Combining CNN Features with Voting Classifiers for Optimizing Performance of Brain Tumor Classification
Abstract
Simple Summary
Abstract
1. Introduction
- This study proposes an ensemble model that utilizes convolutional features from a customized CNN model for predicting brain tumors. The proposed ensemble model is based on logistic regression and a stochastic gradient descent classifier with a voting mechanism for making the final output.
- The impact of the original features is analyzed against the performance of models using convolutional features.
- The performance comparison is performed using various machine learning models including random forest (RF), K-nearest neighbor (k-NN), logistic regression (LR), gradient boosting machine (GBM), decision tree (DT), Gaussian Naive Bayes (GNB), extra tree classifier (ETC), support vector machine (SVM), and stochastic gradient descent (SGD). Moreover, the performance of the proposed model is compared with leading-edge methodologies in terms of accuracy, precision, recall, and F1 score.
2. Materials and Methods
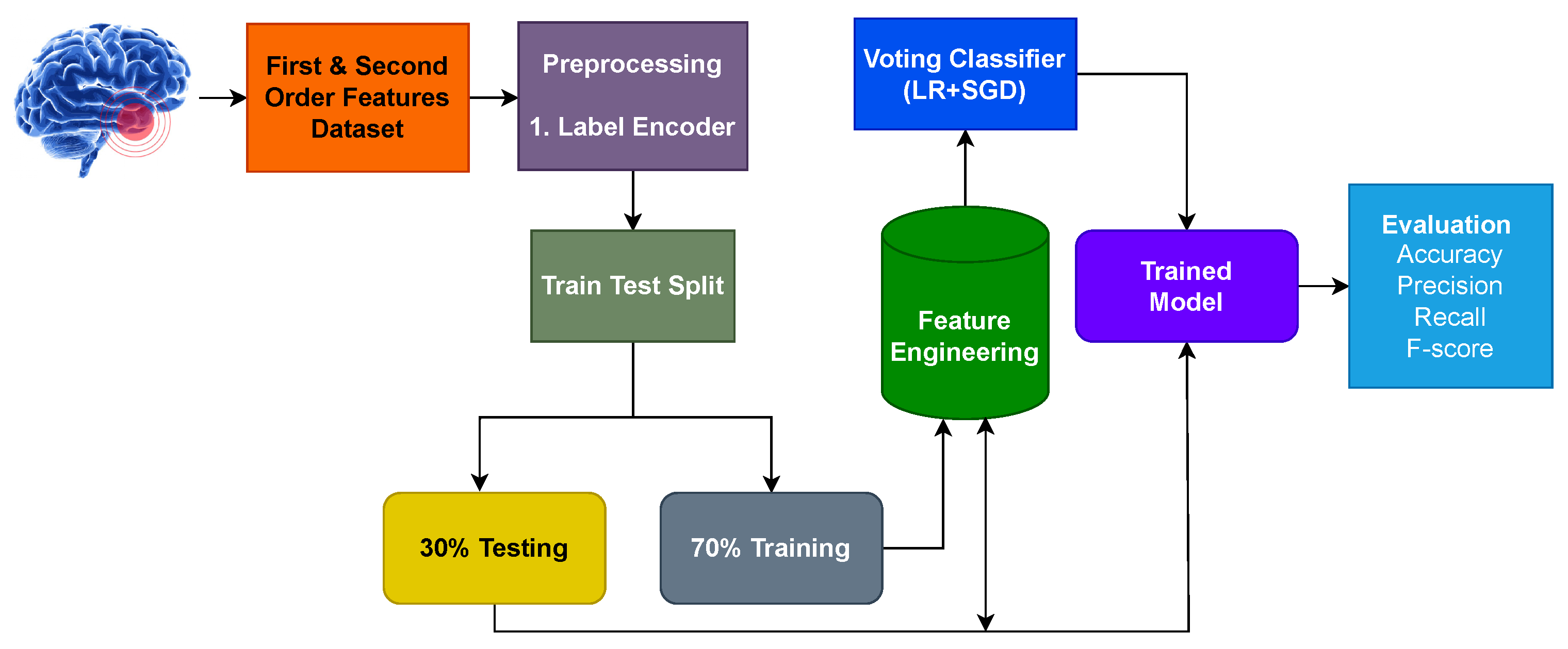
2.1. Dataset
2.2. Machine Learning Models
2.2.1. Random Forest
2.2.2. Decision Tree
- Step 1: determine the target’s entropy.
- Step 2: compute each attribute’s entropy.
2.2.3. K-Nearest Neighbour
2.2.4. Logistic Regression
2.2.5. Support Vector Machine
2.2.6. Gradient Boosting Machine
2.2.7. Extra Tree Classifier
2.2.8. Gaussian Naive Bayes
2.2.9. Stochastic Gradient Decent
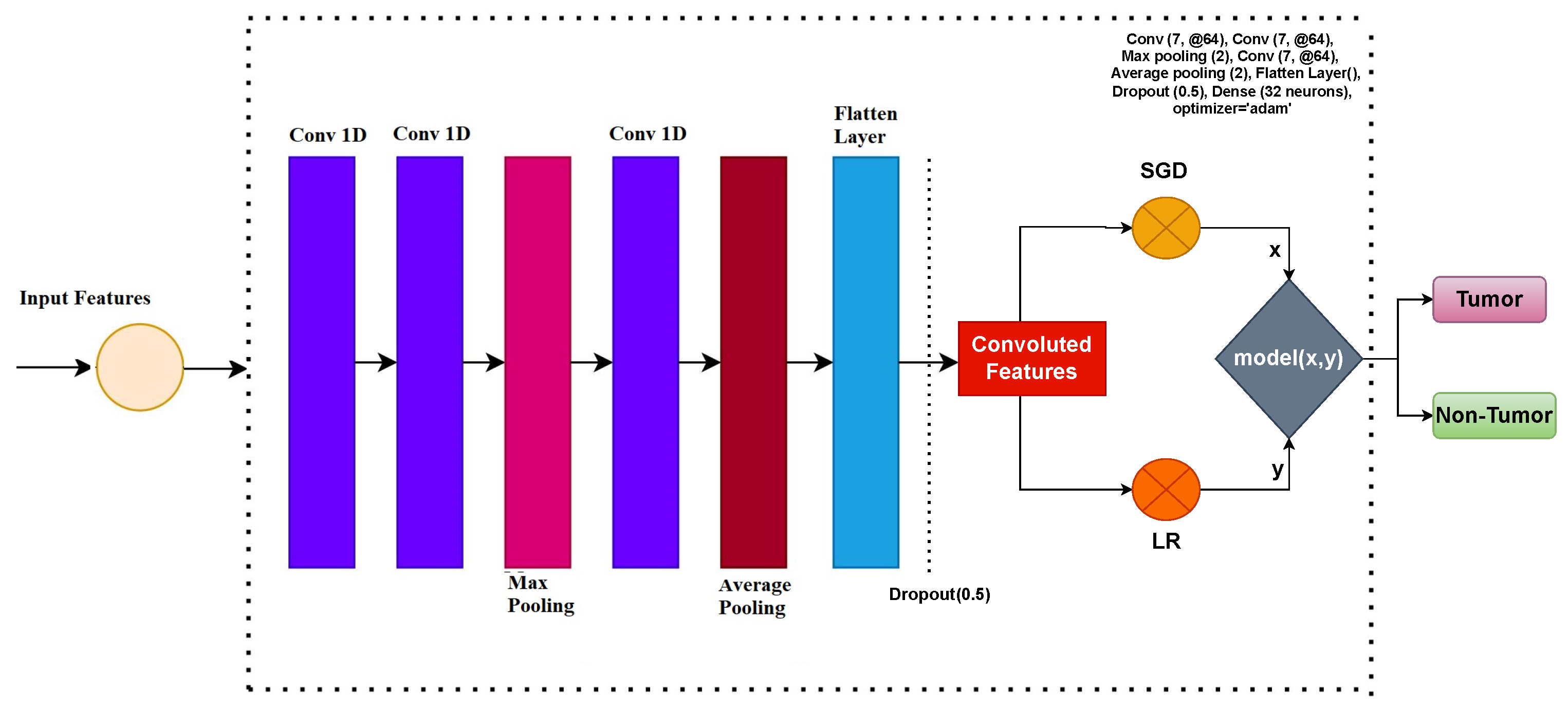
2.3. Convolutional Neural Network for Feature Engineering
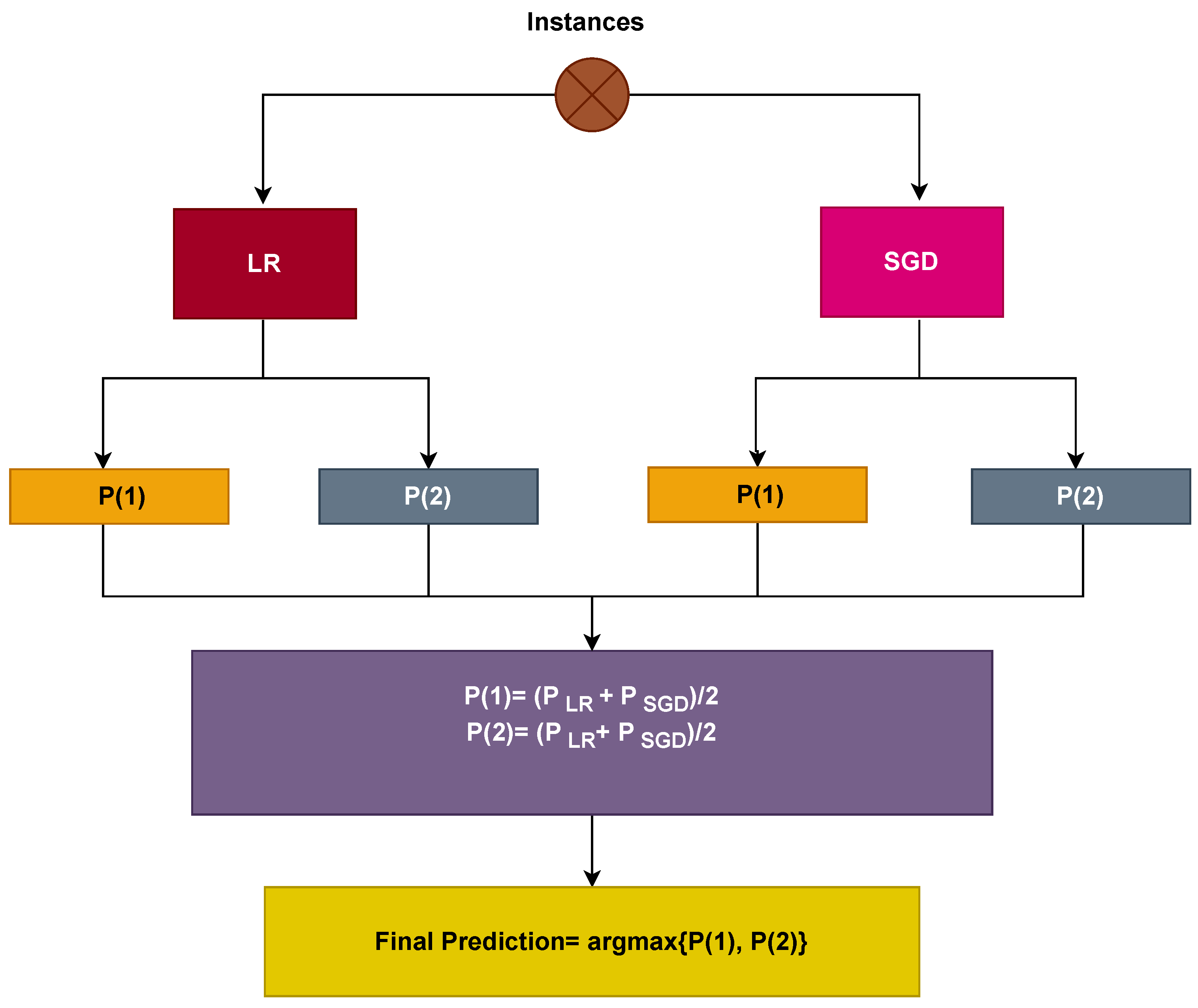
2.4. Proposed Voting Classifier
2.5. Evaluation Metrics
3. Results and Discussion
3.1. Experiment Setup
3.2. Performance of Models Using Original Features
3.3. Results Using CNN Feature Engineering
3.4. Results of K-Fold Cross-Validation
3.5. Performance Comparison with State-of-the-Art Approaches
4. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Umer, M.; Naveed, M.; Alrowais, F.; Ishaq, A.; Hejaili, A.A.; Alsubai, S.; Eshmawi, A.; Mohamed, A.; Ashraf, I. Breast Cancer Detection Using Convoluted Features and Ensemble Machine Learning Algorithm. Cancers 2022, 14, 6015. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Raza, M.; Saba, T.; Anjum, M.A. Brain tumor detection using statistical and machine learning method. Comput. Methods Programs Biomed. 2019, 177, 69–79. [Google Scholar] [CrossRef]
- McFaline-Figueroa, J.R.; Lee, E.Q. Brain tumors. Am. J. Med. 2018, 131, 874–882. [Google Scholar] [CrossRef]
- Arvold, N.D.; Lee, E.Q.; Mehta, M.P.; Margolin, K.; Alexander, B.M.; Lin, N.U.; Anders, C.K.; Soffietti, R.; Camidge, D.R.; Vogelbaum, M.A.; et al. Updates in the management of brain metastases. Neuro-oncology 2016, 18, 1043–1065. [Google Scholar] [CrossRef]
- Saba, T.; Mohamed, A.S.; El-Affendi, M.; Amin, J.; Sharif, M. Brain tumor detection using fusion of hand crafted and deep learning features. Cogn. Syst. Res. 2020, 59, 221–230. [Google Scholar] [CrossRef]
- Soomro, T.A.; Zheng, L.; Afifi, A.J.; Ali, A.; Soomro, S.; Yin, M.; Gao, J. Image Segmentation for MR Brain Tumor Detection Using Machine Learning: A Review. IEEE Rev. Biomed. Eng. 2023, 16, 70–90. [Google Scholar] [CrossRef]
- Khan, A.H.; Abbas, S.; Khan, M.A.; Farooq, U.; Khan, W.A.; Siddiqui, S.Y.; Ahmad, A. Intelligent model for brain tumor identification using deep learning. Appl. Comput. Intell. Soft Comput. 2022, 2022, 8104054. [Google Scholar] [CrossRef]
- Younis, A.; Qiang, L.; Nyatega, C.O.; Adamu, M.J.; Kawuwa, H.B. Brain Tumor Analysis Using Deep Learning and VGG-16 Ensembling Learning Approaches. Appl. Sci. 2022, 12, 7282. [Google Scholar] [CrossRef]
- Zahoor, M.M.; Qureshi, S.A.; Khan, A.; Rehman, A.u.; Rafique, M. A novel dual-channel brain tumor detection system for MR images using dynamic and static features with conventional machine learning techniques. Waves Random Complex Media 2022, 1–20. [Google Scholar] [CrossRef]
- Senan, E.M.; Jadhav, M.E.; Rassem, T.H.; Aljaloud, A.S.; Mohammed, B.A.; Al-Mekhlafi, Z.G. Early Diagnosis of Brain Tumour MRI Images Using Hybrid Techniques between Deep and Machine Learning. Comput. Math. Methods Med. 2022, 2022, 8330833. [Google Scholar] [CrossRef]
- Díaz-Pernas, F.J.; Martínez-Zarzuela, M.; Antón-Rodríguez, M.; González-Ortega, D. A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. Healthcare 2021, 9, 153. [Google Scholar] [CrossRef]
- Budati, A.; Babu, K. An automated brain tumor detection and classification from MRI images using machine learning techniques with IoT. Environ. Dev. Sustain. 2022, 24, 1–15. [Google Scholar] [CrossRef]
- Akinyelu, A.A.; Zaccagna, F.; Grist, J.T.; Castelli, M.; Rundo, L. Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey. J. Imaging 2022, 8, 205. [Google Scholar] [CrossRef]
- Raza, A.; Ayub, H.; Khan, J.A.; Ahmad, I.; Salama, A.S.; Daradkeh, Y.I.; Javeed, D.; Ur Rehman, A.; Hamam, H. A hybrid deep learning-based approach for brain tumor classification. Electronics 2022, 11, 1146. [Google Scholar] [CrossRef]
- Ahmad, S.; Choudhury, P.K. On the Performance of Deep Transfer Learning Networks for Brain Tumor Detection using MR Images. IEEE Access 2022, 10, 59099–59114. [Google Scholar] [CrossRef]
- Amran, G.A.; Alsharam, M.S.; Blajam, A.O.A.; Hasan, A.A.; Alfaifi, M.Y.; Amran, M.H.; Gumaei, A.; Eldin, S.M. Brain Tumor Classification and Detection Using Hybrid Deep Tumor Network. Electronics 2022, 11, 3457. [Google Scholar] [CrossRef]
- Ullah, N.; Khan, J.A.; Khan, M.S.; Khan, W.; Hassan, I.; Obayya, M.; Negm, N.; Salama, A.S. An Effective Approach to Detect and Identify Brain Tumors Using Transfer Learning. Appl. Sci. 2022, 12, 5645. [Google Scholar] [CrossRef]
- Hashmi, A.; Osman, A.H. Brain Tumor Classification Using Conditional Segmentation with Residual Network and Attention Approach by Extreme Gradient Boost. Appl. Sci. 2022, 12, 10791. [Google Scholar] [CrossRef]
- Samee, N.A.; Mahmoud, N.F.; Atteia, G.; Abdallah, H.A.; Alabdulhafith, M.; Al-Gaashani, M.S.; Ahmad, S.; Muthanna, M.S.A. Classification Framework for Medical Diagnosis of Brain Tumor with an Effective Hybrid Transfer Learning Model. Diagnostics 2022, 12, 2541. [Google Scholar] [CrossRef]
- Rasool, M.; Ismail, N.A.; Boulila, W.; Ammar, A.; Samma, H.; Yafooz, W.M.; Emara, A.H.M. A Hybrid Deep Learning Model for Brain Tumour Classification. Entropy 2022, 24, 799. [Google Scholar] [CrossRef]
- DeAngelis, L.M. Brain tumors. N. Engl. J. Med. 2001, 344, 114–123. [Google Scholar] [CrossRef]
- Fathi Kazerooni, A.; Bagley, S.J.; Akbari, H.; Saxena, S.; Bagheri, S.; Guo, J.; Chawla, S.; Nabavizadeh, A.; Mohan, S.; Bakas, S.; et al. Applications of radiomics and radiogenomics in high-grade gliomas in the era of precision medicine. Cancers 2021, 13, 5921. [Google Scholar] [CrossRef]
- Habib, A.; Jovanovich, N.; Hoppe, M.; Ak, M.; Mamindla, P.; Colen, R.R.; Zinn, P.O. MRI-based radiomics and radiogenomics in the management of low-grade gliomas: Evaluating the evidence for a paradigm shift. J. Clin. Med. 2021, 10, 1411. [Google Scholar] [CrossRef]
- Jena, B.; Saxena, S.; Nayak, G.K.; Balestrieri, A.; Gupta, N.; Khanna, N.N.; Laird, J.R.; Kalra, M.K.; Fouda, M.M.; Saba, L.; et al. Brain tumor characterization using radiogenomics in artificial intelligence framework. Cancers 2022, 14, 4052. [Google Scholar] [CrossRef]
- Dutta, S.; Bandyopadhyay, S.K. Revealing brain tumor using cross-validated NGBoost classifier. Int. J. Mach. Learn. Netw. Collab. Eng. 2020, 4, 12–20. [Google Scholar] [CrossRef]
- Methil, A.S. Brain tumor detection using deep learning and image processing. In Proceedings of the 2021 International Conference on Artificial Intelligence and Smart Systems (ICAIS), Tamil Nadu, India, 25–27 March 2021; pp. 100–108. [Google Scholar]
- Shah, H.A.; Saeed, F.; Yun, S.; Park, J.H.; Paul, A.; Kang, J.M. A Robust Approach for Brain Tumor Detection in Magnetic Resonance Images Using Finetuned EfficientNet. IEEE Access 2022, 10, 65426–65438. [Google Scholar] [CrossRef]
- Bohaju, J. Brain Tumor Database. July 2020. Available online: https://www.kaggle.com/datasets/jakeshbohaju/brain-tumor (accessed on 10 January 2023).
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Biau, G.; Scornet, E. A random forest guided tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Manzoor, M.; Umer, M.; Sadiq, S.; Ishaq, A.; Ullah, S.; Madni, H.A.; Bisogni, C. RFCNN: Traffic accident severity prediction based on decision level fusion of machine and deep learning model. IEEE Access 2021, 9, 128359–128371. [Google Scholar] [CrossRef]
- Kotsiantis, S.B. Decision trees: A recent overview. Artif. Intell. Rev. 2013, 39, 261–283. [Google Scholar] [CrossRef]
- Juna, A.; Umer, M.; Sadiq, S.; Karamti, H.; Eshmawi, A.; Mohamed, A.; Ashraf, I. Water Quality Prediction Using KNN Imputer and Multilayer Perceptron. Water 2022, 14, 2592. [Google Scholar] [CrossRef]
- Keller, J.M.; Gray, M.R.; Givens, J.A. A fuzzy k-nearest neighbor algorithm. IEEE Trans. Syst. Man Cybern. 1985, SMC-15, 580–585. [Google Scholar] [CrossRef]
- Besharati, E.; Naderan, M.; Namjoo, E. LR-HIDS: Logistic regression host-based intrusion detection system for cloud environments. J. Ambient Intell. Humaniz. Comput. 2019, 10, 3669–3692. [Google Scholar] [CrossRef]
- Khammassi, C.; Krichen, S. A NSGA2-LR wrapper approach for feature selection in network intrusion detection. Comput. Netw. 2020, 172, 107183. [Google Scholar] [CrossRef]
- Kleinbaum, D.G.; Dietz, K.; Gail, M.; Klein, M.; Klein, M. Logistic Regression; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Ashraf, I.; Narra, M.; Umer, M.; Majeed, R.; Sadiq, S.; Javaid, F.; Rasool, N. A Deep Learning-Based Smart Framework for Cyber-Physical and Satellite System Security Threats Detection. Electronics 2022, 11, 667. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Umer, M.; Sadiq, S.; Nappi, M.; Sana, M.U.; Ashraf, I.; Karamti, H.; Eshmawi, A.A. ETCNN: Extra Tree and Convolutional Neural Network-based Ensemble Model for COVID-19 Tweets Sentiment Classification. Pattern Recognit. Lett. 2022, 164, 224–231. [Google Scholar] [CrossRef]
- Majeed, R.; Abdullah, N.A.; Faheem Mushtaq, M.; Umer, M.; Nappi, M. Intelligent Cyber-Security System for IoT-Aided Drones Using Voting Classifier. Electronics 2021, 10, 2926. [Google Scholar] [CrossRef]
- Rish, I. An empirical study of the naive Bayes classifier. In Proceedings of the IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Seattle, WA, USA, 4–6 August 2001; Volume 3, pp. 41–46. [Google Scholar]
- Umer, M.; Sadiq, S.; Missen, M.M.S.; Hameed, Z.; Aslam, Z.; Siddique, M.A.; Nappi, M. Scientific papers citation analysis using textual features and SMOTE resampling techniques. Pattern Recognit. Lett. 2021, 150, 250–257. [Google Scholar] [CrossRef]
- Bottou, L. Stochastic gradient descent tricks. Neural Networks: Tricks of the Trade, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 421–436. [Google Scholar]
- Hameed, A.; Umer, M.; Hafeez, U.; Mustafa, H.; Sohaib, A.; Siddique, M.A.; Madni, H.A. Skin lesion classification in dermoscopic images using stacked Convolutional Neural Network. J. Ambient. Intell. Humaniz. Comput. 2021, 28, 1–15. [Google Scholar] [CrossRef]
- Rustam, F.; Ishaq, A.; Munir, K.; Almutairi, M.; Aslam, N.; Ashraf, I. Incorporating CNN Features for Optimizing Performance of Ensemble Classifier for Cardiovascular Disease Prediction. Diagnostics 2022, 12, 1474. [Google Scholar] [CrossRef]
| Classifiers | Parameters |
|---|---|
| RF | number of trees = 200, maximum depth = 30, random state = 52 |
| DT | number of trees = 200, maximum depth = 30, random state = 52 |
| k-NN | algorithm = ‘auto’, leaf size = 30, metric = ‘minkowski’, neighbors = 5, weights = ‘uniform’ |
| LR | penalty = ‘l2’, solver = ‘lbfgs’ |
| SVM | C = 2.0, cache size = 200, gamma = ‘auto’, kernel = ‘linear’, maximum iteration = -1, probability = False, random state = 52, tol = 0.001 |
| GBM | number of trees = 200, maximum depth = 30, random state = 52, learning rate = 0.1 |
| ETC | number of trees = 200, maximum depth = 30, random state = 52 |
| GNB | alpha = 1.0, binarize = 0.0 |
| SGD | penalty = ‘l2’, loss = ‘log’ |
| CNN | Conv (7, @64), Conv (7, @64), Max pooling (2), Conv (7, @64), Average pooling (2), Flatten Layer(), Dropout (0.5), Dense (32 neurons), optimizer = ‘adam’ |
| Model | Accuracy | Class | Precision | Recall | F1 Score |
|---|---|---|---|---|---|
| Voting Classifier LR+SGD | 0.845 | Tumour | 0.865 | 0.899 | 0.878 |
| Non-Tumour | 0.748 | 0.799 | 0.776 | ||
| Micro Avg. | 0.824 | 0.858 | 0.856 | ||
| Weighted Avg. | 0.807 | 0.843 | 0.825 | ||
| GBM | 0.805 | Tumour | 0.795 | 0.818 | 0.807 |
| Non-Tumour | 0.818 | 0.818 | 0.818 | ||
| Micro Avg. | 0.805 | 0.819 | 0.827 | ||
| Weighted Avg. | 0.808 | 0.814 | 0.826 | ||
| GNB | 0.769 | Tumour | 0.777 | 0.788 | 0.777 |
| Non-Tumour | 0.744 | 0.766 | 0.755 | ||
| Micro Avg. | 0.766 | 0.777 | 0.766 | ||
| Weighted Avg. | 0.766 | 0.777 | 0.766 | ||
| ETC | 0.829 | Tumour | 0.806 | 0.806 | 0.806 |
| Non-Tumour | 0.815 | 0.815 | 0.815 | ||
| Micro Avg. | 0.805 | 0.805 | 0.805 | ||
| Weighted Avg. | 0.809 | 0.820 | 0.811 | ||
| LR | 0.869 | Tumour | 0.866 | 0.899 | 0.877 |
| Non-Tumour | 0.888 | 0.899 | 0.888 | ||
| M Avg. | 0.855 | 0.902 | 0.883 | ||
| W Avg. | 0.855 | 0.884 | 0.876 | ||
| SGD | 0.881 | Tumour | 0.903 | 0.892 | 0.893 |
| Non-Tumour | 0.923 | 0.924 | 0.922 | ||
| Micro Avg. | 0.922 | 0.922 | 0.911 | ||
| Weighted Avg. | 0.919 | 0.919 | 0.919 | ||
| RF | 0.854 | Tumour | 0.827 | 0.858 | 0.834 |
| Non-Tumour | 0.844 | 0.806 | 0.828 | ||
| Micro Avg. | 0.844 | 0.844 | 0.833 | ||
| Weighted Avg. | 0.833 | 0.833 | 0.833 | ||
| DT | 0.829 | Tumour | 0.806 | 0.822 | 0.811 |
| Non-Tumour | 0.805 | 0.833 | 0.814 | ||
| Micro Avg. | 0.807 | 0.809 | 0.818 | ||
| Weighted Avg. | 0.818 | 0.804 | 0.804 | ||
| SVM | 0.788 | Tumour | 0.788 | 0.800 | 0.799 |
| Non-Tumour | 0.777 | 0.788 | 0.788 | ||
| Micro Avg. | 0.788 | 0.799 | 0.800 | ||
| Weighted Avg. | 0.788 | 0.799 | 0.800 | ||
| k-NN | 0.828 | Tumour | 0.788 | 0.822 | 0.800 |
| Non-Tumour | 0.777 | 0.811 | 0.800 | ||
| Micro Avg. | 0.777 | 0.811 | 0.800 | ||
| Weighted Avg. | 0.799 | 0.824 | 0.824 |
| Model | Accuracy | Class | Precision | Recall | F1 Score |
|---|---|---|---|---|---|
| Voting Classifier LR+SGD | 0.995 | Tumour | 0.999 | 0.999 | 0.999 |
| Non-Tumour | 0.999 | 0.999 | 0.999 | ||
| Micro Avg. | 0.999 | 0.999 | 0.999 | ||
| Weighted Avg. | 0.999 | 0.999 | 0.999 | ||
| GBM | 0.905 | Tumour | 0.928 | 0.944 | 0.926 |
| Non-Tumour | 0.915 | 0.923 | 0.914 | ||
| Micro Avg. | 0.927 | 0.931 | 0.924 | ||
| Weighted Avg. | 0.915 | 0.935 | 0.918 | ||
| GNB | 0.866 | Tumour | 0.877 | 0.888 | 0.877 |
| Non-Tumour | 0.844 | 0.866 | 0.855 | ||
| Micro Avg. | 0.866 | 0.877 | 0.877 | ||
| Weighted Avg. | 0.855 | 0.877 | 0.866 | ||
| ETC | 0.926 | Tumour | 0.907 | 0.903 | 0.905 |
| Non-Tumour | 0.914 | 0.918 | 0.914 | ||
| Micro Avg. | 0.913 | 0.913 | 0.913 | ||
| Weighted Avg. | 0.900 | 0.900 | 0.900 | ||
| LR | 0.989 | Tumour | 0.966 | 0.999 | 0.977 |
| Non-Tumour | 0.988 | 0.999 | 0.988 | ||
| M Avg. | 0.977 | 0.999 | 0.988 | ||
| W Avg. | 0.977 | 0.999 | 0.988 | ||
| SGD | 0.987 | Tumour | 0.985 | 0.997 | 0.986 |
| Non-Tumour | 0.999 | 0.986 | 0.988 | ||
| Micro Avg. | 0.988 | 0.988 | 0.988 | ||
| Weighted Avg. | 0.988 | 0.988 | 0.988 | ||
| RF | 0.958 | Tumour | 0.927 | 0.954 | 0.935 |
| Non-Tumour | 0.944 | 0.960 | 0.952 | ||
| Micro Avg. | 0.944 | 0.960 | 0.952 | ||
| Weighted Avg. | 0.934 | 0.954 | 0.944 | ||
| DT | 0.936 | Tumour | 0.900 | 0.928 | 0.914 |
| Non-Tumour | 0.900 | 0.934 | 0.912 | ||
| Micro Avg. | 0.900 | 0.900 | 0.915 | ||
| Weighted Avg. | 0.914 | 0.900 | 0.900 | ||
| SVM | 0.978 | Tumour | 0.974 | 0.922 | 0.955 |
| Non-Tumour | 0.977 | 0.944 | 0.944 | ||
| Micro Avg. | 0.977 | 0.933 | 0.944 | ||
| Weighted Avg. | 0.988 | 0.955 | 0.966 | ||
| k-NN | 0.982 | Tumour | 0.988 | 0.988 | 0.988 |
| Non-Tumour | 0.977 | 0.977 | 0.977 | ||
| Micro Avg. | 0.966 | 0.966 | 0.966 | ||
| Weighted Avg. | 0.977 | 0.977 | 0.977 |
| Fold Number | Accuracy | Precision | Recall | F-Score |
|---|---|---|---|---|
| Fold-1 | 0.992 | 0.995 | 0.994 | 0.995 |
| Fold-2 | 0.994 | 0.996 | 0.995 | 0.996 |
| Fold-3 | 0.996 | 0.997 | 0.996 | 0.997 |
| Fold-4 | 0.998 | 0.999 | 1.000 | 0.998 |
| Fold-5 | 0.999 | 0.999 | 0.998 | 0.998 |
| Fold-6 | 1.000 | 0.999 | 0.999 | 0.998 |
| Fold-7 | 0.995 | 0.999 | 0.996 | 0.997 |
| Fold-8 | 0.997 | 0.998 | 0.997 | 0.998 |
| Fold-9 | 0.997 | 0.997 | 0.998 | 0.998 |
| Fold-10 | 0.999 | 0.999 | 0.999 | 0.999 |
| Average | 0.996 | 0.998 | 0.998 | 0.997 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alturki, N.; Umer, M.; Ishaq, A.; Abuzinadah, N.; Alnowaiser, K.; Mohamed, A.; Saidani, O.; Ashraf, I. Combining CNN Features with Voting Classifiers for Optimizing Performance of Brain Tumor Classification. Cancers 2023, 15, 1767. https://doi.org/10.3390/cancers15061767
Alturki N, Umer M, Ishaq A, Abuzinadah N, Alnowaiser K, Mohamed A, Saidani O, Ashraf I. Combining CNN Features with Voting Classifiers for Optimizing Performance of Brain Tumor Classification. Cancers. 2023; 15(6):1767. https://doi.org/10.3390/cancers15061767
Chicago/Turabian StyleAlturki, Nazik, Muhammad Umer, Abid Ishaq, Nihal Abuzinadah, Khaled Alnowaiser, Abdullah Mohamed, Oumaima Saidani, and Imran Ashraf. 2023. "Combining CNN Features with Voting Classifiers for Optimizing Performance of Brain Tumor Classification" Cancers 15, no. 6: 1767. https://doi.org/10.3390/cancers15061767
APA StyleAlturki, N., Umer, M., Ishaq, A., Abuzinadah, N., Alnowaiser, K., Mohamed, A., Saidani, O., & Ashraf, I. (2023). Combining CNN Features with Voting Classifiers for Optimizing Performance of Brain Tumor Classification. Cancers, 15(6), 1767. https://doi.org/10.3390/cancers15061767






