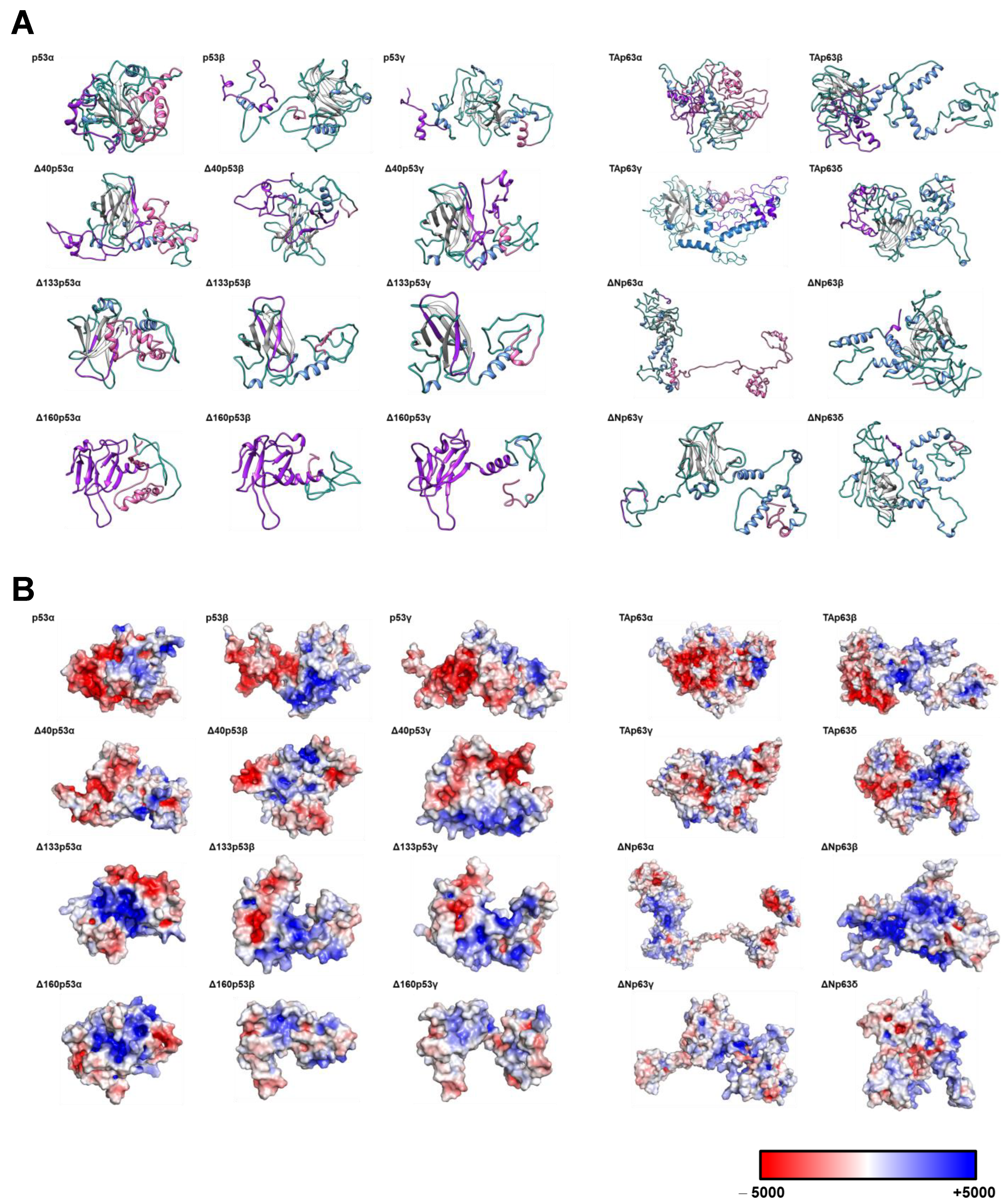
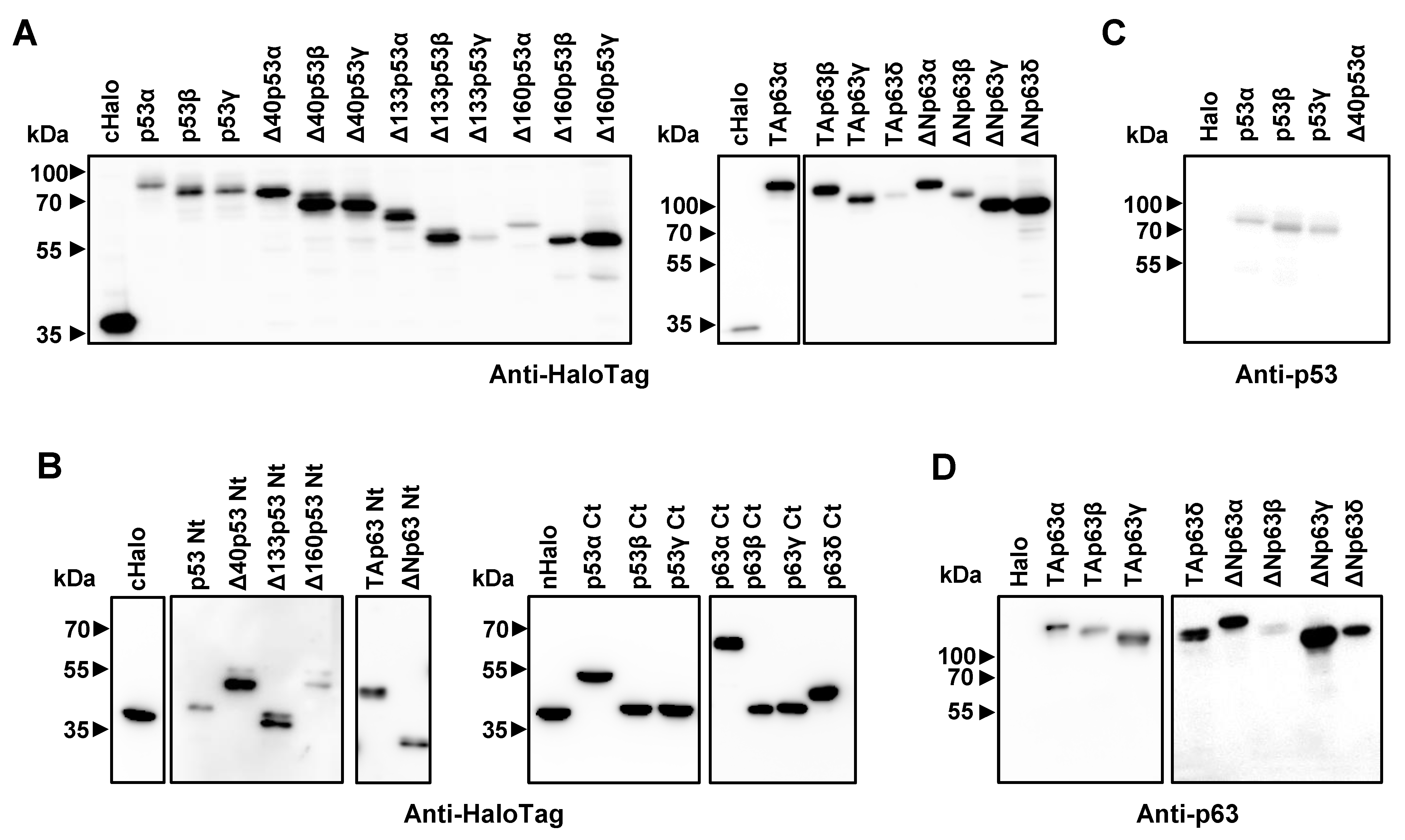
3.1. Cloning, In Vitro Protein Expression of Fusion Proteins, and In Silico Analysis of the Proteoforms
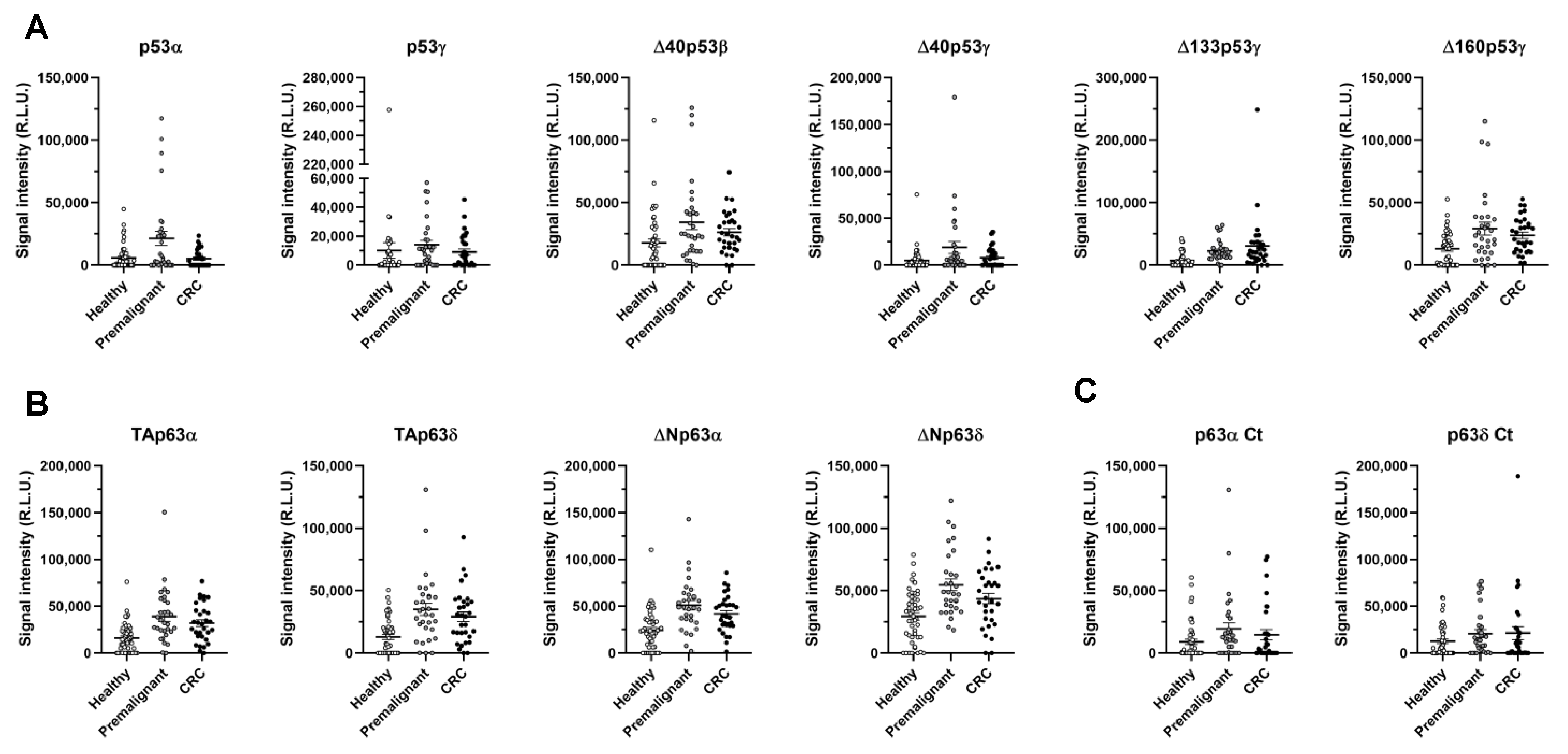
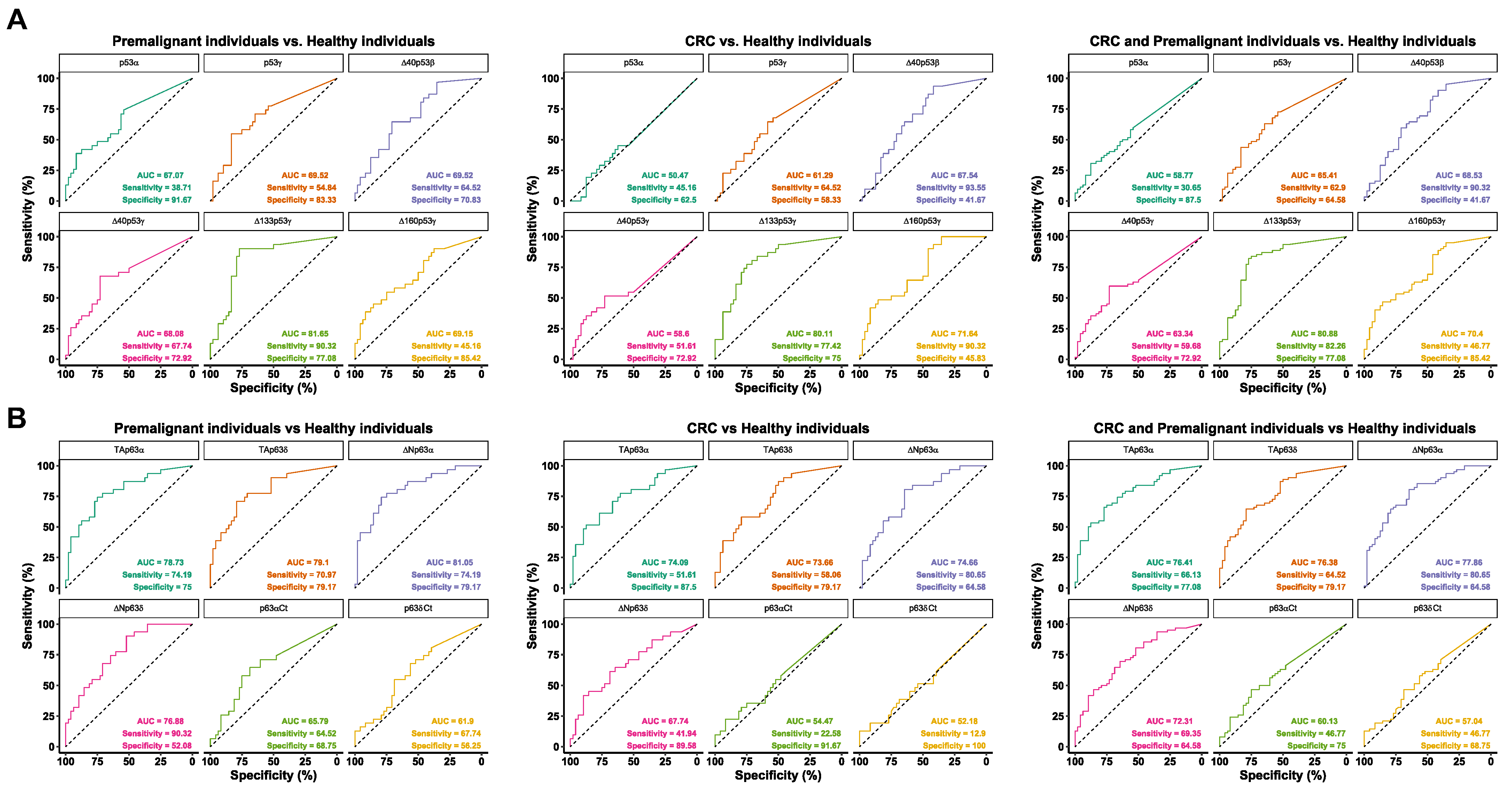
3.2. Evaluation of the Seroreactivity Potential of p53 and p63 Proteoforms in Colorectal Cancer Patients
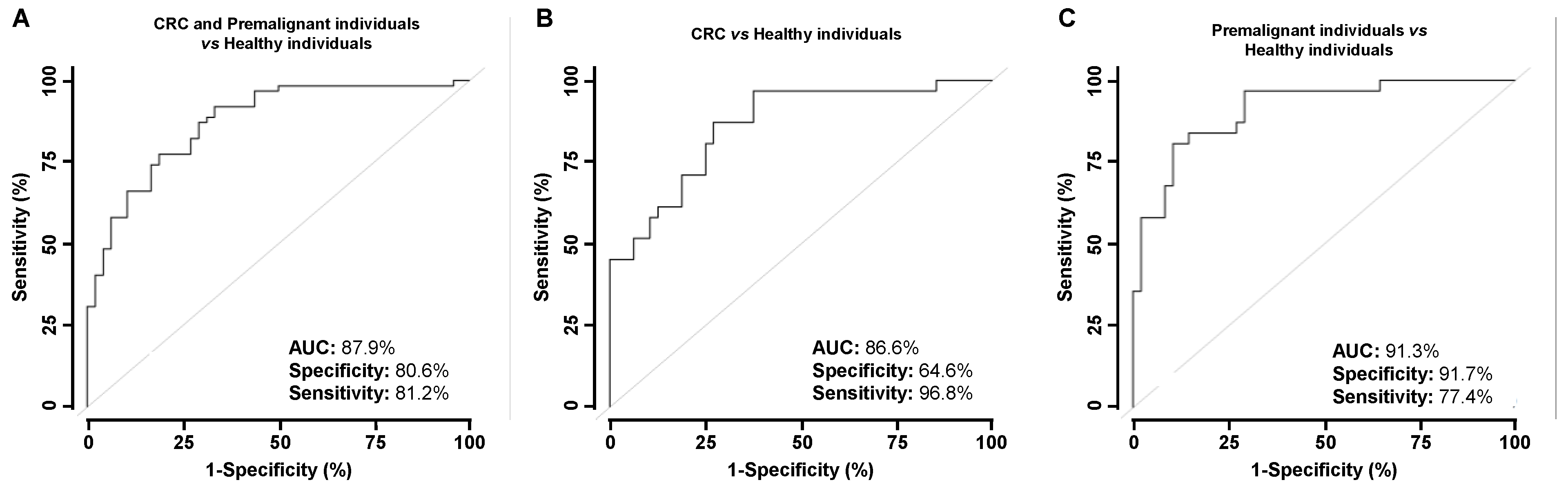
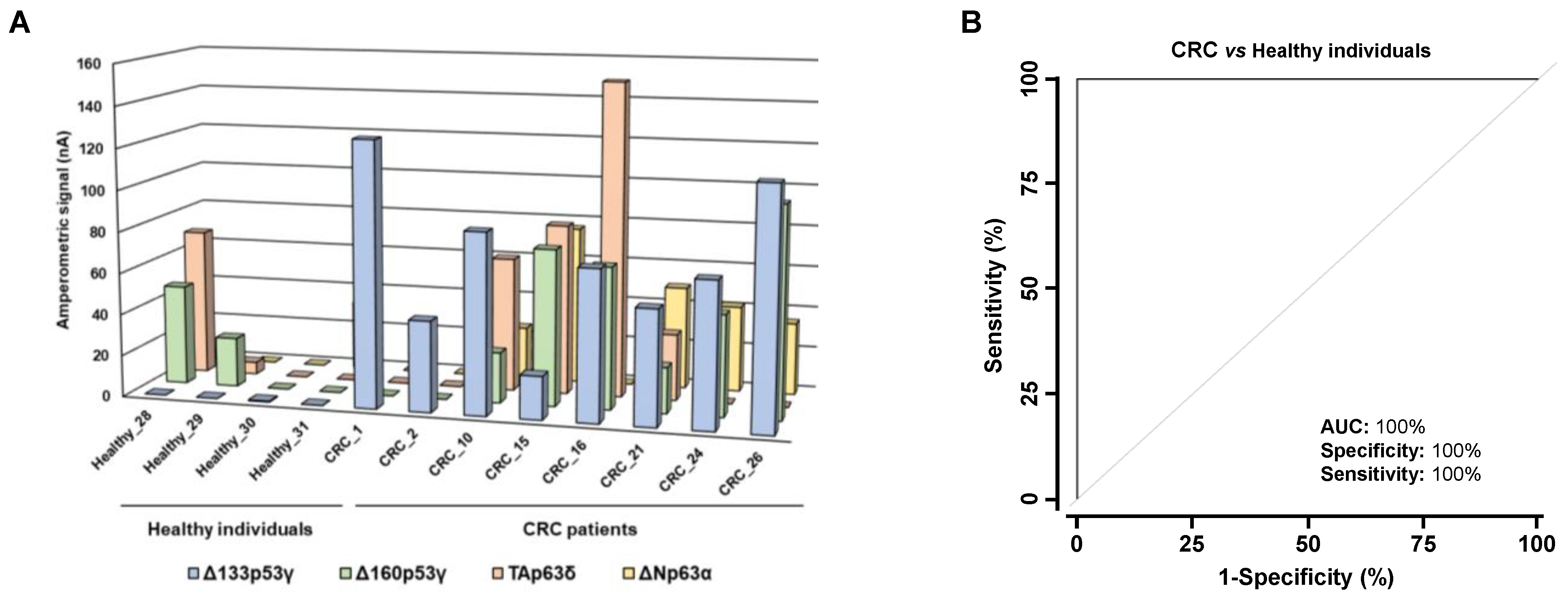
3.3. Analysis of the Diagnostic Value of the p53 and p63 Proteoforms as Targets of CRC Autoantibodies in POC-like Devices
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Klinck, R.; Bramard, A.; Inkel, L.; Dufresne-Martin, G.; Gervais-Bird, J.; Madden, R.; Paquet, E.R.; Koh, C.; Venables, J.P.; Prinos, P.; et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008, 68, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Bourdon, J.C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, M.; Liu, X.; Huang, Y.; Liu, C.; Zhu, J.; Fu, G.; Lei, Z.; Chu, X. Alternative splicing of mRNA in colorectal cancer: New strategies for tumor diagnosis and treatment. Cell Death Dis. 2021, 12, 752. [Google Scholar] [CrossRef]
- Venables, J.P.; Klinck, R.; Koh, C.; Gervais-Bird, J.; Bramard, A.; Inkel, L.; Durand, M.; Couture, S.; Froehlich, U.; Lapointe, E.; et al. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 2009, 16, 670–676. [Google Scholar] [CrossRef]
- Wei, J.; Zaika, E.; Zaika, A. p53 Family: Role of Protein Isoforms in Human Cancer. J. Nucleic. Acids 2012, 2012, 687359. [Google Scholar] [CrossRef]
- Dotsch, V.; Bernassola, F.; Coutandin, D.; Candi, E.; Melino, G. p63 and p73, the ancestors of p53. Cold Spring Harb. Perspect. Biol. 2010, 2, a004887. [Google Scholar] [CrossRef]
- Rutkowski, R.; Hofmann, K.; Gartner, A. Phylogeny and function of the invertebrate p53 superfamily. Cold Spring Harb. Perspect. Biol. 2010, 2, a001131. [Google Scholar] [CrossRef]
- Petre-Lazar, B.; Livera, G.; Moreno, S.G.; Trautmann, E.; Duquenne, C.; Hanoux, V.; Habert, R.; Coffigny, H. The role of p63 in germ cell apoptosis in the developing testis. J. Cell Physiol. 2007, 210, 87–98. [Google Scholar] [CrossRef]
- Lu, W.J.; Amatruda, J.F.; Abrams, J.M. p53 ancestry: Gazing through an evolutionary lens. Nat. Rev. Cancer 2009, 9, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Collavin, L.; Lunardi, A.; Del Sal, G. p53-family proteins and their regulators: Hubs and spokes in tumor suppression. Cell Death Differ. 2010, 17, 901–911. [Google Scholar] [CrossRef]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.M. p53 and autophagy in cancer: Guardian of the genome meets guardian of the proteome. Eur. J. Cancer 2011, 47, 44–50. [Google Scholar] [CrossRef]
- Liu, Y.; Leslie, P.L.; Zhang, Y. Life and Death Decision-Making by p53 and Implications for Cancer Immunotherapy. Trends Cancer 2021, 7, 226–239. [Google Scholar] [CrossRef]
- Oren, M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003, 10, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Bourdon, J.C. p53 Family isoforms. Curr. Pharm. Biotechnol. 2007, 8, 332–336. [Google Scholar] [CrossRef]
- Vieler, M.; Sanyal, S. p53 Isoforms and Their Implications in Cancer. Cancers 2018, 10, 288. [Google Scholar] [CrossRef]
- Mills, A.A. p63: Oncogene or tumor suppressor? Curr. Opin. Genet. Dev. 2006, 16, 38–44. [Google Scholar] [CrossRef]
- Mills, A.A.; Zheng, B.; Wang, X.J.; Vogel, H.; Roop, D.R.; Bradley, A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999, 398, 708–713. [Google Scholar] [CrossRef]
- Yang, A.; McKeon, F. P63 and P73: P53 mimics, menaces and more. Nat. Rev. Mol. Cell Biol. 2000, 1, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Schweitzer, R.; Sun, D.; Kaghad, M.; Walker, N.; Bronson, R.T.; Tabin, C.; Sharpe, A.; Caput, D.; Crum, C.; et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999, 398, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Pozniak, C.D.; Radinovic, S.; Yang, A.; McKeon, F.; Kaplan, D.R.; Miller, F.D. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 2000, 289, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Garranzo-Asensio, M.; Rodriguez-Cobos, J.; San Millan, C.; Poves, C.; Fernandez-Acenero, M.J.; Pastor-Morate, D.; Vinal, D.; Montero-Calle, A.; Solis-Fernandez, G.; Ceron, M.A.; et al. In-depth proteomics characterization of ∆Np73 effectors identifies key proteins with diagnostic potential implicated in lymphangiogenesis, vasculogenesis and metastasis in colorectal cancer. Mol. Oncol. 2022, 16, 2672–2692. [Google Scholar] [CrossRef]
- Rodriguez, N.; Pelaez, A.; Barderas, R.; Dominguez, G. Clinical implications of the deregulated TP73 isoforms expression in cancer. Clin. Transl. Oncol. 2018, 20, 827–836. [Google Scholar] [CrossRef]
- Rodriguez-Cobos, J.; Vinal, D.; Poves, C.; Fernandez-Acenero, M.J.; Peinado, H.; Pastor-Morate, D.; Prieto, M.I.; Barderas, R.; Rodriguez-Salas, N.; Dominguez, G. DeltaNp73, TAp73 and Delta133p53 Extracellular Vesicle Cargo as Early Diagnosis Markers in Colorectal Cancer. Cancers 2021, 13, 2240. [Google Scholar] [CrossRef]
- Deyoung, M.P.; Ellisen, L.W. p63 and p73 in human cancer: Defining the network. Oncogene 2007, 26, 5169–5183. [Google Scholar] [CrossRef]
- Anderson, K.S.; LaBaer, J. The sentinel within: Exploiting the immune system for cancer biomarkers. J. Proteome. Res. 2005, 4, 1123–1133. [Google Scholar] [CrossRef]
- Pedersen, J.W.; Gentry-Maharaj, A.; Fourkala, E.O.; Dawnay, A.; Burnell, M.; Zaikin, A.; Pedersen, A.E.; Jacobs, I.; Menon, U.; Wandall, H.H. Early detection of cancer in the general population: A blinded case-control study of p53 autoantibodies in colorectal cancer. Br. J. Cancer 2013, 108, 107–114. [Google Scholar] [CrossRef]
- Garranzo-Asensio, M.; Guzman-Aranguez, A.; Poves, C.; Fernandez-Acenero, M.J.; Torrente-Rodriguez, R.M.; Ruiz-Valdepenas Montiel, V.; Dominguez, G.; Frutos, L.S.; Rodriguez, N.; Villalba, M.; et al. Toward Liquid Biopsy: Determination of the Humoral Immune Response in Cancer Patients Using HaloTag Fusion Protein-Modified Electrochemical Bioplatforms. Anal. Chem. 2016, 88, 12339–12345. [Google Scholar] [CrossRef]
- Katchman, B.A.; Barderas, R.; Alam, R.; Chowell, D.; Field, M.S.; Esserman, L.J.; Wallstrom, G.; LaBaer, J.; Cramer, D.W.; Hollingsworth, M.A.; et al. Proteomic mapping of p53 immunogenicity in pancreatic, ovarian, and breast cancers. Proteomics Clin. Appl. 2016, 10, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Garranzo-Asensio, M.; Guzman-Aranguez, A.; Poves, C.; Fernandez-Acenero, M.J.; Montero-Calle, A.; Ceron, M.A.; Fernandez-Diez, S.; Rodriguez, N.; Gomez de Cedron, M.; Ramirez de Molina, A.; et al. The specific seroreactivity to ∆Np73 isoforms shows higher diagnostic ability in colorectal cancer patients than the canonical p73 protein. Sci. Rep. 2019, 9, 13547. [Google Scholar] [CrossRef] [PubMed]
- Garranzo-Asensio, M.; San Segundo-Acosta, P.; Poves, C.; Fernandez-Acenero, M.J.; Martinez-Useros, J.; Montero-Calle, A.; Solis-Fernandez, G.; Sanchez-Martinez, M.; Rodriguez, N.; Ceron, M.A.; et al. Identification of tumor-associated antigens with diagnostic ability of colorectal cancer by in-depth immunomic and seroproteomic analysis. J. Proteomics 2020, 214, 103635. [Google Scholar] [CrossRef] [PubMed]
- San Segundo-Acosta, P.; Montero-Calle, A.; Fuentes, M.; Rabano, A.; Villalba, M.; Barderas, R. Identification of Alzheimer’s Disease Autoantibodies and Their Target Biomarkers by Phage Microarrays. J. Proteome. Res. 2019, 18, 2940–2953. [Google Scholar] [CrossRef]
- Garranzo-Asensio, M.; Solis-Fernandez, G.; Montero-Calle, A.; Garcia-Martinez, J.M.; Fiuza, M.C.; Pallares, P.; Palacios-Garcia, N.; Garcia-Jimenez, C.; Guzman-Aranguez, A.; Barderas, R. Seroreactivity against Tyrosine Phosphatase PTPRN Links Type 2 Diabetes and Colorectal Cancer and Identifies a Potential Diagnostic and Therapeutic Target. Diabetes 2022, 71, 497–510. [Google Scholar] [CrossRef]
- Garranzo-Asensio, M.; Guzman-Aranguez, A.; Povedano, E.; Ruiz-Valdepenas Montiel, V.; Poves, C.; Fernandez-Acenero, M.J.; Montero-Calle, A.; Solis-Fernandez, G.; Fernandez-Diez, S.; Camps, J.; et al. Multiplexed monitoring of a novel autoantibody diagnostic signature of colorectal cancer using HaloTag technology-based electrochemical immunosensing platform. Theranostics 2020, 10, 3022–3034. [Google Scholar] [CrossRef]
- Montero-Calle, A.; Aranguren-Abeigon, I.; Garranzo-Asensio, M.; Poves, C.; Fernández-Aceñero, M.J.; Martínez-Useros, J.; Sanz, R.; Dziaková, J.; Rodriguez-Cobos, J.; Solis-Fernandez, G.; et al. Multiplexed biosensing diagnostic platforms detecting autoantibodies to tumor-associated antigens from exosomes released by CRC cells and tissue samples showed high diagnostic ability for colorectal cancer. Engineering 2021, 7, 1393–1412. [Google Scholar] [CrossRef]
- Valverde, A.; Montero-Calle, A.; Arévalo, B.; San Segundo-Acosta, P.; Serafín, V.; Alonso-Navarro, M.; Solís-Fernández, G.; Pingarrón, J.M.; Campuzano, S.; Barderas, R. Phage-Derived and Aberrant HaloTag Peptides Immobilized on Magnetic Microbeads for Amperometric Biosensing of Serum Autoantibodies and Alzheimer’s Disease Diagnosis. Anal. Sens. 2021, 1, 161–165. [Google Scholar] [CrossRef]
- Marrugo-Ramirez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef]
- Lokshin, A.; Bast, R.C.; Rodland, K. Circulating Cancer Biomarkers. Cancers 2021, 13, 802. [Google Scholar] [CrossRef]
- Macdonald, I.K.; Parsy-Kowalska, C.B.; Chapman, C.J. Autoantibodies: Opportunities for Early Cancer Detection. Trends Cancer 2017, 3, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Heo, C.K.; Bahk, Y.Y.; Cho, E.W. Tumor-associated autoantibodies as diagnostic and prognostic biomarkers. BMB Rep. 2012, 45, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.A.; O’Connell, D.J.; O’Kane, S.L.; O’Brien, J.K.; O’Toole, S.; Martin, C.; Sheils, O.; O’Leary, J.J.; Cahill, D.J. Epitope presentation is an important determinant of the utility of antigens identified from protein arrays in the development of autoantibody diagnostic assays. J. Proteomics 2012, 75, 4668–4675. [Google Scholar] [CrossRef]
- Barderas, R.; Villar-Vazquez, R.; Fernandez-Acenero, M.J.; Babel, I.; Pelaez-Garcia, A.; Torres, S.; Casal, J.I. Sporadic colon cancer murine models demonstrate the value of autoantibody detection for preclinical cancer diagnosis. Sci. Rep. 2013, 3, 2938. [Google Scholar] [CrossRef]
- Zaenker, P.; Gray, E.S.; Ziman, M.R. Autoantibody Production in Cancer--The Humoral Immune Response toward Autologous Antigens in Cancer Patients. Autoimmun. Rev. 2016, 15, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Babel, I.; Barderas, R.; Diaz-Uriarte, R.; Moreno, V.; Suarez, A.; Fernandez-Acenero, M.J.; Salazar, R.; Capella, G.; Casal, J.I. Identification of MST1/STK4 and SULF1 proteins as autoantibody targets for the diagnosis of colorectal cancer by using phage microarrays. Mol. Cell Proteom. 2011, 10, M110.001784. [Google Scholar] [CrossRef]
- Barderas, R.; Babel, I.; Casal, J.I. Colorectal cancer proteomics, molecular characterization and biomarker discovery. Proteomics Clin. Appl. 2010, 4, 159–178. [Google Scholar] [CrossRef]
- Babel, I.; Barderas, R.; Diaz-Uriarte, R.; Martinez-Torrecuadrada, J.L.; Sanchez-Carbayo, M.; Casal, J.I. Identification of tumor-associated autoantigens for the diagnosis of colorectal cancer in serum using high density protein microarrays. Mol. Cell Proteom. 2009, 8, 2382–2395. [Google Scholar] [CrossRef]
- Rodriguez Calleja, L.; Lavaud, M.; Tesfaye, R.; Brounais-Le-Royer, B.; Baud’huin, M.; Georges, S.; Lamoureux, F.; Verrecchia, F.; Ory, B. The p53 Family Members p63 and p73 Roles in the Metastatic Dissemination: Interactions with microRNAs and TGFbeta Pathway. Cancers 2022, 14, 5948. [Google Scholar] [CrossRef]
- May, P.; May, E. Twenty years of p53 research: Structural and functional aspects of the p53 protein. Oncogene 1999, 18, 7621–7636. [Google Scholar] [CrossRef]
- Marcel, V.; Hainaut, P. p53 isoforms—A conspiracy to kidnap p53 tumor suppressor activity? Cell Mol. Life Sci. 2009, 66, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, J.; Xiao, Z.X. Role of p63 in Development, Tumorigenesis and Cancer Progression. Cancer Microenviron. 2012, 5, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Campbell, H.; Fleming, N.; Roth, I.; Mehta, S.; Wiles, A.; Williams, G.; Vennin, C.; Arsic, N.; Parkin, A.; Pajic, M.; et al. ∆133p53 isoform promotes tumour invasion and metastasis via interleukin-6 activation of JAK-STAT and RhoA-ROCK signalling. Nat. Commun. 2018, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tan, Q.; Song, Y.; Shi, Y.; Han, X. Anti-p53 autoantibody in blood as a diagnostic biomarker for colorectal cancer: A meta-analysis. Scand J. Immunol. 2020, 91, e12829. [Google Scholar] [CrossRef]
| Sample | Number (n) | Age Average ± SD (Years) | Age Range (Years) | Gender (n) | CRC Stage (n) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | I | II | III | IV | |||||
| Luminescence assays | Healthy individuals | 48 | 67.08 ± 17.60 | 36–101 | 23 | 25 | - | |||
| Premalignant lesions | 31 | 63.77 ± 6.50 | 54–79 | 21 | 10 | - | ||||
| CRC patients | 31 | 70.90 ± 13.95 | 38–86 | 15 | 16 | 3 | 2 | 15 | 11 | |
| Electrochemical biosensing | Healthy individuals | 4 | 67.50 ± 6.45 | 63–74 | 2 | 2 | - | |||
| Premalignant lesions | 4 | 65.25 ± 1.71 | 63–67 | 2 | 2 | - | ||||
| CRC patients | 8 | 71.00 ± 11.70 | 59–86 | 1 | 7 | 2 | - | 3 | 3 | |
| Healthy | Premalignant | CRC | Pathology | Threshold ^ | p-Value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteoform | Mean | SEM | Mean | SEM | Mean | SEM | Mean | SEM | (H vs. Pre) * | (H vs. CRC) * | (H vs. P) * | (H vs. Pre) * | (H vs. CRC) * | (H vs. P) * |
| p53α | 5793 | 1433 | 21,387 | 5721 | 5135 | 1255 | 13,261 | 3085 | 22,773 | 3361 | 13,280 | 0.0080 | 0.9432 | 0.0980 |
| p53β | 16,312 | 4724 | 17,134 | 5269 | 12,151 | 3947 | 14,642 | 3280 | 23,059 | 4743 | 47,192 | 0.9025 | 0.8287 | 0.8359 |
| p53γ | 10,090 | 5399 | 14,142 | 2986 | 9113 | 2072 | 11,628 | 1831 | 10,016 | 1396 | 2383 | 0.0026 | 0.0784 | 0.0045 |
| Δ40p53α | 20,732 | 5262 | 12,965 | 8135 | 10,503 | 3186 | 11,734 | 4335 | 12,547 | 314 | 12,547 | 0.0613 | 0.5312 | 0.1342 |
| Δ40p53β | 17,802 | 3195 | 34,370 | 5965 | 26,365 | 2994 | 30,368 | 3349 | 21,224 | 6920 | 6920 | 0.0034 | 0.0084 | 0.0009 |
| Δ40p53γ | 4960 | 1687 | 18,959 | 6372 | 7982 | 1829 | 13,470 | 3361 | 3289 | 3152 | 3152 | 0.0053 | 0.1745 | 0.0131 |
| Δ133p53α | 27,121 | 13,961 | 17,050 | 3921 | 15,271 | 3010 | 16,160 | 2454 | 20,620 | 10,045 | 20,620 | 0.5712 | 0.5765 | 0.4984 |
| Δ133p53β | 18,875 | 3200 | 20,586 | 3906 | 20,598 | 3608 | 20,592 | 2637 | 5794 | 10,265 | 5794 | 0.7425 | 0.6167 | 0.6164 |
| Δ133p53γ | 7295 | 1616 | 22,708 | 3020 | 30,647 | 8125 | 26,678 | 4328 | 9474 | 8230 | 9474 | 0.0000 | 0.0000 | 0.0000 |
| Δ160p53α | 41,082 | 8238 | 28,720 | 3545 | 27,919 | 4281 | 28,319 | 2757 | 8925 | 18,463 | 18,167 | 0.8327 | 0.8326 | 1.0000 |
| Δ160p53β | 40,157 | 6085 | 31,251 | 5123 | 34,769 | 6249 | 33,010 | 4013 | 33,587 | 65,132 | 33,587 | 0.4304 | 0.7823 | 0.5207 |
| Δ160p53γ | 13,117 | 1870 | 29,256 | 5160 | 23,874 | 2577 | 26,565 | 2881 | 26,147 | 7124 | 26,147 | 0.0041 | 0.0012 | 0.0003 |
| TAp63α | 16,148 | 2226 | 39,160 | 5211 | 32,113 | 3633 | 35,637 | 3182 | 23,630 | 30,240 | 24,128 | 0.0000 | 0.0003 | 0.0000 |
| TAp63β | 28,778 | 4809 | 17,873 | 4547 | 35,166 | 12,679 | 26,520 | 6771 | 26,860 | 16,164 | 28,653 | 0.1680 | 0.9878 | 0.4050 |
| TAp63γ | 31,057 | 6352 | 37,917 | 8951 | 34,715 | 10,443 | 36,316 | 6824 | 35,915 | 38,003 | 31,196 | 0.7591 | 0.9069 | 0.7972 |
| TAp63δ | 12,998 | 2064 | 35,005 | 4944 | 29,176 | 3879 | 32,091 | 3138 | 22,093 | 21,769 | 21,769 | 0.0000 | 0.0004 | 0.0000 |
| ΔNp63α | 24,323 | 3057 | 51,002 | 4959 | 41,914 | 3554 | 46,458 | 3081 | 36,392 | 27,973 | 27,973 | 0.0000 | 0.0002 | 0.0000 |
| ΔNp63β | 20,404 | 4843 | 27,926 | 7202 | 20,176 | 4397 | 24,051 | 4214 | 2135 | 37,008 | 15,429 | 0.7190 | 0.5817 | 0.5823 |
| ΔNp63γ | 23,568 | 4232 | 34,393 | 12,519 | 32,923 | 6528 | 33,658 | 7002 | 13,056 | 8122 | 7230 | 0.9143 | 0.1754 | 0.3773 |
| ΔNp63δ | 29,457 | 2959 | 54,744 | 4673 | 43,705 | 4122 | 49,225 | 3170 | 32,007 | 52,889 | 37,134 | 0.0001 | 0.0081 | 0.0001 |
| p53Nt | 29,293 | 3665 | 35,447 | 9143 | 40,625 | 10,808 | 38,036 | 7028 | 33,231 | 12,771 | 32,674 | 0.4847 | 0.7745 | 0.8069 |
| Δ40p53Nt | 47,605 | 9055 | 49,939 | 17,142 | 30,812 | 4116 | 40,376 | 8828 | 30,280 | 38,814 | 38,847 | 0.5943 | 0.2497 | 0.3108 |
| Δ133p53Nt | 55,418 | 7128 | 46,807 | 7534 | 52,115 | 10,629 | 49,461 | 6469 | 32,579 | 64,396 | 64,396 | 0.6192 | 0.8803 | 0.6952 |
| Δ160p53Nt | 85,145 | 18,721 | 53,696 | 6032 | 47,763 | 4415 | 50,729 | 3726 | 81,010 | 76,414 | 81,010 | 0.6192 | 0.4218 | 0.4333 |
| p53αCt | 6277 | 1710 | 10,212 | 2900 | 4125 | 1944 | 7169 | 1774 | 13,882 | 42,926 | 12,780 | 0.6080 | 0.1955 | 0.6544 |
| p53βCt | 5645 | 1870 | 5098 | 1741 | 12,408 | 7642 | 8753 | 3915 | 15,463 | 82,634 | 15,463 | 0.8585 | 0.3421 | 0.4952 |
| p53γCt | 11,121 | 3310 | 17,343 | 3987 | 9852 | 3586 | 13,598 | 2702 | 7471 | 38,470 | 4373 | 0.1598 | 0.5660 | 0.6056 |
| TAp63Nt | 28,242 | 5042 | 33,052 | 6496 | 29,976 | 5025 | 31,514 | 4077 | 24,042 | 8922 | 23,957 | 0.5538 | 0.3720 | 0.3720 |
| ΔNp63Nt | 33,492 | 5719 | 31,462 | 6993 | 46,664 | 10,231 | 39,063 | 6222 | 14,546 | 38,591 | 34,511 | 0.7488 | 0.5865 | 0.8955 |
| p63αCt | 9109 | 2140 | 19,383 | 4923 | 14,656 | 4140 | 17,019 | 3204 | 8884 | 30,323 | 10,516 | 0.0151 | 0.4862 | 0.0608 |
| p63βCt | 20,646 | 6678 | 17,065 | 2935 | 18,355 | 4458 | 17,710 | 2648 | 12,826 | 233 | 24,377 | 0.3322 | 0.6048 | 0.7836 |
| p63γCt | 10,515 | 1793 | 12,553 | 3423 | 13,825 | 3720 | 13,189 | 2508 | 4896 | 934 | 4896 | 0.9629 | 0.7655 | 0.8339 |
| p63δCt | 12,734 | 2402 | 20,809 | 4205 | 21,350 | 6924 | 21,079 | 4017 | 7405 | 65,040 | 13,241 | 0.0717 | 0.7403 | 0.1990 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montero-Calle, A.; Garranzo-Asensio, M.; Torrente-Rodríguez, R.M.; Ruiz-Valdepeñas Montiel, V.; Poves, C.; Dziaková, J.; Sanz, R.; Díaz del Arco, C.; Pingarrón, J.M.; Fernández-Aceñero, M.J.; et al. p53 and p63 Proteoforms Derived from Alternative Splicing Possess Differential Seroreactivity in Colorectal Cancer with Distinct Diagnostic Ability from the Canonical Proteins. Cancers 2023, 15, 2102. https://doi.org/10.3390/cancers15072102
Montero-Calle A, Garranzo-Asensio M, Torrente-Rodríguez RM, Ruiz-Valdepeñas Montiel V, Poves C, Dziaková J, Sanz R, Díaz del Arco C, Pingarrón JM, Fernández-Aceñero MJ, et al. p53 and p63 Proteoforms Derived from Alternative Splicing Possess Differential Seroreactivity in Colorectal Cancer with Distinct Diagnostic Ability from the Canonical Proteins. Cancers. 2023; 15(7):2102. https://doi.org/10.3390/cancers15072102
Chicago/Turabian StyleMontero-Calle, Ana, María Garranzo-Asensio, Rebeca M. Torrente-Rodríguez, Víctor Ruiz-Valdepeñas Montiel, Carmen Poves, Jana Dziaková, Rodrigo Sanz, Cristina Díaz del Arco, José Manuel Pingarrón, María Jesús Fernández-Aceñero, and et al. 2023. "p53 and p63 Proteoforms Derived from Alternative Splicing Possess Differential Seroreactivity in Colorectal Cancer with Distinct Diagnostic Ability from the Canonical Proteins" Cancers 15, no. 7: 2102. https://doi.org/10.3390/cancers15072102
APA StyleMontero-Calle, A., Garranzo-Asensio, M., Torrente-Rodríguez, R. M., Ruiz-Valdepeñas Montiel, V., Poves, C., Dziaková, J., Sanz, R., Díaz del Arco, C., Pingarrón, J. M., Fernández-Aceñero, M. J., Campuzano, S., & Barderas, R. (2023). p53 and p63 Proteoforms Derived from Alternative Splicing Possess Differential Seroreactivity in Colorectal Cancer with Distinct Diagnostic Ability from the Canonical Proteins. Cancers, 15(7), 2102. https://doi.org/10.3390/cancers15072102










