Extracellular Vesicles as Biomarkers in Head and Neck Squamous Cell Carcinoma: From Diagnosis to Disease-Free Survival
Abstract
Simple Summary
Abstract
1. Introduction to Extracellular Vesicles and Head and Neck Squamous Cell Carcinoma
1.1. Head and Neck Squamous Cell Carcinoma
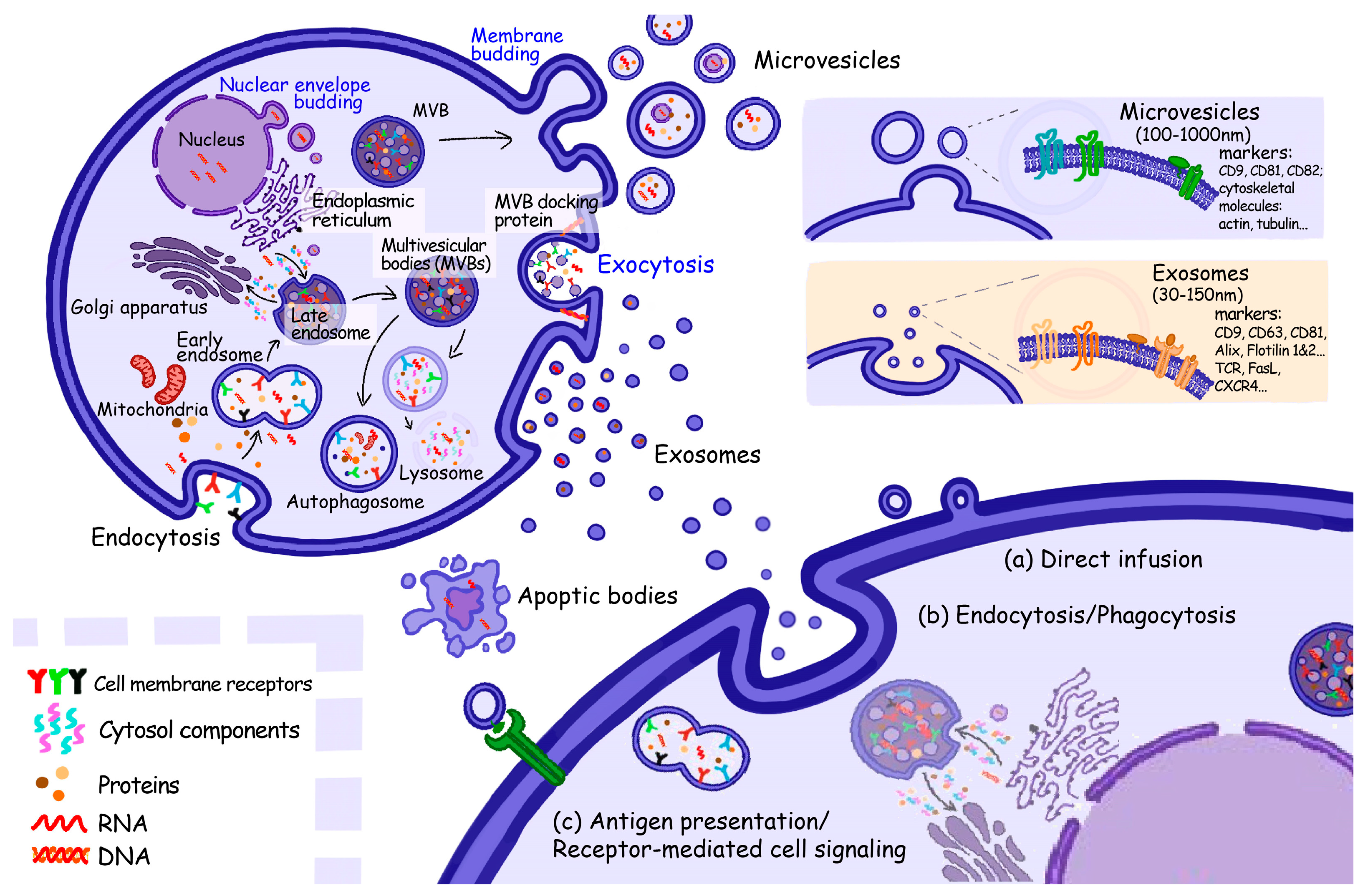
1.2. Extracellular Vesicles
2. Overview of Biomarkers in HNSCC—cfDNA, Circulating Tumor Cells, EVs
2.1. Cell-Free DNA
2.2. Circulating Tumor Cells, CTCs
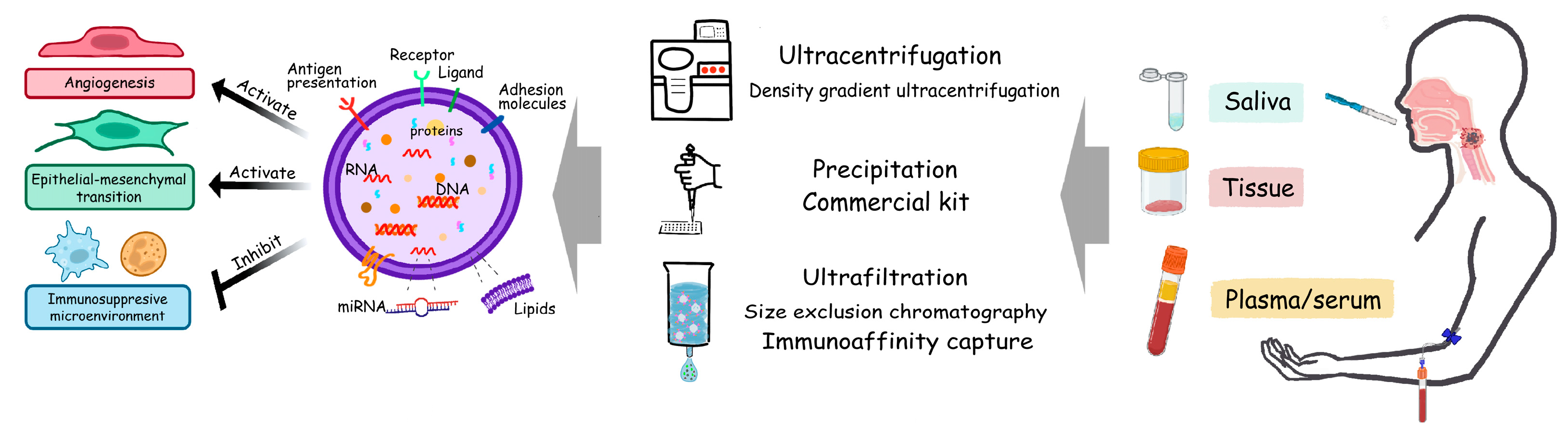
2.3. Extracellular Vesicles, EVs
3. EV Cargoes and Their Instructive Effects in HNSCC
3.1. DNA
3.2. RNA
| First Author | Year | Cancer Type | RNA | Sample Type | Grouping | EVs Isolation Method | RNA Detection Method | Clinical Use | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Wang | 2014 | LSCC * | miR-21 | Serum | LSCC (n = 52); benign laryngeal disease (n = 49) | Precipitation | qRT-PCR | Diagnosis, prognosis | [151] |
| Li | 2016 | OSCC * | miR-21 | Serum | OSCC (n = 108); healthy control (n = 108) | ExoQuick kit (SBI)/precipitation | qRT-PCR | Progression (metastasis) | [152] |
| Ye | 2016 | NPC * | miR-24-3p | Plasma | NPC (n = 65); healthy control (n = 20) | Ultra-centrifugation (UC) | qRT-PCR array, qRT-PCR | Prognosis | [153] |
| Langevin | 2017 | HNSCC * | miR-486-5p, miR-486-3p, miR-10b-5p, miR-31 | Saliva | HNSCC (n = 11); healthy control (n = 9) | UC | Sequencing, ddPCR | Diagnosis | [154] |
| Ramayanti | 2019 | NPC | BART7-3p, BART9-3p, BART13-3p | Serum | NPC (n = 39); healthy control (n = 33) | SEC Sepharose CL-2B column | LightCycler stem-loop RT-PCR | Diagnosis | [155] |
| Momen-Heravi | 2018 | OSCC | miR-21, miR-27, miR-412-3p, miR-512-3p | Saliva | OSCC (n = 24); healthy control (n = 10) | ExoQuick kit (SBI)/precipitation | qRT-PCR and TaqMan miRNA assay | Diagnosis | [156] |
| Bao | 2018 | NPC | miR-23a | Tissue, serum | NPC (n = 150) | UC | qRT-PCR | Progression (metastasis), prognosis | [157] |
| Gai | 2018 | OSCC | miR-512-3p, miR-412-3p, miR-302b-3p, miR-517b-3p, miR-21, HOTAIR | Saliva | OSCC (n = 21); healthy control (n = 11) | Precipitation | qRT-PCR array, qRT-PCR | Diagnosis | [112] |
| Lu | 2018 | NPC | miR-9 | Plasma | NPC (n = 110); healthy control (n = 60) | UC | qRT-PCR | Diagnosis, prognosis | [158] |
| Xie | 2019 | OSCC | miR-101-3p | Tissue | OSCC (n = 36, tumor tissue vs. adjacent normal tissue) | UC | qRT-PCR | Progression (stage) | [159] |
| Qin | 2019 | HNSCC | miR-196a | Plasma | HNSCC (n = 74); healthy control (n = 30) | ExoQuick kit (SBI), precipitation | qRT-PCR | Diagnosis, prognosis, treatment response (cisplatin) | [160] |
| Sun | 2019 | OSCC | miR-382-5p | Cocultured tissue | 47 OSCC patients (CAF-rich vs. CAF-poor) | Precipitation | qRT-PCR | Progression | [161] |
| Mayne | 2020 | OPSCC * | 11-miR (ratio) panel | Serum | OPSCC (n = 40); GORD (n = 20); healthy control (n = 20) | Precipitation | Microarray | Diagnosis | [162] |
| Wang | 2020 | OSCC | miR-210-3p | Tissue | OSCC (n = 80); healthy control (n = 7) | UC | qRT-PCR | Progression (stage) | [163] |
| He | 2020 | OSCC | miR-24-3p | Saliva | HNSCC (n = 49); healthy control (n = 14) | ExoQuick kit (SBI), precipitation | Microarray, qRT-PCR | Diagnosis | [164] |
| Shuang | 2022 | LSCC | miR-27a | Plasma | LSCC (n = 62); healthy control uk | ExoQuick kit (SBI), precipitation | qRT-PCR array, qRT-PCR | Diagnosis, prognosis | [165] |
| Yin | 2021 | NPC | miR-433-3p | Tissue | LSCC (n = 13, tumor tissue vs. adjacent normal tissue) | UC | qRT-PCR | Progression (stage), prognosis | [166] |
| Tong | 2020 | HNSCC | miR-9 | Tissue | HNSCC (n = 303) | UC | qRT-PCR | Prognosis, treatment response (RT *) | [167] |
| Jiang | 2021 | NPC | miR-134-5p, miR-205-5p, miR-409-3p | Plasma | NPC (n = 16); healthy control (n = 5) | Precipitation | Nanoparticle tracking analysis (NTA) and qRT-PCR | Diagnosis | [168] |
| Chen | 2021 | OSCC | miR-155, miR-21, miR-126 | Serum | OSCC (n = 35); healthy control (n = 11) | Exosome Isolation Kit | Microarray, qRT-PCR | Progression (stage), prognosis | [169] |
| Liu | 2021 | NPC | miR-181a | Tissue | NPC (n = 134) | UC | qRT-PCR | Progression (metastasis, staging) | [170] |
| Bigagli | 2021 | OSCC | miR-210 | Plasma | OSCC (n = 30); healthy control (n = 14) | Precipitation | qRT-PCR | Diagnosis, prognosis | [138] |
| Panvongsa | 2021 | HNSCC | miR-491-5p | Plasma | HNSCC (n = 73); healthy control (n = 20) | ExoQuick kit (SBI), precipitation | miRNA microarray analysis, qRT-PCR | Diagnosis, prognosis | [171] |
| Yu | 2022 | LSCC | miR-15b-5p | Cocultured tissue | LSCC (n = 16, tumor tissue vs. adjacent normal tissue) | ExoQuick kit (SBI), precipitation | qRT-PCR | Progression (LNM *) | [172] |
| Jiang | 2022 | NPC | miR-197.3p | Tissue | NPC (n = 40, with good vs. poor RT effects) | ExoQuick kit (SBI), precipitation | qRT-PCR | Treatment (RT *) | [173] |
| Lou | 2022 | OSCC | miR-626 | Serum, tissue | OSCC (n = 65, tumor and serum; n = 10, tumor tissue vs. adjacent normal tissue) | UC | qRT-PCR | Prognosis | [174] |
3.3. Protein
4. EV as Prospective Biomarkers in HNSCC
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Castellsagué, X.; Quintana, M.J.; Martínez, M.C.; Nieto, A.; Sánchez, M.J.; Juan, A.; Monner, A.; Carrera, M.; Agudo, A.; Quer, M.; et al. The role of type of tobacco and type of alcoholic beverage in oral carcinogenesis. Int. J. Cancer 2004, 108, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Aupérin, A. Epidemiology of head and neck cancers: An update. Curr. Opin. Oncol. 2020, 32, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Livingston, M.; Room, R.; Gan, Y.; English, D.; Chenhall, R. Can public health policies on alcohol and tobacco reduce a cancer epidemic? Australia’s experience. BMC Med. 2019, 17, 213. [Google Scholar] [CrossRef]
- Marur, S.; D’Souza, G.; Westra, W.H.; Forastiere, A.A. HPV-associated head and neck cancer: A virus-related cancer epidemic. Lancet Oncol. 2010, 11, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Rassy, E.; Nicolai, P.; Pavlidis, N. Comprehensive management of HPV-related squamous cell carcinoma of the head and neck of unknown primary. Head Neck 2019, 41, 3700–3711. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Bell, R.B.; Bifulco, C.B.; Burtness, B.; Gillison, M.L.; Harrington, K.J.; Le, Q.T.; Lee, N.Y.; Leidner, R.; Lewis, R.L.; et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC). J. Immunother. Cancer 2019, 7, 184. [Google Scholar] [CrossRef]
- Marur, S.; Forastiere, A.A. Head and neck cancer: Changing epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2008, 83, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Bhat, G.R.; Hyole, R.G.; Li, J. Head and neck cancer: Current challenges and future perspectives. Adv. Cancer Res. 2021, 152, 67–102. [Google Scholar] [CrossRef]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Azarnia, N.; Shin, D.M.; Cohen, R.B.; Jones, C.U.; Sur, R.; Raben, D.; Jassem, J.; et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2006, 354, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Seiwert, T.Y.; Burtness, B.; Mehra, R.; Weiss, J.; Berger, R.; Eder, J.P.; Heath, K.; McClanahan, T.; Lunceford, J.; Gause, C.; et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): An open-label, multicentre, phase 1b trial. Lancet Oncol. 2016, 17, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Schwarz, M.A. Cell-Cell Communication Breakdown and Endothelial Dysfunction. Crit. Care Clin. 2020, 36, 189–200. [Google Scholar] [CrossRef] [PubMed]
- O’Loughlin, A.J.; Woffindale, C.A.; Wood, M.J. Exosomes and the emerging field of exosome-based gene therapy. Curr. Gene Ther. 2012, 12, 262–274. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef]
- Müller Bark, J.; Kulasinghe, A.; Amenábar, J.M.; Punyadeera, C. Exosomes in cancer. Adv. Clin. Chem. 2021, 101, 1–40. [Google Scholar] [CrossRef]
- Vago, R.; Salonia, A. Re: Circulating Extracellular Vesicles in Human Disease. Eur. Urol. 2019, 75, 342–343. [Google Scholar] [CrossRef]
- Willis, G.R.; Kourembanas, S.; Mitsialis, S.A. Toward Exosome-Based Therapeutics: Isolation, Heterogeneity, and Fit-for-Purpose Potency. Front. Cardiovasc. Med. 2017, 4, 63. [Google Scholar] [CrossRef]
- D’Acunzo, P.; Hargash, T.; Pawlik, M.; Goulbourne, C.N.; Pérez-González, R.; Levy, E. Enhanced generation of intraluminal vesicles in neuronal late endosomes in the brain of a Down syndrome mouse model with endosomal dysfunction. Dev. Neurobiol. 2019, 79, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Elkin, S.R.; Lakoduk, A.M.; Schmid, S.L. Endocytic pathways and endosomal trafficking: A primer. Wien Med. Wochenschr. 2016, 166, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Pollet, H.; Conrard, L.; Cloos, A.S.; Tyteca, D. Plasma Membrane Lipid Domains as Platforms for Vesicle Biogenesis and Shedding? Biomolecules 2018, 8, 94. [Google Scholar] [CrossRef]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, J.; Yu, P.; Guo, L.; Mao, X.; Wang, J.; Miao, S.; Sun, J. The roles of extracellular vesicles in the development, microenvironment, anticancer drug resistance, and therapy of head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 35. [Google Scholar] [CrossRef]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef]
- Visan, K.S.; Wu, L.Y.; Voss, S.; Wuethrich, A.; Möller, A. Status quo of Extracellular Vesicle isolation and detection methods for clinical utility. Semin. Cancer Biol. 2023, 88, 157–171. [Google Scholar] [CrossRef]
- Woo, H.K.; Sunkara, V.; Park, J.; Kim, T.H.; Han, J.R.; Kim, C.J.; Choi, H.I.; Kim, Y.K.; Cho, Y.K. Exodisc for Rapid, Size-Selective, and Efficient Isolation and Analysis of Nanoscale Extracellular Vesicles from Biological Samples. ACS Nano 2017, 11, 1360–1370. [Google Scholar] [CrossRef]
- Xu, H.; Liao, C.; Zuo, P.; Liu, Z.; Ye, B.C. Magnetic-Based Microfluidic Device for On-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal. Chem. 2018, 90, 13451–13458. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Yang, F.; Zhang, Y.; Liu, L.; Zhou, Y.; Wang, F.; Zhang, G.J. ExoAPP: Exosome-Oriented, Aptamer Nanoprobe-Enabled Surface Proteins Profiling and Detection. Anal. Chem. 2018, 90, 14402–14411. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin. Immunol. 2018, 35, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Szczepanski, M.J.; Szajnik, M.; Welsh, A.; Whiteside, T.L.; Boyiadzis, M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica 2011, 96, 1302–1309. [Google Scholar] [CrossRef]
- Whiteside, T.L. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin. Exp. Immunol. 2017, 189, 259–267. [Google Scholar] [CrossRef]
- Boyiadzis, M.; Whiteside, T.L. Information transfer by exosomes: A new frontier in hematologic malignancies. Blood Rev. 2015, 29, 281–290. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5, e10250. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Zou, L.; Zhu, Z. Role of Exosomes in Crosstalk between Cancer-Associated Fibroblasts and Cancer Cells. Front. Oncol. 2019, 9, 356. [Google Scholar] [CrossRef]
- Zhu, T.; Hu, Z.; Wang, Z.; Ding, H.; Li, R.; Wang, J.; Wang, G. microRNA-301b-3p from mesenchymal stem cells-derived extracellular vesicles inhibits TXNIP to promote multidrug resistance of gastric cancer cells. Cell Biol. Toxicol. 2022, 1–15. [Google Scholar] [CrossRef]
- Weng, Z.; Zhang, B.; Wu, C.; Yu, F.; Han, B.; Li, B.; Li, L. Therapeutic roles of mesenchymal stem cell-derived extracellular vesicles in cancer. J. Hematol. Oncol. 2021, 14, 136. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, T.; Ge, S.; Liu, Y.; Bai, M.; Zhu, K.; Fan, Q.; Li, J.; Ning, T.; Tian, F.; et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 2019, 38, 2844–2859. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.N.; Cheng, L.C.; Kuo, C.L.; Lo, Y.K.; Chou, H.Y.; Chen, C.H.; Wang, Y.H.; Chuang, T.H.; Cheng, S.J.; Lee, A.Y. Mitochondrial Lon-induced mtDNA leakage contributes to PD-L1-mediated immunoescape via STING-IFN signaling and extracellular vesicles. J. Immunother. Cancer 2020, 8, e001372. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Lee, J.E.; Sung, K.J.; Sung, Y.H.; Pack, C.G.; Jung, M.K.; Han, B.; et al. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Song, C.; Zheng, L.; Xia, L.; Li, Y.; Zhou, Y. The roles of extracellular vesicles in gastric cancer development, microenvironment, anti-cancer drug resistance, and therapy. Mol. Cancer 2019, 18, 62. [Google Scholar] [CrossRef]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yu, X.; Zhou, Z.; Li, B.; Peng, J.; Wu, X.; Luo, X.; Yang, L. LMP1-positive extracellular vesicles promote radioresistance in nasopharyngeal carcinoma cells through P38 MAPK signaling. Cancer Med. 2019, 8, 6082–6094. [Google Scholar] [CrossRef] [PubMed]
- EL Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Besse, B.; Charrier, M.; Lapierre, V.; Dansin, E.; Lantz, O.; Planchard, D.; Le Chevalier, T.; Livartoski, A.; Barlesi, F.; Laplanche, A.; et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology 2016, 5, e1071008. [Google Scholar] [CrossRef]
- Dai, S.; Wei, D.; Wu, Z.; Zhou, X.; Wei, X.; Huang, H.; Li, G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol. Ther. 2008, 16, 782–790. [Google Scholar] [CrossRef]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef]
- Li, S.; Han, Y.; Lu, M.; Liu, Z.; Jin, J.; Guo, Q.; Wang, Y.; Liu, H. Mesenchymal stem cell-exosome-mediated matrix metalloproteinase 1 participates in oral leukoplakia and carcinogenesis by inducing angiogenesis. J. Oral Pathol. Med. 2022, 51, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer 2021, 144, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Kubo, H. Extracellular Vesicles in Lung Disease. Chest 2018, 153, 210–216. [Google Scholar] [CrossRef]
- Wu, H.; Fu, M.; Liu, J.; Chong, W.; Fang, Z.; Du, F.; Liu, Y.; Shang, L.; Li, L. The role and application of small extracellular vesicles in gastric cancer. Mol. Cancer 2021, 20, 71. [Google Scholar] [CrossRef]
- Zheng, X.; Xu, K.; Zhou, B.; Chen, T.; Huang, Y.; Li, Q.; Wen, F.; Ge, W.; Wang, J.; Yu, S.; et al. A circulating extracellular vesicles-based novel screening tool for colorectal cancer revealed by shotgun and data-independent acquisition mass spectrometry. J. Extracell. Vesicles 2020, 9, 1750202. [Google Scholar] [CrossRef] [PubMed]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.J.; Erice, O.; Gonzalez, E.; Jimenez-Agüero, R.; Lacasta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef]
- Thietart, S.; Rautou, P.E. Extracellular vesicles as biomarkers in liver diseases: A clinician’s point of view. J. Hepatol. 2020, 73, 1507–1525. [Google Scholar] [CrossRef]
- Logozzi, M.; Angelini, D.F.; Iessi, E.; Mizzoni, D.; Di Raimo, R.; Federici, C.; Lugini, L.; Borsellino, G.; Gentilucci, A.; Pierella, F.; et al. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett. 2017, 403, 318–329. [Google Scholar] [CrossRef]
- Leung, L.L.; Riaz, M.K.; Qu, X.; Chan, J.; Meehan, K. Profiling of extracellular vesicles in oral cancer, from transcriptomics to proteomics. Semin. Cancer Biol. 2021, 74, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Leung, T.C.N.; Ngai, S.M.; Tsai, S.N.; Thakur, A.; Li, W.K.; Lee, Y.; Leung, L.; Ng, T.H.; Yam, J.; et al. Proteomic Analysis of Circulating Extracellular Vesicles Identifies Potential Biomarkers for Lymph Node Metastasis in Oral Tongue Squamous Cell Carcinoma. Cells 2021, 10, 2179. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Li, J.W.; Chan, J.; Meehan, K. Extracellular Vesicles in Head and Neck Cancer: A Potential New Trend in Diagnosis, Prognosis, and Treatment. Int. J. Mol. Sci. 2020, 21, 8260. [Google Scholar] [CrossRef]
- Galle, P.R.; Foerster, F.; Kudo, M.; Chan, S.L.; Llovet, J.M.; Qin, S.; Schelman, W.R.; Chintharlapalli, S.; Abada, P.B.; Sherman, M.; et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019, 39, 2214–2229. [Google Scholar] [CrossRef] [PubMed]
- Saini, S. PSA and beyond: Alternative prostate cancer biomarkers. Cell Oncol. 2016, 39, 97–106. [Google Scholar] [CrossRef]
- Vaitukaitis, J.L. Human chorionic gonadotropin—A hormone secreted for many reasons. N. Engl. J. Med. 1979, 301, 324–326. [Google Scholar] [CrossRef]
- Izuhara, K.; Yamaguchi, Y.; Ohta, S.; Nunomura, S.; Nanri, Y.; Azuma, Y.; Nomura, N.; Noguchi, Y.; Aihara, M. Squamous Cell Carcinoma Antigen 2 (SCCA2, SERPINB4): An Emerging Biomarker for Skin Inflammatory Diseases. Int. J. Mol. Sci. 2018, 19, 1102. [Google Scholar] [CrossRef]
- Kommu, S.; Hadway, P.; Watkin, N. Squamous cell carcinoma antigen as a biomarker for penile cancer. BJU Int. 2005, 95, 478–479. [Google Scholar] [CrossRef]
- Sun, Z.; Shi, X.; Wang, Y.; Zhao, Y. Serum Squamous Cell Carcinoma Antigen in Psoriasis: A Potential Quantitative Biomarker for Disease Severity. Dermatology 2018, 234, 120–126. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear Acids In Human Blood Plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, F.S.; Barauna, V.G.; Dos Santos, L.; Costa, G.; Vassallo, P.F.; Campos, L.C.G. Properties and Application of Cell-Free DNA as a Clinical Biomarker. Int. J. Mol. Sci. 2021, 22, 9110. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Corcoran, R.B. Liquid biopsy versus tumor biopsy for clinical-trial recruitment. Nat. Med. 2020, 26, 1815–1816. [Google Scholar] [CrossRef]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Nakamura, Y.; Taniguchi, H.; Ikeda, M.; Bando, H.; Kato, K.; Morizane, C.; Esaki, T.; Komatsu, Y.; Kawamoto, Y.; Takahashi, N.; et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat. Med. 2020, 26, 1859–1864. [Google Scholar] [CrossRef]
- Yang, X.; Song, H.; Ji, T.; Du, G.; Liu, W. The implications of gene mutations in salivary DNA for noninvasive diagnosis of head and neck cancer with a focus on oral cancer. Oral. Oncol. 2022, 130, 105924. [Google Scholar] [CrossRef] [PubMed]
- Arantes, L.; De Carvalho, A.C.; Melendez, M.E.; Lopes Carvalho, A. Serum, plasma and saliva biomarkers for head and neck cancer. Expert. Rev. Mol. Diagn. 2018, 18, 85–112. [Google Scholar] [CrossRef] [PubMed]
- Langevin, S.M.; Stone, R.A.; Bunker, C.H.; Grandis, J.R.; Sobol, R.W.; Taioli, E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis 2010, 31, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.A.; Beck, J.; Leu, M.; Oellerich, M.; Rave-Fränk, M.; Walson, P.D.; Schütz, E.; Canis, M. Cell-Free Plasma DNA for Disease Stratification and Prognosis in Head and Neck Cancer. Clin. Chem. 2018, 64, 959–970. [Google Scholar] [CrossRef]
- Wilson, H.L.; D’Agostino, R.B., Jr.; Meegalla, N.; Petro, R.; Commander, S.; Topaloglu, U.; Zhang, W.; Porosnicu, M. The Prognostic and Therapeutic Value of the Mutational Profile of Blood and Tumor Tissue in Head and Neck Squamous Cell Carcinoma. Oncologist 2021, 26, e279–e289. [Google Scholar] [CrossRef]
- Wuerdemann, N.; Jain, R.; Adams, A.; Speel, E.M.; Wagner, S.; Joosse, S.A.; Klussmann, J.P. Cell-Free HPV-DNA as a Biomarker for Oropharyngeal Squamous Cell Carcinoma-A Step Towards Personalized Medicine? Cancers 2020, 12, 2997. [Google Scholar] [CrossRef] [PubMed]
- Gkountela, S.; Castro-Giner, F.; Szczerba, B.M.; Vetter, M.; Landin, J.; Scherrer, R.; Krol, I.; Scheidmann, M.C.; Beisel, C.; Stirnimann, C.U.; et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019, 176, 98–112.e14. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Toner, M.; Maheswaran, S.; Haber, D.A. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends Cancer 2015, 1, 44–52. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef]
- Hristozova, T.; Konschak, R.; Stromberger, C.; Fusi, A.; Liu, Z.; Weichert, W.; Stenzinger, A.; Budach, V.; Keilholz, U.; Tinhofer, I. The presence of circulating tumor cells (CTCs) correlates with lymph node metastasis in nonresectable squamous cell carcinoma of the head and neck region (SCCHN). Ann. Oncol. 2011, 22, 1878–1885. [Google Scholar] [CrossRef] [PubMed]
- Strati, A.; Koutsodontis, G.; Papaxoinis, G.; Angelidis, I.; Zavridou, M.; Economopoulou, P.; Kotsantis, I.; Avgeris, M.; Mazel, M.; Perisanidis, C.; et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann. Oncol. 2017, 28, 1923–1933. [Google Scholar] [CrossRef]
- De Wit, S.; van Dalum, G.; Lenferink, A.T.; Tibbe, A.G.; Hiltermann, T.J.; Groen, H.J.; van Rijn, C.J.; Terstappen, L.W. The detection of EpCAM(+) and EpCAM(−) circulating tumor cells. Sci. Rep. 2015, 5, 12270. [Google Scholar] [CrossRef]
- Mikolajczyk, S.D.; Millar, L.S.; Tsinberg, P.; Coutts, S.M.; Zomorrodi, M.; Pham, T.; Bischoff, F.Z.; Pircher, T.J. Detection of EpCAM-Negative and Cytokeratin-Negative Circulating Tumor Cells in Peripheral Blood. J. Oncol. 2011, 2011, 252361. [Google Scholar] [CrossRef] [PubMed]
- Pauken, C.M.; Kenney, S.R.; Brayer, K.J.; Guo, Y.; Brown-Glaberman, U.A.; Marchetti, D. Heterogeneity of Circulating Tumor Cell Neoplastic Subpopulations Outlined by Single-Cell Transcriptomics. Cancers 2021, 13, 4885. [Google Scholar] [CrossRef]
- Mego, M.; Cierna, Z.; Janega, P.; Karaba, M.; Minarik, G.; Benca, J.; Sedlácková, T.; Sieberova, G.; Gronesova, P.; Manasova, D.; et al. Relationship between circulating tumor cells and epithelial to mesenchymal transition in early breast cancer. BMC Cancer 2015, 15, 533. [Google Scholar] [CrossRef]
- Smolkova, B.; Cierna, Z.; Kalavska, K.; Miklikova, S.; Plava, J.; Minarik, G.; Sedlackova, T.; Cholujova, D.; Gronesova, P.; Cihova, M.; et al. Increased Stromal Infiltrating Lymphocytes Are Associated with the Risk of Disease Progression in Mesenchymal Circulating Tumor Cell-Positive Primary Breast Cancer Patients. Int. J. Mol. Sci. 2020, 21, 9460. [Google Scholar] [CrossRef]
- Deutsch, T.M.; Riethdorf, S.; Nees, J.; Hartkopf, A.D.; Schönfisch, B.; Domschke, C.; Sprick, M.R.; Schütz, F.; Brucker, S.Y.; Stefanovic, S.; et al. Impact of apoptotic circulating tumor cells (aCTC) in metastatic breast cancer. Breast Cancer Res. Treat. 2016, 160, 277–290. [Google Scholar] [CrossRef]
- Kallergi, G.; Konstantinidis, G.; Markomanolaki, H.; Papadaki, M.A.; Mavroudis, D.; Stournaras, C.; Georgoulias, V.; Agelaki, S. Apoptotic circulating tumor cells in early and metastatic breast cancer patients. Mol. Cancer Ther. 2013, 12, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Strilic, B.; Offermanns, S. Intravascular Survival and Extravasation of Tumor Cells. Cancer Cell 2017, 32, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Dopico, P.; Varillas, J.; Zhang, J.; George, T.J.; Fan, Z.H. Integration of Lateral Filter Arrays with Immunoaffinity for Circulating-Tumor-Cell Isolation. Angew. Chem. Int. Ed. Engl. 2019, 58, 7606–7610. [Google Scholar] [CrossRef]
- Cho, H.; Kim, J.; Song, H.; Sohn, K.Y.; Jeon, M.; Han, K.H. Microfluidic technologies for circulating tumor cell isolation. Analyst 2018, 143, 2936–2970. [Google Scholar] [CrossRef]
- Gabriel, M.T.; Calleja, L.R.; Chalopin, A.; Ory, B.; Heymann, D. Circulating Tumor Cells: A Review of Non-EpCAM-Based Approaches for Cell Enrichment and Isolation. Clin. Chem. 2016, 62, 571–581. [Google Scholar] [CrossRef]
- Lin, M.; Chen, J.F.; Lu, Y.T.; Zhang, Y.; Song, J.; Hou, S.; Ke, Z.; Tseng, H.R. Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc. Chem. Res. 2014, 47, 2941–2950. [Google Scholar] [CrossRef]
- Cohen, E.N.; Jayachandran, G.; Moore, R.G.; Cristofanilli, M.; Lang, J.E.; Khoury, J.D.; Press, M.F.; Kim, K.K.; Khazan, N.; Zhang, Q.; et al. A Multi-Center Clinical Study to Harvest and Characterize Circulating Tumor Cells from Patients with Metastatic Breast Cancer Using the Parsortix(®) PC1 System. Cancers 2022, 14, 5238. [Google Scholar] [CrossRef] [PubMed]
- Garrel, R.; Mazel, M.; Perriard, F.; Vinches, M.; Cayrefourcq, L.; Guigay, J.; Digue, L.; Aubry, K.; Alfonsi, M.; Delord, J.P.; et al. Circulating Tumor Cells as a Prognostic Factor in Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma: The CIRCUTEC Prospective Study. Clin. Chem. 2019, 65, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Tinhofer, I.; Konschak, R.; Stromberger, C.; Raguse, J.D.; Dreyer, J.H.; Jöhrens, K.; Keilholz, U.; Budach, V. Detection of circulating tumor cells for prediction of recurrence after adjuvant chemoradiation in locally advanced squamous cell carcinoma of the head and neck. Ann. Oncol. 2014, 25, 2042–2047. [Google Scholar] [CrossRef]
- Wang, H.M.; Wu, M.H.; Chang, P.H.; Lin, H.C.; Liao, C.D.; Wu, S.M.; Hung, T.M.; Lin, C.Y.; Chang, T.C.; Tzu-Tsen, Y.; et al. The change in circulating tumor cells before and during concurrent chemoradiotherapy is associated with survival in patients with locally advanced head and neck cancer. Head Neck 2019, 41, 2676–2687. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Y.; Feng, L.F.; Meng, X.; Chen, R.; Xu, W.H.; Hou, J.; Xu, T.; Zhang, L. Liquid biopsy in head and neck squamous cell carcinoma: Circulating tumor cells, circulating tumor DNA, and exosomes. Expert Rev. Mol. Diagn. 2020, 20, 1213–1227. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef]
- Ahn, S.M.; Chan, J.Y.; Zhang, Z.; Wang, H.; Khan, Z.; Bishop, J.A.; Westra, W.; Koch, W.M.; Califano, J.A. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol. Head Neck Surg. 2014, 140, 846–854. [Google Scholar] [CrossRef]
- Chiabotto, G.; Gai, C.; Deregibus, M.C.; Camussi, G. Salivary Extracellular Vesicle-Associated exRNA as Cancer Biomarker. Cancers 2019, 11, 891. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.; Camussi, F.; Broccoletti, R.; Gambino, A.; Cabras, M.; Molinaro, L.; Carossa, S.; Camussi, G.; Arduino, P.G. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 2018, 18, 439. [Google Scholar] [CrossRef]
- Nonaka, T.; Wong, D.T.W. Saliva-Exosomics in Cancer: Molecular Characterization of Cancer-Derived Exosomes in Saliva. Enzymes 2017, 42, 125–151. [Google Scholar] [CrossRef]
- Topkas, E.; Keith, P.; Dimeski, G.; Cooper-White, J.; Punyadeera, C. Evaluation of saliva collection devices for the analysis of proteins. Clin. Chim. Acta 2012, 413, 1066–1070. [Google Scholar] [CrossRef]
- Zlotogorski-Hurvitz, A.; Dekel, B.Z.; Malonek, D.; Yahalom, R.; Vered, M. FTIR-based spectrum of salivary exosomes coupled with computational-aided discriminating analysis in the diagnosis of oral cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 685–694. [Google Scholar] [CrossRef]
- Fontana, S.; Mauceri, R.; Novara, M.E.; Alessandro, R.; Campisi, G. Protein Cargo of Salivary Small Extracellular Vesicles as Potential Functional Signature of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 11160. [Google Scholar] [CrossRef]
- Kim, J.W.; Wieckowski, E.; Taylor, D.D.; Reichert, T.E.; Watkins, S.; Whiteside, T.L. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin. Cancer Res. 2005, 11, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, C.; Strauss, L.; Wieckowski, E.; Czystowska, M.; Albers, A.; Wang, Y.; Zeidler, R.; Lang, S.; Whiteside, T.L. Tumor-derived microvesicles in sera of patients with head and neck cancer and their role in tumor progression. Head Neck 2009, 31, 371–380. [Google Scholar] [CrossRef]
- Bigler, L.R.; Streckfus, C.F.; Dubinsky, W.P. Salivary biomarkers for the detection of malignant tumors that are remote from the oral cavity. Clin. Lab. Med. 2009, 29, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.L.; Khosroheidari, M.; Kanchi Ravi, R.; DiStefano, J.K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012, 82, 1024–1032. [Google Scholar] [CrossRef]
- Nguyen, B.; Wong, N.C.; Semple, T.; Clark, M.; Wong, S.Q.; Leslie, C.; Mirzai, B.; Millward, M.; Meehan, K.; Lim, A.M. Low-coverage whole-genome sequencing of extracellular vesicle-associated DNA in patients with metastatic cancer. Sci. Rep. 2021, 11, 4016. [Google Scholar] [CrossRef]
- Bauer, M.; Jasinski-Bergner, S.; Mandelboim, O.; Wickenhauser, C.; Seliger, B. Epstein-Barr Virus-Associated Malignancies and Immune Escape: The Role of the Tumor Microenvironment and Tumor Cell Evasion Strategies. Cancers 2021, 13, 5189. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Meckes, D.G., Jr. Methodological Approaches to Study Extracellular Vesicle miRNAs in Epstein-Barr Virus-Associated Cancers. Int. J. Mol. Sci. 2018, 19, 2810. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.W.; Tsang, C.M.; Lo, K.W. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160270. [Google Scholar] [CrossRef]
- Dunmire, S.K.; Hogquist, K.A.; Balfour, H.H. Infectious Mononucleosis. Curr. Top. Microbiol. Immunol. 2015, 390, 211–240. [Google Scholar] [CrossRef] [PubMed]
- Dunmire, S.K.; Verghese, P.S.; Balfour, H.H., Jr. Primary Epstein-Barr virus infection. J. Clin. Virol. 2018, 102, 84–92. [Google Scholar] [CrossRef]
- Grande, B.M.; Gerhard, D.S.; Jiang, A.; Griner, N.B.; Abramson, J.S.; Alexander, T.B.; Allen, H.; Ayers, L.W.; Bethony, J.M.; Bhatia, K.; et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood 2019, 133, 1313–1324. [Google Scholar] [CrossRef]
- Xiong, J.; Cui, B.W.; Wang, N.; Dai, Y.T.; Zhang, H.; Wang, C.F.; Zhong, H.J.; Cheng, S.; Ou-Yang, B.S.; Hu, Y.; et al. Genomic and Transcriptomic Characterization of Natural Killer T Cell Lymphoma. Cancer Cell 2020, 37, 403–419.e6. [Google Scholar] [CrossRef]
- Png, Y.T.; Yang, A.Z.Y.; Lee, M.Y.; Chua, M.J.M.; Lim, C.M. The Role of NK Cells in EBV Infection and EBV-Associated NPC. Viruses 2021, 13, 300. [Google Scholar] [CrossRef]
- Cheerathodi, M.; Nkosi, D.; Cone, A.S.; York, S.B.; Meckes, D.G., Jr. Epstein-Barr Virus LMP1 Modulates the CD63 Interactome. Viruses 2021, 13, 675. [Google Scholar] [CrossRef]
- Teow, S.Y.; Liew, K.; Khoo, A.S.; Peh, S.C. Pathogenic Role of Exosomes in Epstein-Barr Virus (EBV)-Associated Cancers. Int. J. Biol. Sci. 2017, 13, 1276–1286. [Google Scholar] [CrossRef]
- Higgs, P.G.; Lehman, N. The RNA World: Molecular cooperation at the origins of life. Nat. Rev. Genet. 2015, 16, 7–17. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Saw, P.E.; Xu, X.; Chen, J.; Song, E.W. Non-coding RNAs: The new central dogma of cancer biology. Sci. China Life Sci. 2021, 64, 22–50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liang, C.; Wang, G.; Jia, S.; Zhao, Q.; Xiang, Z.; Li, Y.; Cho, W.C.; Pestell, R.G.; Liang, L.; et al. MicroRNA-mediated cancer metastasis regulation via heterotypic signals in the microenvironment. Curr. Pharm. Biotechnol. 2014, 15, 455–458. [Google Scholar] [CrossRef]
- Flemming, J.P.; Hill, B.L.; Haque, M.W.; Raad, J.; Bonder, C.S.; Harshyne, L.A.; Rodeck, U.; Luginbuhl, A.; Wahl, J.K., 3rd; Tsai, K.Y.; et al. miRNA- and cytokine-associated extracellular vesicles mediate squamous cell carcinomas. J. Extracell. Vesicles 2020, 9, 1790159. [Google Scholar] [CrossRef]
- Bigagli, E.; Locatello, L.G.; Di Stadio, A.; Maggiore, G.; Valdarnini, F.; Bambi, F.; Gallo, O.; Luceri, C. Extracellular vesicles miR-210 as a potential biomarker for diagnosis and survival prediction of oral squamous cell carcinoma patients. J. Oral. Pathol. Med. 2022, 51, 350–357. [Google Scholar] [CrossRef]
- Zhan, C.; Yang, X.; Yin, X.; Hou, J. Exosomes and other extracellular vesicles in oral and salivary gland cancers. Oral. Dis. 2020, 26, 865–875. [Google Scholar] [CrossRef]
- Ludwig, S.; Sharma, P.; Wise, P.; Sposto, R.; Hollingshead, D.; Lamb, J.; Lang, S.; Fabbri, M.; Whiteside, T.L. mRNA and miRNA Profiles of Exosomes from Cultured Tumor Cells Reveal Biomarkers Specific for HPV16-Positive and HPV16-Negative Head and Neck Cancer. Int. J. Mol. Sci. 2020, 21, 8570. [Google Scholar] [CrossRef] [PubMed]
- Meckes, D.G., Jr.; Shair, K.H.; Marquitz, A.R.; Kung, C.P.; Edwards, R.H.; Raab-Traub, N. Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. USA 2010, 107, 20370–20375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.L.; Liu, Y.; Jiang, J.; Tang, Y.J.; Tang, Y.L.; Liang, X.H. Extracellular vesicle long non-coding RNA-mediated crosstalk in the tumor microenvironment: Tiny molecules, huge roles. Cancer Sci. 2020, 111, 2726–2735. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhou, L.; Shen, N.; Ning, X.; Wu, D.; Jiang, K.; Huang, X. M1 macrophage-derived exosomes and their key molecule lncRNA HOTTIP suppress head and neck squamous cell carcinoma progression by upregulating the TLR5/NF-κB pathway. Cell Death Dis. 2022, 13, 183. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Ye, S.; Yao, Y.; Zhang, C.; Liu, W. Oral Cancer Stem Cell-Derived Small Extracellular Vesicles Promote M2 Macrophage Polarization and Suppress CD4(+) T-Cell Activity by Transferring UCA1 and Targeting LAMC2. Stem. Cells Int. 2022, 2022, 5817684. [Google Scholar] [CrossRef]
- Ye, D.; Gong, M.; Deng, Y.; Fang, S.; Cao, Y.; Xiang, Y.; Shen, Z. Roles and clinical application of exosomal circRNAs in the diagnosis and treatment of malignant tumors. J. Transl. Med. 2022, 20, 161. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Zheng, X.; Dai, F.; Lu, Y.; Dai, L.; Niu, M.; Guo, H.; Li, W.; Xue, X.; et al. Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Mol. Cancer 2020, 19, 99. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, A.; Yan, A.; Yao, J.; Huang, H.; Gao, Z.; Han, T.; Gu, J.; Li, N.; Wu, H.; et al. Circular RNA MTCL1 promotes advanced laryngeal squamous cell carcinoma progression by inhibiting C1QBP ubiquitin degradation and mediating beta-catenin activation. Mol. Cancer 2022, 21, 92. [Google Scholar] [CrossRef]
- Gao, W.; Guo, H.; Niu, M.; Zheng, X.; Zhang, Y.; Xue, X.; Bo, Y.; Guan, X.; Li, Z.; Guo, Y.; et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol. Cancer 2020, 19, 166. [Google Scholar] [CrossRef]
- Verduci, L.; Ferraiuolo, M.; Sacconi, A.; Ganci, F.; Vitale, J.; Colombo, T.; Paci, P.; Strano, S.; Macino, G.; Rajewsky, N.; et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017, 18, 237. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Y.; Lu, J.; Sun, Y.; Xiao, H.; Liu, M.; Tian, L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med. Oncol. 2014, 31, 148. [Google Scholar] [CrossRef]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016, 76, 1770–1780. [Google Scholar] [CrossRef]
- Ye, S.B.; Zhang, H.; Cai, T.T.; Liu, Y.N.; Ni, J.J.; He, J.; Peng, J.Y.; Chen, Q.Y.; Mo, H.Y.; Jun, C.; et al. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J. Pathol. 2016, 240, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Langevin, S.; Kuhnell, D.; Parry, T.; Biesiada, J.; Huang, S.; Wise-Draper, T.; Casper, K.; Zhang, X.; Medvedovic, M.; Kasper, S. Comprehensive microRNA-sequencing of exosomes derived from head and neck carcinoma cells in vitro reveals common secretion profiles and potential utility as salivary biomarkers. Oncotarget 2017, 8, 82459–82474. [Google Scholar] [CrossRef] [PubMed]
- Ramayanti, O.; Verkuijlen, S.; Novianti, P.; Scheepbouwer, C.; Misovic, B.; Koppers-Lalic, D.; van Weering, J.; Beckers, L.; Adham, M.; Martorelli, D.; et al. Vesicle-bound EBV-BART13-3p miRNA in circulation distinguishes nasopharyngeal from other head and neck cancer and asymptomatic EBV-infections. Int. J. Cancer 2019, 144, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Bala, S. Extracellular vesicles in oral squamous carcinoma carry oncogenic miRNA profile and reprogram monocytes via NF-κB pathway. Oncotarget 2018, 9, 34838–34854. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; You, B.; Shi, S.; Shan, Y.; Zhang, Q.; Yue, H.; Zhang, J.; Zhang, W.; Shi, Y.; Liu, Y.; et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 2018, 37, 2873–2889. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Liu, Q.H.; Wang, F.; Tan, J.J.; Deng, Y.Q.; Peng, X.H.; Liu, X.; Zhang, B.; Xu, X.; Li, X.P. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 147. [Google Scholar] [CrossRef]
- Xie, C.; Du, L.Y.; Guo, F.; Li, X.; Cheng, B. Exosomes derived from microRNA-101-3p-overexpressing human bone marrow mesenchymal stem cells suppress oral cancer cell proliferation, invasion, and migration. Mol. Cell Biochem. 2019, 458, 11–26. [Google Scholar] [CrossRef]
- Qin, X.; Guo, H.; Wang, X.; Zhu, X.; Yan, M.; Wang, X.; Xu, Q.; Shi, J.; Lu, E.; Chen, W.; et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019, 20, 12. [Google Scholar] [CrossRef]
- Sun, L.P.; Xu, K.; Cui, J.; Yuan, D.Y.; Zou, B.; Li, J.; Liu, J.L.; Li, K.Y.; Meng, Z.; Zhang, B. Cancer-associated fibroblast-derived exosomal miR-382-5p promotes the migration and invasion of oral squamous cell carcinoma. Oncol. Rep. 2019, 42, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Mayne, G.C.; Woods, C.M.; Dharmawardana, N.; Wang, T.; Krishnan, S.; Hodge, J.C.; Foreman, A.; Boase, S.; Carney, A.S.; Sigston, E.A.W.; et al. Cross validated serum small extracellular vesicle microRNAs for the detection of oropharyngeal squamous cell carcinoma. J. Transl. Med. 2020, 18, 280. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Zhou, X.; Luo, X.; Liu, K.; Jiang, E.; Chen, Y.; Shao, Z.; Shang, Z. OSCC Exosomes Regulate miR-210-3p Targeting EFNA3 to Promote Oral Cancer Angiogenesis through the PI3K/AKT Pathway. Biomed. Res. Int. 2020, 2020, 2125656. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2020, 121, 109553. [Google Scholar] [CrossRef]
- Shuang, Y.; Yao, X.; Liu, J.; Niu, J.; Guo, W.; Li, C. Serum-derived extracellular vesicles mediate Smad4 expression through shuttling microRNA-27a in the progression of laryngeal squamous cell carcinoma. Hum. Cell 2022, 35, 1084–1099. [Google Scholar] [CrossRef]
- Yin, H.; Qiu, X.; Shan, Y.; You, B.; Xie, L.; Zhang, P.; Zhao, J.; You, Y. HIF-1α downregulation of miR-433-3p in adipocyte-derived exosomes contributes to NPC progression via targeting SCD1. Cancer Sci. 2021, 112, 1457–1470. [Google Scholar] [CrossRef]
- Tong, F.; Mao, X.; Zhang, S.; Xie, H.; Yan, B.; Wang, B.; Sun, J.; Wei, L. HPV + HNSCC-derived exosomal miR-9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer Lett. 2020, 478, 34–44. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, Y.; Li, B.; Kang, M.; Yang, Z.; Lin, C.; Hu, K.; Wei, Z.; Xu, M.; Mi, J.; et al. miRNAs derived from circulating small extracellular vesicles as diagnostic biomarkers for nasopharyngeal carcinoma. Cancer Sci. 2021, 112, 2393–2404. [Google Scholar] [CrossRef]
- Chen, C.M.; Chu, T.H.; Chou, C.C.; Chien, C.Y.; Wang, J.S.; Huang, C.C. Exosome-derived microRNAs in oral squamous cell carcinomas impact disease prognosis. Oral. Oncol. 2021, 120, 105402. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, M.; Tang, Q. Human umbilical cord mesenchymal stem cells-derived exosomal microRNA-181a retards nasopharyngeal carcinoma development by mediating KDM5C. J. Cancer Res. Clin. Oncol. 2021, 147, 2867–2877. [Google Scholar] [CrossRef]
- Panvongsa, W.; Siripoon, T.; Worakitchanon, W.; Arsa, L.; Trachu, N.; Jinawath, N.; Ngamphaiboon, N.; Chairoungdua, A. Plasma extracellular vesicle microRNA-491-5p as diagnostic and prognostic marker for head and neck squamous cell carcinoma. Cancer Sci. 2021, 112, 4257–4269. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lin, Y.; Tan, G.; Ai, M.; Gong, H.; Liu, W.; Huang, J.; Zou, Z. Tumor-derived exosomal microRNA-15b-5p augments laryngeal cancer by targeting TXNIP. Cell Cycle 2022, 21, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Tang, Q.; Gong, J.; Jiang, W.; Chen, Y.; Zhou, Q.; Aldeen, A.; Wang, S.; Li, C.; Lv, W.; et al. Radiosensitizer EXO-miR-197-3p Inhibits Nasopharyngeal Carcinoma Progression and Radioresistance by Regulating the AKT/mTOR Axis and HSPA5-mediated Autophagy. Int. J. Biol. Sci. 2022, 18, 1878–1895. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Shi, J.; Xu, Q. Exosomal miR-626 promotes the malignant behavior of oral cancer cells by targeting NFIB. Mol. Biol. Rep. 2022, 49, 4829–4840. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.; Routray, S. Assessing the analytical efficacy of TEX in diagnosing oral cancer using a systematic review approach. J. Oral. Pathol. Med. 2021, 50, 123–128. [Google Scholar] [CrossRef]
- Yu, Z.; Ni, F.; Chen, Y.; Zhang, J.; Cai, J.; Shi, W. miR-125b suppresses cell proliferation and metastasis by targeting HAX-1 in esophageal squamous cell carcinoma. Pathol. Res. Pract. 2020, 216, 152792. [Google Scholar] [CrossRef]
- You, B.; Cao, X.; Shao, X.; Ni, H.; Shi, S.; Shan, Y.; Gu, Z.; You, Y. Clinical and biological significance of HAX-1 overexpression in nasopharyngeal carcinoma. Oncotarget 2016, 7, 12505–12524. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef]
- Theodoraki, M.N.; Laban, S.; Jackson, E.K.; Lotfi, R.; Schuler, P.J.; Brunner, C.; Hoffmann, T.K.; Whiteside, T.L.; Hofmann, L. Changes in circulating exosome molecular profiles following surgery/(chemo)radiotherapy: Early detection of response in head and neck cancer patients. Br. J. Cancer 2021, 125, 1677–1686. [Google Scholar] [CrossRef]
- Kannan, A.; Hertweck, K.L.; Philley, J.V.; Wells, R.B.; Dasgupta, S. Genetic Mutation and Exosome Signature of Human Papilloma Virus Associated Oropharyngeal Cancer. Sci. Rep. 2017, 7, 46102. [Google Scholar] [CrossRef]
- Visan, K.S.; Lobb, R.J.; Wen, S.W.; Bedo, J.; Lima, L.G.; Krumeich, S.; Palma, C.; Ferguson, K.; Green, B.; Niland, C.; et al. Blood-Derived Extracellular Vesicle-Associated miR-3182 Detects Non-Small Cell Lung Cancer Patients. Cancers 2022, 14, 257. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.H.; Zhang, J.; Rima, X.Y.; Wang, X.; Kwak, K.J.; Okimoto, T.; Amann, J.; Yoon, M.J.; Shukuya, T.; Chiang, C.L.; et al. An immunogold single extracellular vesicular RNA and protein ((Au) SERP) biochip to predict responses to immunotherapy in non-small cell lung cancer patients. J. Extracell. Vesicles 2022, 11, e12258. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.X.; Yu, R.; Wu, X.; Wu, S.Y.; Pi, C.; Chen, Z.H.; Zhang, X.C.; Gao, C.Y.; Shao, Y.W.; Liu, L.; et al. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J. Immunother. Cancer 2020, 8, e000376. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.; Leung, L.L.; Qu, X.; Chan, J.Y.-K. Extracellular Vesicles as Biomarkers in Head and Neck Squamous Cell Carcinoma: From Diagnosis to Disease-Free Survival. Cancers 2023, 15, 1826. https://doi.org/10.3390/cancers15061826
Chen B, Leung LL, Qu X, Chan JY-K. Extracellular Vesicles as Biomarkers in Head and Neck Squamous Cell Carcinoma: From Diagnosis to Disease-Free Survival. Cancers. 2023; 15(6):1826. https://doi.org/10.3390/cancers15061826
Chicago/Turabian StyleChen, Bojie, Leanne Lee Leung, Xinyu Qu, and Jason Ying-Kuen Chan. 2023. "Extracellular Vesicles as Biomarkers in Head and Neck Squamous Cell Carcinoma: From Diagnosis to Disease-Free Survival" Cancers 15, no. 6: 1826. https://doi.org/10.3390/cancers15061826
APA StyleChen, B., Leung, L. L., Qu, X., & Chan, J. Y.-K. (2023). Extracellular Vesicles as Biomarkers in Head and Neck Squamous Cell Carcinoma: From Diagnosis to Disease-Free Survival. Cancers, 15(6), 1826. https://doi.org/10.3390/cancers15061826






