Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
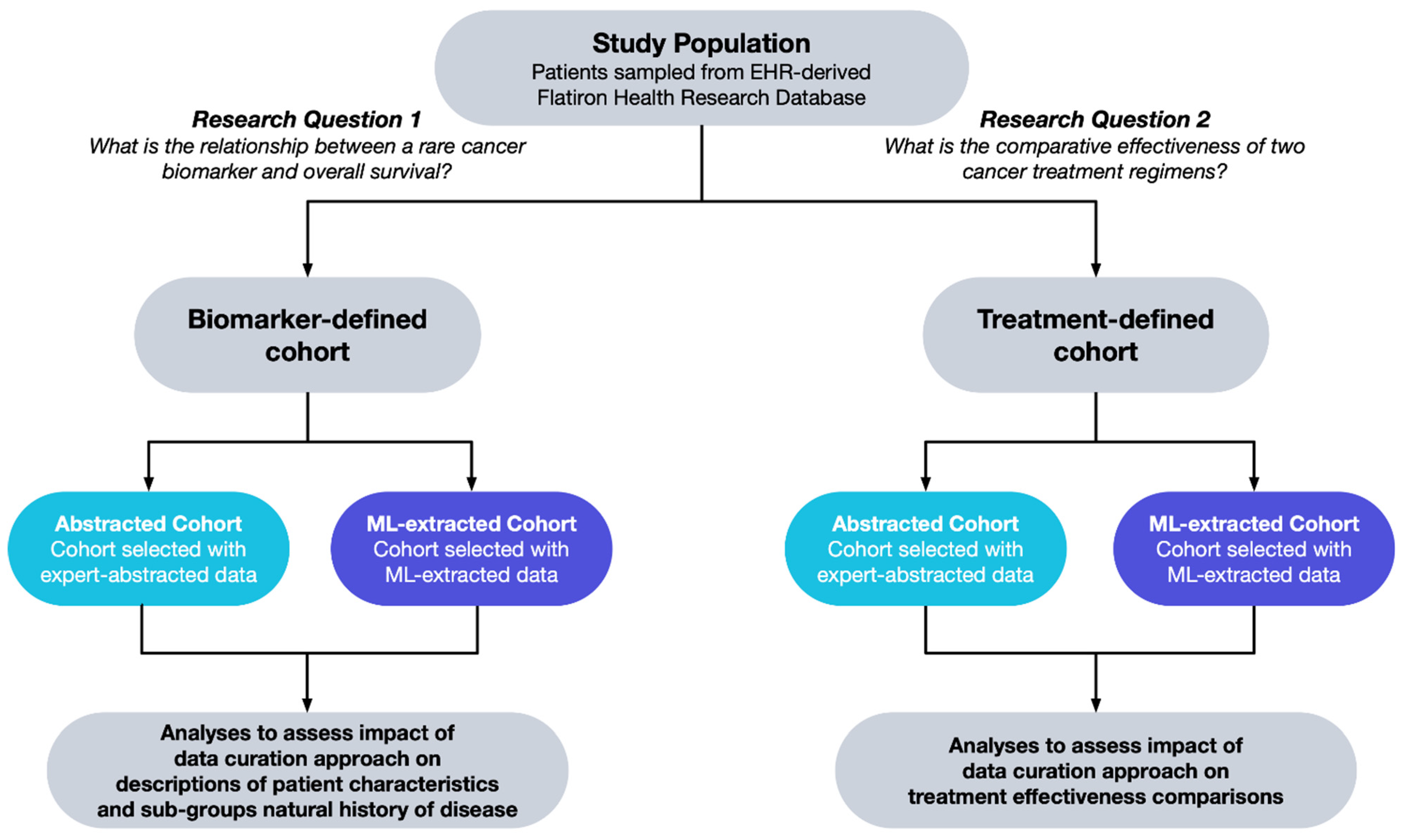
- What is the relationship between a rare cancer biomarker alteration and patient survival?
- What is the comparative effectiveness of two cancer treatment regimens?
2.1. Data Source
2.1.1. Expert Abstraction
2.1.2. Machine Learning Extraction
2.2. Study Population
2.2.1. Biomarker-Defined Cohort
2.2.2. Treatment-Defined Cohort
2.3. Statistical Analysis
- Defining baseline characteristics;
- Describing natural history of disease in biomarker sub-groups;
- Balancing populations;
- Measuring treatment comparative effectiveness.
2.3.1. Defining Baseline Characteristics
2.3.2. Natural History of Disease in Biomarker Sub-Groups
2.3.3. Balancing Populations
2.3.4. Comparative Effectiveness Analysis
3. Results
3.1. Biomarker-Defined Cohort
3.1.1. Defining Baseline Characteristics
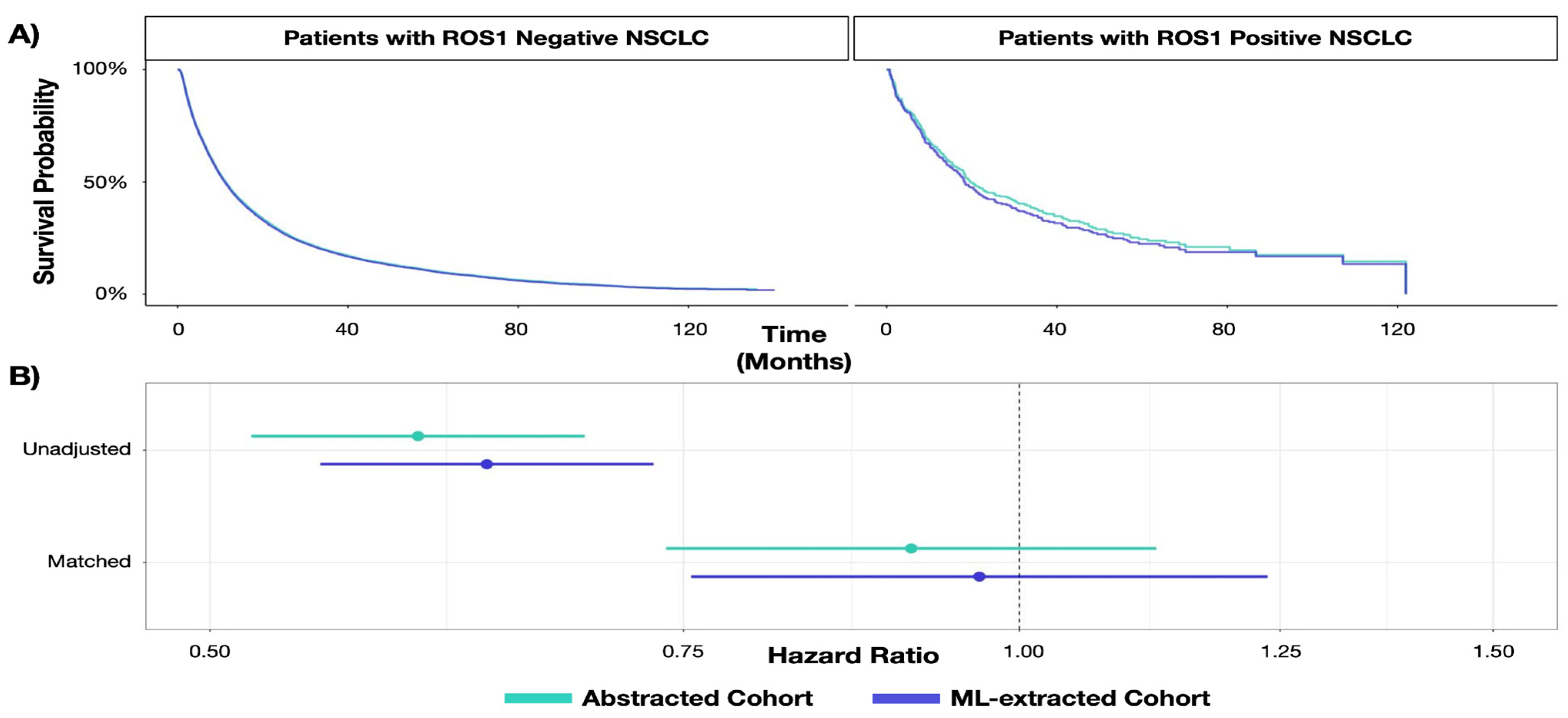
3.1.2. Describing Natural History of Disease in Biomarker Sub-Groups
3.2. Treatment-Defined Cohort
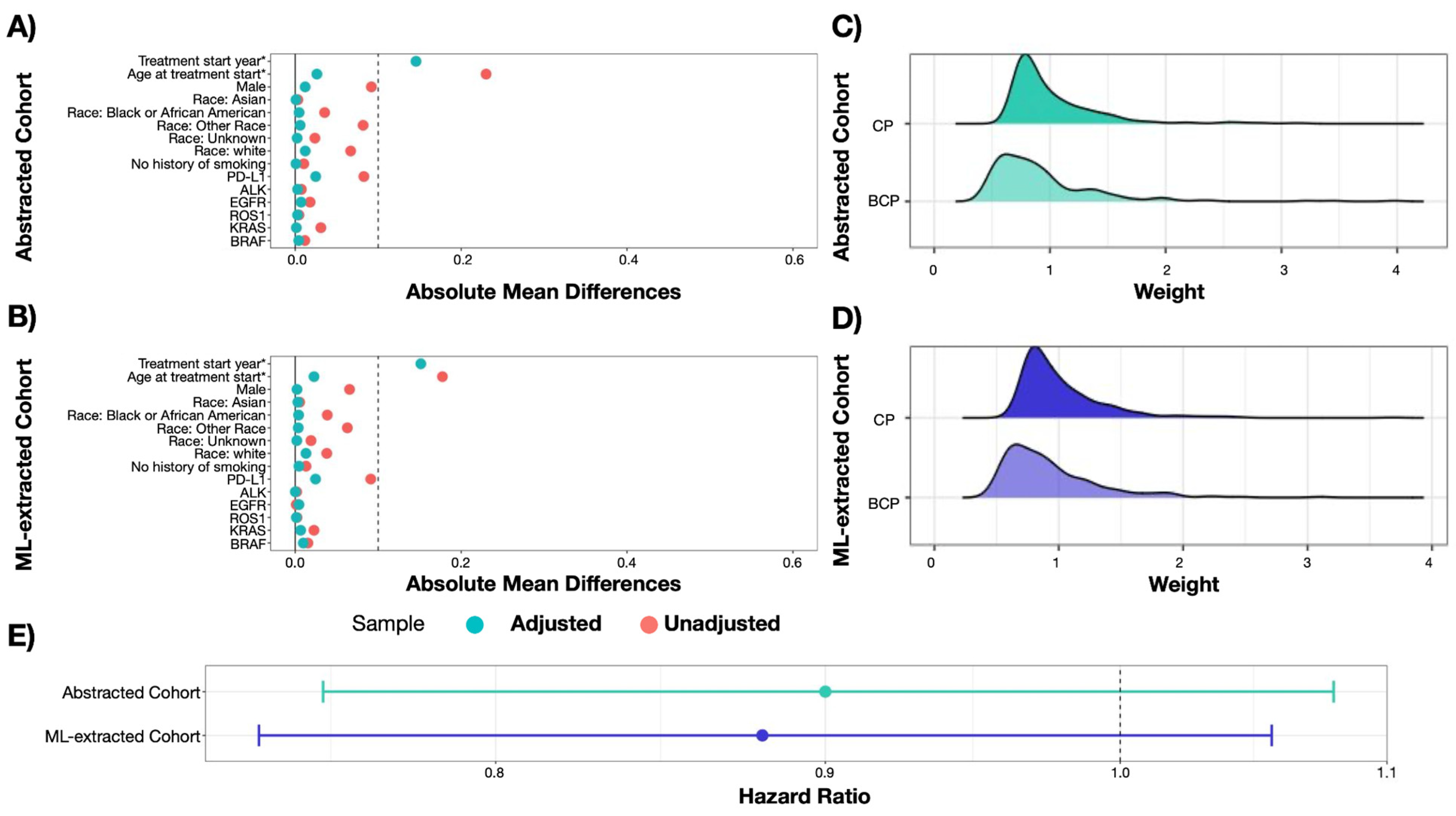
3.2.1. Balancing Populations
3.2.2. Measuring Treatment Comparative Effectiveness
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guinn, D.; Wilhelm, E.E.; Lieberman, G.; Khozin, S. Assessing function of electronic health records for real-world data generation. BMJ Evid.-Based Med. 2019, 24, 95–98. [Google Scholar] [CrossRef]
- Stark, P. Congressional intent for the HITECH Act. Am. J. Manag. Care 2010, 16, SP24–SP28. Available online: https://www.ncbi.nlm.nih.gov/pubmed/21314216 (accessed on 12 January 2023).
- Stewart, M.; Norden, A.D.; Dreyer, N.; Henk, H.J.; Abernethy, A.P.; Chrischilles, E.; Kushi, L.; Mansfield, A.S.; Khozin, S.; Sharon, E.; et al. An Exploratory Analysis of Real-World End Points for Assessing Outcomes Among Immunotherapy-Treated Patients with Advanced Non–Small-Cell Lung Cancer. JCO Clin. Cancer Inform. 2019, 3, 1–15. [Google Scholar] [CrossRef]
- Zhang, J.; Symons, J.; Agapow, P.; Teo, J.T.; Paxton, C.A.; Abdi, J.; Mattie, H.; Davie, C.; Torres, A.Z.; Folarin, A.; et al. Best practices in the real-world data life cycle. PLoS Digit. Health 2022, 1, e0000003. [Google Scholar] [CrossRef]
- Birnbaum, B.; Nussbaum, N.; Seidl-Rathkopf, K.; Agrawal, M.; Estevez, M.; Estola, E.; Haimson, J.; He, L.; Larson, P.; Richardson, P. Model-assisted cohort selection with bias analysis for generating large-scale cohorts from the EHR for oncology research. arXiv 2020, arXiv:2001.09765. [Google Scholar] [CrossRef]
- Estevez, M.; Benedum, C.M.; Jiang, C.; Cohen, A.B.; Phadke, S.; Sarkar, S.; Bozkurt, S. Considerations for the Use of Machine Learning Extracted Real-World Data to Support Evidence Generation: A Research-Centric Evaluation Framework. Cancers 2022, 14, 3063. [Google Scholar] [CrossRef]
- Koskimaki, J.; Hu, J.; Zhang, Y.; Mena, J.; Jones, N.; Lipschultz, E.; Vaidya, V.P.; Altay, G.; Erese, V.A.; Swaminathan, K.K.; et al. Natural language processing-optimized case selection for real-world evidence studies. JCO 2022, 40, 1556. [Google Scholar] [CrossRef]
- Padula, W.V.; Kreif, N.; Vanness, D.J.; Adamson, B.; Rueda, J.-D.; Felizzi, F.; Jonsson, P.; Ijzerman, M.J.; Butte, A.; Crown, W. Machine Learning Methods in Health Economics and Outcomes Research—The PALISADE Checklist: A Good Practices Report of an ISPOR Task Force. Value Health 2022, 25, 1063–1080. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Good Machine Learning Practice for Medical Device Development: Guiding Principles. 2021. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/good-machine-learning-practice-medical-device-development-guiding-principles.http://elsibi.hypotheses.org/3154 (accessed on 2 November 2022).
- NICE Real-World Evidence Framework. National Institute for Health and Care Excellence Web Site. Available online: https://www.nice.org.uk/corporate/ecd9/chapter/overview (accessed on 27 February 2023).
- Real-World Data: Assessing Electronic Health Records and Medical Claims Data To Support Regulatory Decision-Making for Drug and Biological Products; Draft Guidance for Industry; Availability. U.S. Food & Drug Administration Documents/FIND. 2021. Available online: https://www.fda.gov/media/152503/download (accessed on 2 November 2022).
- Schurman, B. The Framework for FDA’s Real-World Evidence Program. Appl. Clin. Trials 2019, 28, 15–17. Available online: https://search.proquest.com/docview/2228576959 (accessed on 2 November 2022).
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef]
- Maarseveen, T.D.; Maurits, M.P.; Niemantsverdriet, E.; Mil, A.H.M.V.D.H.-V.; Huizinga, T.W.J.; Knevel, R. Handwork vs machine: A comparison of rheumatoid arthritis patient populations as identified from EHR free-text by diagnosis extraction through machine-learning or traditional criteria-based chart review. Arthritis Res. Ther. 2021, 23, 174. [Google Scholar] [CrossRef]
- Ma, X.; Long, L.; Moon, S.; Adamson, B.J.S.; Baxi, S.S. Comparison of Population Characteristics in Real-World Clinical Oncology Databases in the US: Flatiron Health, SEER, and NPCR. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Q.; Gossai, A.; Monroe, S.; Nussbaum, N.C.; Parrinello, C.M. Validation analysis of a composite real-world mortality endpoint for patients with cancer in the United States. Health Serv. Res. 2021, 56, 1281–1287. [Google Scholar] [CrossRef]
- Adamson, B.J.; Waskom, M.; Blarre, A.; Kelly, J.; Krismer, K.; Nemeth, S.; Gippetti, J.; Ritten, J.; Harrison, K.; Ho, G.; et al. Approach to Machine Learning for Extraction of Real-World Data Variables from Electronic Health Records. medRxiv 2023. [Google Scholar] [CrossRef]
- Austin, P.C. An Introduction to Propensity Score Methods for Reducing the Effects of Confounding in Observational Studies. Multivar. Behav. Res. 2011, 46, 399–424. [Google Scholar] [CrossRef]
- Tsai, W.-Y.; Jewell, N.P.; Wang, M.-C. A note on the product-limit estimator under right censoring and left truncation. Biometrika 1987, 74, 883–886. [Google Scholar] [CrossRef]
- Sondhi, A. Estimating survival parameters under conditionally independent left truncation. Pharm. Stat. 2022, 21, 895–906. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, Y.-L.; Chen, G.; Liu, X.; Zhu, Y.; Lu, S.; Feng, J.; He, J.; Han, B.; Wang, J.; et al. BEYOND: A Randomized, Double-Blind, Placebo-Controlled, Multicenter, Phase III Study of First-Line Carboplatin/Paclitaxel Plus Bevacizumab or Placebo in Chinese Patients With Advanced or Recurrent Nonsquamous Non–Small-Cell Lung Cancer. JCO 2015, 33, 2197–2204. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 2 November 2022).
- Doebele, R.C.; Perez, L.; Trinh, H.; Martinec, M.; Martina, R.; Riehl, T.; Krebs, M.G.; Meropol, N.J.; Wong, W.B.; Crane, G. Comparative effectiveness analysis between entrectinib clinical trial and crizotinib real-world data in ROS1 + NSCLC. J. Comp. Eff. Res. 2021, 10, 1271–1282. [Google Scholar] [CrossRef]
- Ahmadzada, T.; Kao, S.; Reid, G.; Boyer, M.; Mahar, A.; Cooper, W.A. An Update on Predictive Biomarkers for Treatment Selection in Non-Small Cell Lung Cancer. J. Clin. Med. 2018, 7, 153. [Google Scholar] [CrossRef]
- Gadgeel, S.M.; Thakur, M.K. Predictive and Prognostic Biomarkers in Non-Small Cell Lung Cancer. Semin. Respir. Crit. Care Med. 2016, 37, 760. [Google Scholar] [CrossRef]
- Thunnissen, E.; Van Der Oord, K.; Bakker, M.D. Prognostic and predictive biomarkers in lung cancer. A review. Virchows Arch. 2014, 464, 347–358. [Google Scholar] [CrossRef]
- Tu, H.; Wu, M.; Huang, W.; Wang, L. Screening of potential biomarkers and their predictive value in early stage non-small cell lung cancer: A bioinformatics analysis. Transl. Lung Cancer Res. 2019, 8, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.-M.; Wang, R. Effectiveness and Safety of Adding Bevacizumab to Platinum-Based Chemotherapy as First-Line Treatment for Advanced Non-Small-Cell Lung Cancer: A Meta-Analysis. Front. Med. 2021, 8, 616380. [Google Scholar] [CrossRef]
- Sandler, A.; Gray, R.; Perry, M.C.; Brahmer, J.; Schiller, J.H.; Dowlati, A.; Lilenbaum, R.; Johnson, D.H. Paclitaxel–Carboplatin Alone or with Bevacizumab for Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2006, 355, 2542–2550. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, J.; Chubak, J.; Hubbard, R.A. Inflation of type I error rates due to differential misclassification in EHR-derived outcomes: Empirical illustration using breast cancer recurrence. Pharmacoepidemiol. Drug Saf. 2019, 28, 264–268. [Google Scholar] [CrossRef]
- Van Smeden, M.; Lash, T.L.; Groenwold, R.H.H. Reflection on modern methods: Five myths about measurement error in epidemiological research. Int. J. Epidemiol. 2020, 49, 338–347. [Google Scholar] [CrossRef]
- Lash, T.L.; Fox, M.P.; MacLehose, R.F.; Maldonado, G.; McCandless, L.C.; Greenland, S. Good practices for quantitative bias analysis. Int. J. Epidemiol. 2014, 43, 1969–1985. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; McCormick, T.H.; Leek, J.T. Methods for correcting inference based on outcomes predicted by machine learning. Proc. Natl. Acad. Sci. USA 2020, 117, 30266–30275. [Google Scholar] [CrossRef]
- Richardson, S.; Gilks, W.R. A Bayesian Approach to Measurement Error Problems in Epidemiology Using Conditional Independence Models. Am. J. Epidemiol. 1993, 138, 430–442. [Google Scholar] [CrossRef]
- Berger, M.L.; Curtis, M.D.; Smith, G.; Harnett, J.; Abernethy, A.P. Opportunities and challenges in leveraging electronic health record data in oncology. Futur. Oncol. 2016, 12, 1261–1274. [Google Scholar] [CrossRef]
- Tayefi, M.; Ngo, P.; Chomutare, T.; Dalianis, H.; Salvi, E.; Budrionis, A.; Godtliebsen, F. Challenges and opportunities beyond structured data in analysis of electronic health records. WIREs Comput. Stat. 2021, 13, e1549. [Google Scholar] [CrossRef]
- Hernandez-Boussard, T.; Bozkurt, S.; Ioannidis, J.P.A.; Shah, N.H. MINIMAR (MINimum Information for Medical AI Reporting): Developing reporting standards for artificial intelligence in health care. J. Am. Med. Inform. Assoc. 2020, 27, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Coquet, J.; Bievre, N.; Billaut, V.; Seneviratne, M.; Magnani, C.J.; Bozkurt, S.; Brooks, J.D.; Hernandez-Boussard, T. Assessment of a Clinical Trial–Derived Survival Model in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Netw. Open 2021, 4, e2031730. [Google Scholar] [CrossRef] [PubMed]
- Mansournia, M.A.; Hernán, M.A.; Greenland, S. Matched Designs and Causal Diagrams. Int. J. Epidemiol. 2013, 42, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Stuart, E.A. Matching Methods for Causal Inference: A Review and a Look Forward. Stat. Sci. 2010, 25, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Vander Weele, T.J.; Shpitser, I. A New Criterion for Confounder Selection. Biometrics 2011, 67, 1406–1413. [Google Scholar] [CrossRef]
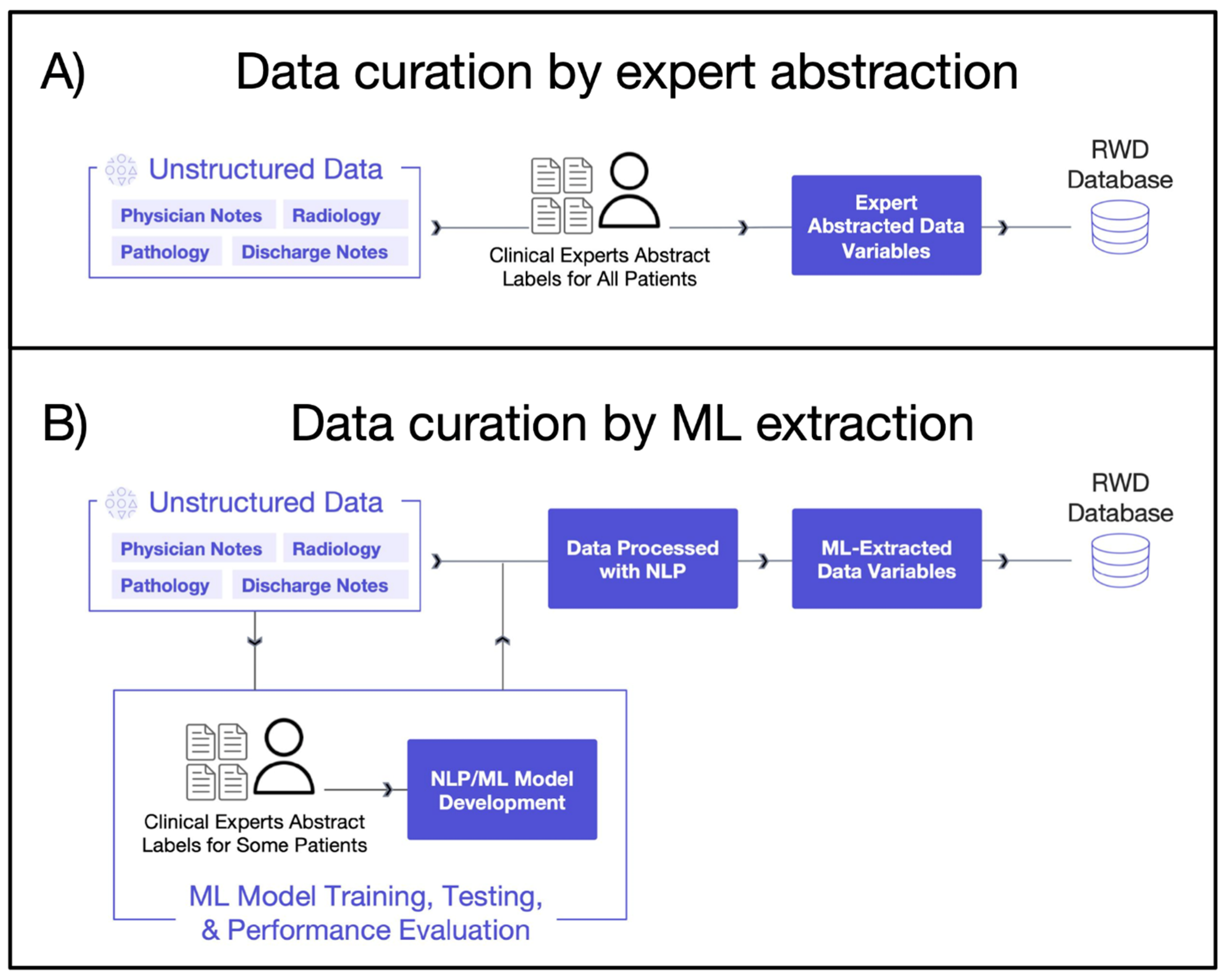
| EHR Source Information Type | Variables Needed for Analysis | Curation Approaches |
|---|---|---|
| Structured data (e.g., date of birth) |
| Transformation, harmonization, and deduplication |
| Unstructured data (e.g., clinic notes, PDF lab reports, radiology images, etc.) |
| Expert abstraction OR ML-extraction |
| ROS1-Positive | ROS1-Negative | |||||
|---|---|---|---|---|---|---|
| Abstracted Cohort | ML-Extracted Cohort | aSMD | Abstracted Cohort | ML-Extracted Cohort | aSMD | |
| N | 349 | 367 | 27,478 | 29,219 | ||
| Practice Type, n (%) | 0.02 | 0.01 | ||||
| Academic | 94 (26.9%) | 102 (27.8%) | 3907 (14.2%) | 4032 (13.8%) | ||
| Community | 255 (73.1%) | 265 (72.2%) | 23,571 (85.8%) | 25,187 (86.2%) | ||
| Gender, n (%) | 0.06 | 0.00 | ||||
| Female | 217 (62.2%) | 218 (59.4%) | 12,966 (47.2%) | 13,790 (47.2%) | ||
| Male | 132 (37.8%) | 149 (40.6%) | 14,510 (52.8%) | 15,427 (52.8%) | ||
| Race/ethnicity, n (%) | 0.03 | 0.01 | ||||
| Black or African American | 38 (10.9%) | 42 (11.4%) | 2419 (8.8%) | 2568 (8.8%) | ||
| Other race a | 64 (18.3%) | 70 (19.1%) | 3637 (13.2%) | 3901 (13.4%) | ||
| Unknown | 32 (9.2%) | 35 (9.5%) | 2794 (10.2%) | 3009 (10.3%) | ||
| White | 215 (61.6%) | 220 (59.9%) | 18,628 (67.8%) | 19,741 (67.6%) | ||
| Age at advanced diagnosis, median [IQR] | 65 (55, 75) | 65 (54, 74) | 0.02 | 69 (62, 76) | 69 (62, 76) | 0.00 |
| Advanced diagnosis year, n (%) | 0.16 | 0.04 | ||||
| 2011 | 3 (0.9%) | 4 (1.1%) | 95 (0.3%) | 105 (0.4%) | ||
| 2012 | 10 (2.9%) | 7 (1.9%) | 253 (0.9%) | 272 (0.9%) | ||
| 2013 | 12 (3.4%) | 16 (4.4%) | 638 (2.3%) | 676 (2.3%) | ||
| 2014 | 18 (5.2%) | 18 (4.9%) | 1147 (4.2%) | 1229 (4.2%) | ||
| 2015 | 15 (4.3%) | 14 (3.8%) | 2025 (7.4%) | 2091 (7.2%) | ||
| 2016 | 37 (10.6%) | 34 (9.3%) | 2647 (9.6%) | 2791 (9.6%) | ||
| 2017 | 56 (16.0%) | 52 (14.2%) | 3487 (12.7%) | 3719 (12.7%) | ||
| 2018 | 44 (12.6%) | 46 (12.5%) | 3726 (13.6%) | 3949 (13.5%) | ||
| 2019 | 47 (13.5%) | 44 (12.0%) | 3811 (13.9%) | 3997 (13.7%) | ||
| 2020 | 36 (10.3%) | 49 (13.4%) | 3713 (13.5%) | 3812 (13.0%) | ||
| 2021 | 49 (14.0%) | 53 (14.4%) | 3708 (13.5%) | 3917 (13.4%) | ||
| 2022 | 22 (6.3%) | 30 (8.2%) | 2228 (8.1%) | 2661 (9.1%) | ||
| Group stage, n (%) | 0.10 | 0.06 | ||||
| Stage I | 16 (4.6%) | 17 (4.6%) | 2331 (8.5%) | 2582 (8.8%) | ||
| Stage II | 5 (1.4%) | 5 (1.4%) | 1387 (5.0%) | 1438 (4.9%) | ||
| Stage III | 60 (17.2%) | 53 (14.4%) | 5514 (20.1%) | 5832 (20.0%) | ||
| Stage IV | 262 (75.1%) | 288 (78.5%) | 17,692 (64.4%) | 18,999 (65.0%) | ||
| Group stage is not reported | 6 (1.7%) | 4 (1.1%) | 554 (2.0%) | 368 (1.3%) | ||
| Histology, n (%) | 0.08 | 0.04 | ||||
| Non-squamous cell carcinoma | 313 (89.7%) | 334 (91.0%) | 20,266 (73.8%) | 21,880 (74.9%) | ||
| NSCLC histology NOS | 12 (3.4%) | 8 (2.2%) | 1274 (4.6%) | 1155 (4.0%) | ||
| Squamous cell carcinoma | 24 (6.9%) | 25 (6.8%) | 5938 (21.6%) | 6184 (21.2%) | ||
| ECOG PS at advanced diagnosis, n (%) | 0.10 | 0.02 | ||||
| 0 | 86 (24.6%) | 82 (22.3%) | 5549 (20.2%) | 5985 (20.5%) | ||
| 1 | 99 (28.4%) | 99 (27.0%) | 7762 (28.2%) | 8405 (28.8%) | ||
| 2 | 18 (5.2%) | 17 (4.6%) | 2588 (9.4%) | 2788 (9.5%) | ||
| 3 | 5 (1.4%) | 4 (1.1%) | 618 (2.2%) | 632 (2.2%) | ||
| 4 | 0 (0.0%) | 0 (0.0%) | 32 (0.1%) | 34 (0.1%) | ||
| Missing/not documented | 141 (40.4%) | 165 (45.0%) | 10,929 (39.8%) | 11,375 (38.9%) | ||
| PD-L1 status, n (%) | 0.09 | 0.07 | ||||
| Negative | 57 (16.3%) | 56 (15.3%) | 6548 (23.8%) | 6878 (23.5%) | ||
| Positive | 178 (51.0%) | 189 (51.5%) | 12,500 (45.5%) | 13,162 (45.0%) | ||
| Unknown | 21 (6.0%) | 30 (8.2%) | 1117 (4.1%) | 1614 (5.5%) | ||
| Not tested | 93 (26.6%) | 92 (25.1%) | 7313 (26.6%) | 7565 (25.9%) | ||
| Treatment received, n (%) | 0.13 | 0.03 | ||||
| Non-oral antineoplastic | 51 (14.6%) | 68 (18.5%) | 19,505 (71.0%) | 20,662 (70.7%) | ||
| Other oral therapy | 36 (10.3%) | 33 (9.0%) | 2691 (9.8%) | 2674 (9.2%) | ||
| ROS1 inhibitor | 224 (64.2%) | 220 (59.9%) | 159 (0.6%) | 139 (0.5%) | ||
| No treatment documented | 38 (10.9%) | 46 (12.5%) | 5123 (18.6%) | 5744 (19.7%) | ||
| RWD Curation Approach | Biomarker Overall Survival HR (95% CI) | SE | p-Value | |
|---|---|---|---|---|
| Unadjusted analysis | Expert-abstracted data | 0.60 (0.52, 0.69) | 0.073 | p < 0.001 |
| ML-extracted data | 0.63 (0.55, 0.73) | 0.073 | p < 0.001 | |
| Adjusted analysis | Expert-abstracted data | 0.91(0.74, 1.12) | 0.107 | 0.387 |
| ML-extracted data | 0.97 (0.76, 1.24) | 0.126 | 0.785 |
| RWD Curation Approach | Treatment Effectiveness HR (95% CI) | SE | p-Value |
|---|---|---|---|
| Expert-abstracted data | 0.90 (0.75, 1.08) | 0.092 | 0.258 |
| ML-extracted data | 0.88 (0.74, 1.06) | 0.092 | 0.170 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benedum, C.M.; Sondhi, A.; Fidyk, E.; Cohen, A.B.; Nemeth, S.; Adamson, B.; Estévez, M.; Bozkurt, S. Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning. Cancers 2023, 15, 1853. https://doi.org/10.3390/cancers15061853
Benedum CM, Sondhi A, Fidyk E, Cohen AB, Nemeth S, Adamson B, Estévez M, Bozkurt S. Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning. Cancers. 2023; 15(6):1853. https://doi.org/10.3390/cancers15061853
Chicago/Turabian StyleBenedum, Corey M., Arjun Sondhi, Erin Fidyk, Aaron B. Cohen, Sheila Nemeth, Blythe Adamson, Melissa Estévez, and Selen Bozkurt. 2023. "Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning" Cancers 15, no. 6: 1853. https://doi.org/10.3390/cancers15061853
APA StyleBenedum, C. M., Sondhi, A., Fidyk, E., Cohen, A. B., Nemeth, S., Adamson, B., Estévez, M., & Bozkurt, S. (2023). Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning. Cancers, 15(6), 1853. https://doi.org/10.3390/cancers15061853









