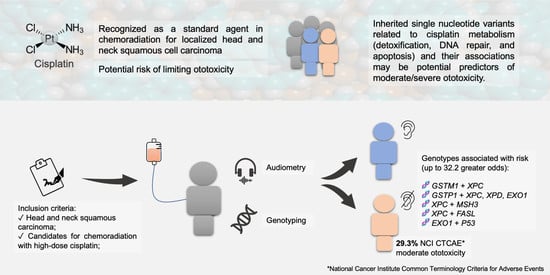
Association of Clinical Aspects and Genetic Variants with the Severity of Cisplatin-Induced Ototoxicity in Head and Neck Squamous Cell Carcinoma: A Prospective Cohort Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
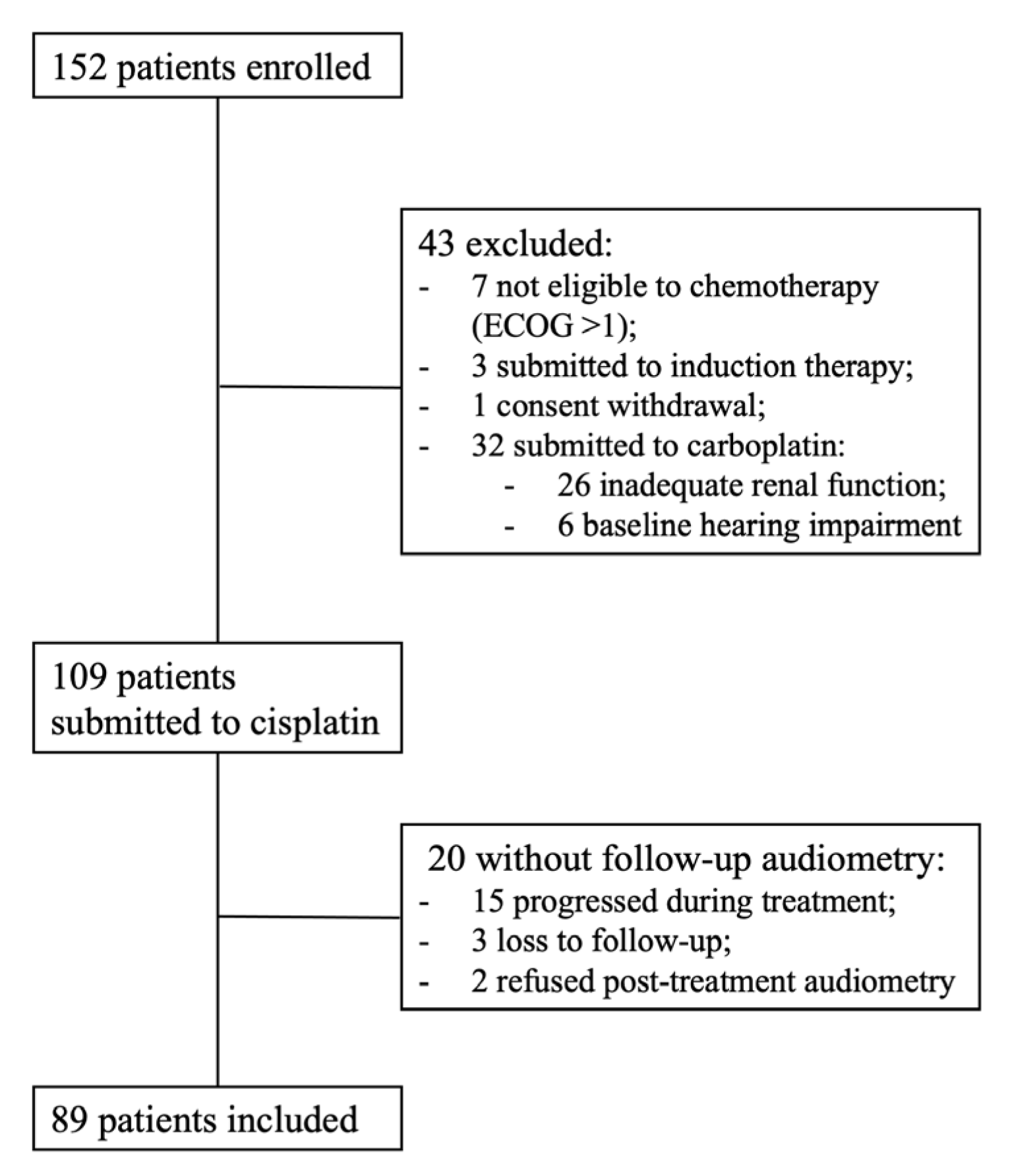
2.1. Study Population
2.2. Clinical Data
2.3. Hearing Assessment
2.3.1. Pure Tone Audiometry
2.3.2. Speech Audiometry
2.4. Hearing Loss Classification
2.4.1. Global Burden of Disease (GBD) Hearing Loss Classification
2.4.2. National Cancer Institute Common Terminology Criteria for Adverse Events (NCI CTCAE)
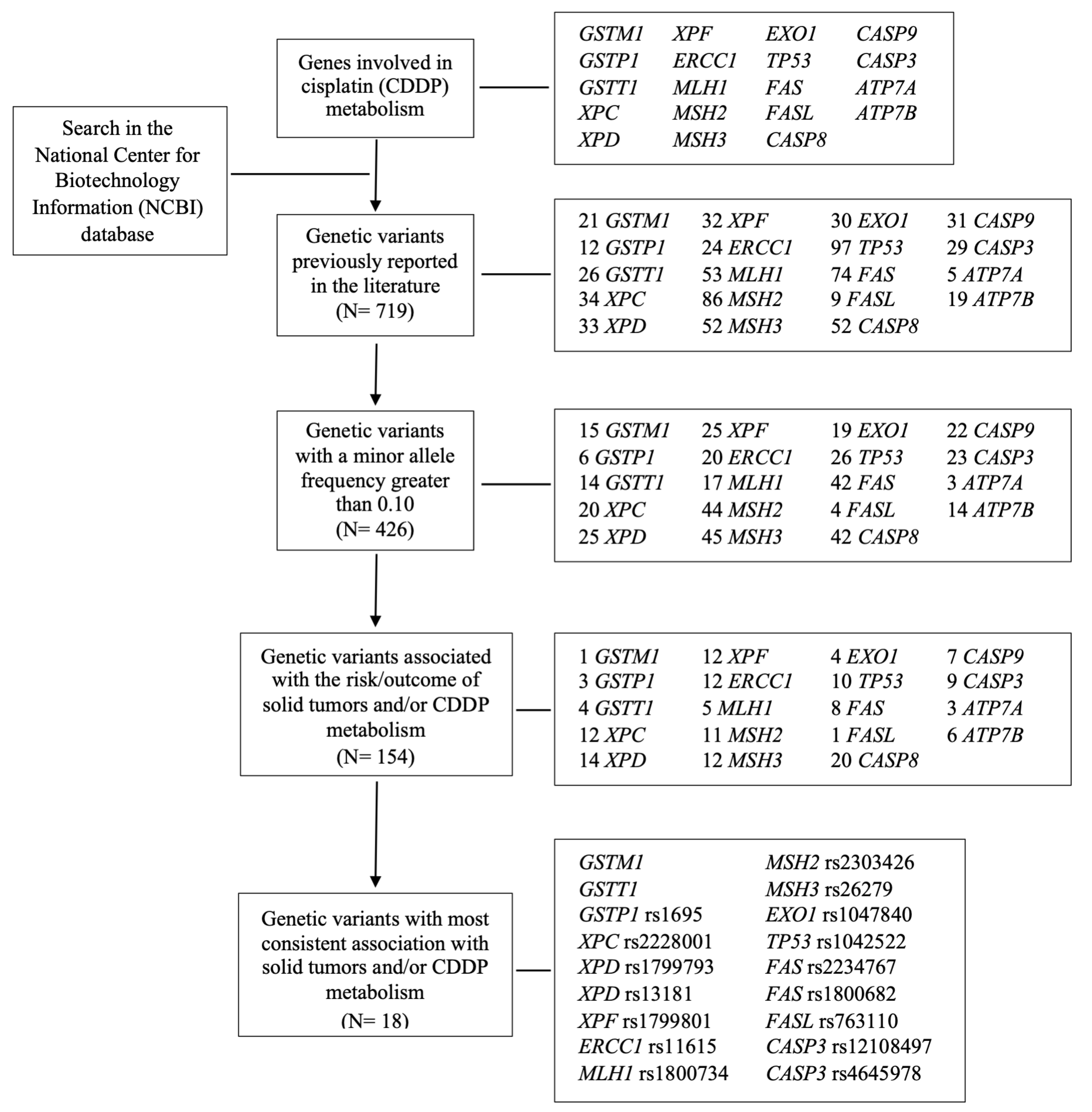
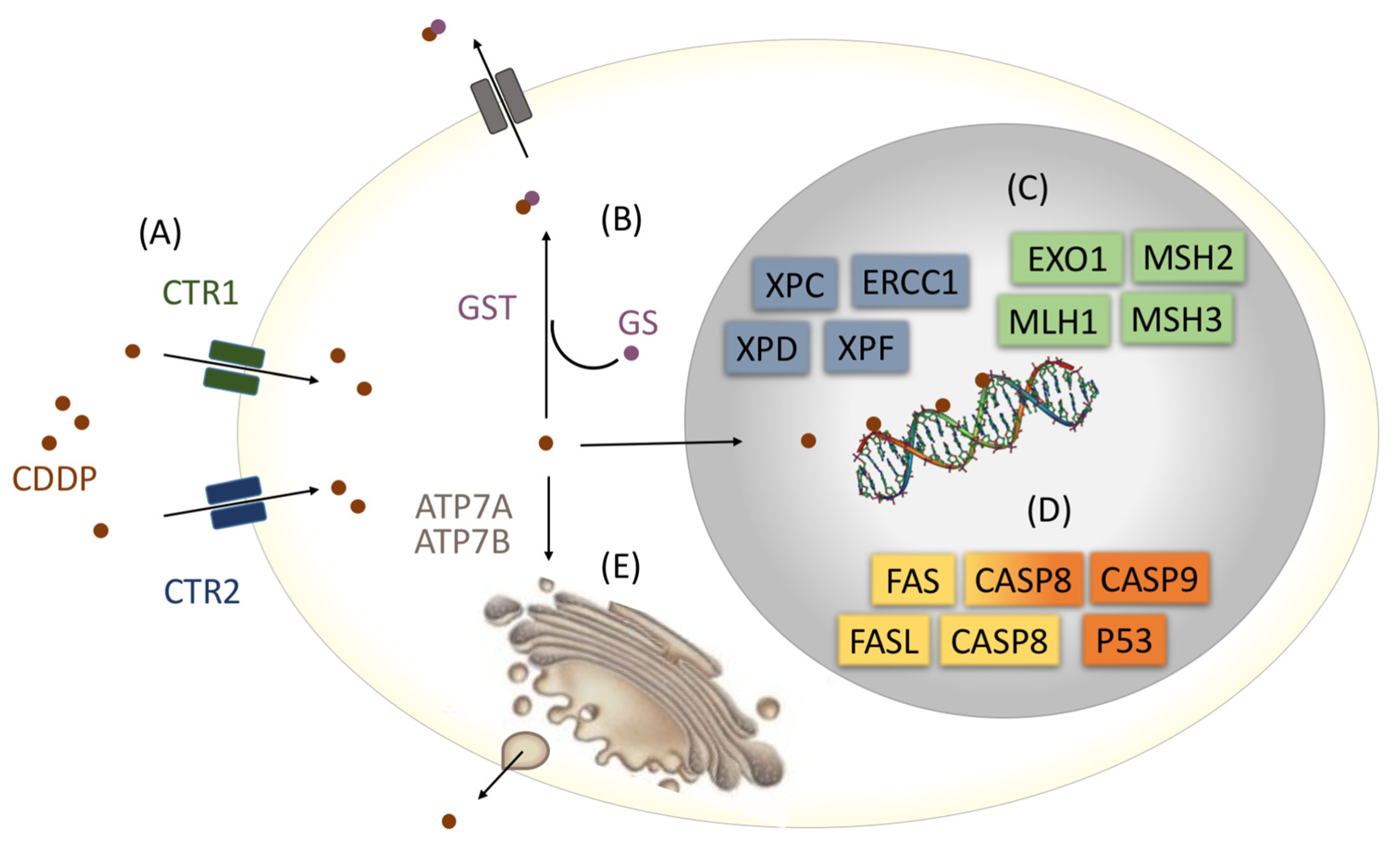
2.5. Genetic Variants Analysis
2.6. Statistical Analysis
3. Results
3.1. Study Population
3.2. Hearing Impairment in Monitoring Audiometry
3.2.1. Pure Tone Audiometry
3.2.2. Speech Audiometry
3.2.3. GBD Classification for Hearing Loss
3.2.4. Hearing Impairment in Monitoring Audiometry According to NCI CTCAE Criteria
3.3. Clinicopathological Aspects and Genotypes in Hearing Impairment
3.3.1. Average of Minimum Threshold for Pure Tone Air Conduction Audiometry at High Frequencies (3, 4, 6, and 8 kHz)
3.3.2. Hearing Impairment in Monitoring Audiometry According to NCI CTCAE Criteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| SNV | SNV ID (rs) | Ch Region | Gene Location | Coding Change | Allele | Function | Allele | Function | Assay | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| GSTM1 | NA | 1p13.3 | NA | Deletion | Present | Normal detoxification | Null | No detoxification | Luciferase | [62] |
| GSTT1 | NA | 22q11.23 | NA | Deletion | Present | Normal detoxification | Null | No detoxification | Luciferase | [62] |
| GSTP1 c.313A>G | 1695 | 11q13.2 | Exon 5 | Ile105Val | A | Normal detoxification | G | Reduced detoxification | IH | [63] |
| XPC c.2815A>C | 2228001 | 3p25.1 | Exon 15 | Lys939Gln | A | Normal repair | C | Reduced repair | Comet assay | [64] |
| XPD c.934G>A | 1799793 | 19q13.32 | Exon 10 | Asp312Asn | G | Normal repair | A | Reduced repair | HCR | [65] |
| XPD c.2251A>C | 13181 | 19q13.32 | Exon 23 | Lys751Gln | A | Normal repair | C | Reduced repair | HCR | [65] |
| XPF c.2505T>C | 1799801 | 16p13.12 | Exon 11 | Ser835Ser | T | Normal repair | C | Reduced repair | IH | [66] |
| ERCC1 c.354C>T | 11615 | 19q13.32 | Exon 4 | Asn118Asn | C | Normal repair | T | Reduced repair | AAS | [67] |
| MLH1 c.-93G>A | 1800734 | 3p22.2 | Promoter | NA | G | Normal repair | A | Reduced repair | Luciferase | [68] |
| MSH2 c.211+9G>C | 2303426 | 2p21 | Intron | NA | G | Reduced repair | C | Normal repair | Western-Blot | [69] |
| MSH3 c.3133G>A | 26279 | 5q14.1 | Exon 23 | Ala1045Thr | G | Normal repair | A | Reduced repair | Expression by qPCR | [70] |
| EXO1 c.1762G>A | 1047840 | 1q43 | Exon 12 | Glu267Lys | G | Normal repair | A | Reduced repair | Western-Blot | [71] |
| TP53 c.215G>C | 1042522 | 17p13.1 | Exon 4 | Arg72Pro | G | Normal apoptosis | C | Reduced apoptosis | Western-Blot | [72] |
| FAS c.-1378G>A | 2234767 | 10q23.31 | Promoter | NA | G | Normal apoptosis | A | Reduced apoptosis | EMSA | [73] |
| FAS c.-671A>G | 1800682 | 10q23.31 | Intron | NA | A | Normal apoptosis | G | Reduced apoptosis | Luciferase | [74] |
| FASL c.-844C>T | 763110 | 1q24.3 | Promoter | NA | C | Normal apoptosis | T | Reduced apoptosis | Luciferase | [74] |
| CASP3 c.-1191A>G | 12108497 | 4q35.1 | Promoter | NA | A | Normal apoptosis | G | Reduced apoptosis | Expression by qPCR | [75] |
| CASP3 c.-182-247G>T | 4647601 | 4q35.1 | Intron | NA | G | Normal apoptosis | T | Reduced apoptosis | Expression by qPCR | [76] |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Vermorken, J.B.; Remenar, E.; van Herpen, C.; Gorlia, T.; Mesia, R.; Degardin, M.; Stewart, J.S.; Jelic, S.; Betka, J.; Preiss, J.H.; et al. Cisplatin, Fluorouracil, and Docetaxel in Unresectable Head and Neck Cancer. N. Engl. J. Med. 2007, 357, 1695–1704. [Google Scholar] [CrossRef]
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and Neck Cancer. Lancet Lond. Engl. 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Azarnia, N.; Shin, D.M.; Cohen, R.B.; Jones, C.U.; Sur, R.; Raben, D.; Jassem, J.; et al. Radiotherapy plus Cetuximab for Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2006, 354, 567–578. [Google Scholar] [CrossRef]
- Vermorken, J.B.; Mesia, R.; Rivera, F.; Remenar, E.; Kawecki, A.; Rottey, S.; Erfan, J.; Zabolotnyy, D.; Kienzer, H.-R.; Cupissol, D.; et al. Platinum-Based Chemotherapy plus Cetuximab in Head and Neck Cancer. N. Engl. J. Med. 2008, 359, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab Alone or with Chemotherapy versus Cetuximab with Chemotherapy for Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (KEYNOTE-048): A Randomised, Open-Label, Phase 3 Study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Albers, J.W.; Chaudhry, V.; Cavaletti, G.; Donehower, R.C. Interventions for Preventing Neuropathy Caused by Cisplatin and Related Compounds. Cochrane Database Syst. Rev. 2014, CD005228. [Google Scholar] [CrossRef]
- Pincinato, E.C.; Visacri, M.B.; de Souza, C.M.; Tuan, B.T.; Ferrari, G.B.; de Oliveira, D.N.; Barbosa, C.R.; Rodrigues, R.F.; Granja, S.; Ambrósio, R.F.L.; et al. Impact of Drug Formulation and Free Platinum/Cisplatin Ratio on Hypersensitivity Reactions to Cisplatin: Formulation Matters. J. Clin. Pharm. Ther. 2015, 40, 41–47. [Google Scholar] [CrossRef]
- Ganesan, P.; Schmiedge, J.; Manchaiah, V.; Swapna, S.; Dhandayutham, S.; Kothandaraman, P.P. Ototoxicity: A Challenge in Diagnosis and Treatment. J. Audiol. Otol. 2018, 22, 59–68. [Google Scholar] [CrossRef]
- Laurell, G. Pharmacological Intervention in the Field of Ototoxicity. HNO 2019, 67, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Bokemeyer, C.; Berger, C.; Hartmann, J.; Kollmannsberger, C.; Schmoll, H.; Kuczyk, M.; Kanz, L. Analysis of Risk Factors for Cisplatin-Induced Ototoxicity in Patients with Testicular Cancer. Br. J. Cancer 1998, 77, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Frisina, R.D.; Wheeler, H.E.; Fossa, S.D.; Kerns, S.L.; Fung, C.; Sesso, H.D.; Monahan, P.O.; Feldman, D.R.; Hamilton, R.; Vaughn, D.J.; et al. Comprehensive Audiometric Analysis of Hearing Impairment and Tinnitus After Cisplatin-Based Chemotherapy in Survivors of Adult-Onset Cancer. J. Clin. Oncol. 2016, 34, 2712–2720. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.; Jackson, A.; Budnick, A.S.; Pfister, D.G.; Kraus, D.H.; Hunt, M.A.; Stambuk, H.; Levegrun, S.; Wolden, S.L. Sensorineural Hearing Loss in Combined Modality Treatment of Nasopharyngeal Carcinoma. Cancer 2006, 106, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Caballero, M.; Mackers, P.; Reig, O.; Buxo, E.; Navarrete, P.; Blanch, J.L.; Grau, J.J. The Role of Audiometry Prior to High-Dose Cisplatin in Patients with Head and Neck Cancer. Oncology 2017, 93, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Pearson, S.E.; Taylor, J.; Patel, P.; Baguley, D.M. Cancer Survivors Treated with Platinum-Based Chemotherapy Affected by Ototoxicity and the Impact on Quality of Life: A Narrative Synthesis Systematic Review. Int. J. Audiol. 2019, 58, 685–695. [Google Scholar] [CrossRef]
- Rademaker-Lakhai, J.M.; Crul, M.; Zuur, L.; Baas, P.; Beijnen, J.H.; Simis, Y.J.W.; van Zandwijk, N.; Schellens, J.H.M. Relationship Between Cisplatin Administration and the Development of Ototoxicity. J. Clin. Oncol. 2006, 24, 918–924. [Google Scholar] [CrossRef]
- Talach, T.; Rottenberg, J.; Gal, B.; Kostrica, R.; Jurajda, M.; Kocak, I.; Lakomy, R.; Vogazianos, E. Genetic Risk Factors of Cisplatin Induced Ototoxicity in Adult Patients. Neoplasma 2016, 63, 263–268. [Google Scholar] [CrossRef]
- Silva Nogueira, G.A.; Dias Costa, E.F.; Lopes-Aguiar, L.; Penna Lima, T.R.; Visacri, M.B.; Pincinato, E.C.; Lourenço, G.J.; Calonga, L.; Mariano, F.V.; Messias de Almeida Milani Altemani, A.; et al. Polymorphisms in DNA Mismatch Repair Pathway Genes Predict Toxicity and Response to Cisplatin Chemoradiation in Head and Neck Squamous Cell Carcinoma Patients. Oncotarget 2018, 9, 29538–29547. [Google Scholar] [CrossRef]
- Clemens, E.; Broer, L.; Langer, T.; Uitterlinden, A.G.; de Vries, A.C.H.; van Grotel, M.; Pluijm, S.F.M.; Binder, H.; Byrne, J.; Broeder, E. van D.; et al. Genetic Variation of Cisplatin-Induced Ototoxicity in Non-Cranial-Irradiated Pediatric Patients Using a Candidate Gene Approach: The International PanCareLIFE Study. Pharm. J. 2020, 20, 294–305. [Google Scholar] [CrossRef]
- Siddik, Z.H. Cisplatin: Mode of Cytotoxic Action and Molecular Basis of Resistance. Oncogene 2003, 22, 7265–7279. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.T.; Chen, H.H.W.; Song, I.-S.; Savaraj, N.; Ishikawa, T. The Roles of Copper Transporters in Cisplatin Resistance. Cancer Metastasis Rev. 2007, 26, 71–83. [Google Scholar] [CrossRef]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione Transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Wright, C.; Ishizuka, H. GS-X Pump Is Functionally Overexpressed in Cis-Diamminedichloroplatinum (II)-Resistant Human Leukemia HL-60 Cells and down-Regulated by Cell Differentiation. J. Biol. Chem. 1994, 269, 29085–29093. [Google Scholar] [CrossRef] [PubMed]
- Shuck, S.C.; Short, E.A.; Turchi, J.J. Eukaryotic Nucleotide Excision Repair: From Understanding Mechanisms to Influencing Biology. Cell Res. 2008, 18, 64–72. [Google Scholar] [CrossRef]
- Khan, S.G. A New Xeroderma Pigmentosum Group C Poly(AT) Insertion/Deletion Polymorphism. Carcinogenesis 2000, 21, 1821–1825. [Google Scholar] [CrossRef]
- Lainé, J.P.; Mocquet, V.; Bonfanti, M.; Braun, C.; Egly, J.M.; Brousset, P. Common XPD (ERCC2) Polymorphisms Have No Measurable Effect on Nucleotide Excision Repair and Basal Transcription. DNA Repair 2007, 6, 1264–1270. [Google Scholar] [CrossRef]
- Goode, E.L.; Ulrich, C.M.; Potter, J.D. Polymorphisms in DNA Repair Genes and Associations with Cancer Risk. Cancer Epidemiol. Biomark. Prev. 2002, 11, 1513. [Google Scholar]
- Martin, L.P.; Hamilton, T.C.; Schilder, R.J. Platinum Resistance: The Role of DNA Repair Pathways. Clin. Cancer Res. 2008, 14, 1291–1295. [Google Scholar] [CrossRef]
- Mello, J.A.; Acharya, S.; Fishe, R.; Essigmann’, J.M. The Mismatch-Repair Protein HMSH2 Binds Selectively to DNA Adducts of the Anticancer Drug Cisplatin. Chem. Biol. 1996, 3, 579–589. [Google Scholar] [CrossRef]
- Singh, S. Cytoprotective and Regulatory Functions of Glutathione S-Transferases in Cancer Cell Proliferation and Cell Death. Cancer Chemother. Pharmacol. 2015, 75, 1–15. [Google Scholar] [CrossRef]
- De Zio, D.; Cianfanelli, V.; Cecconi, F. New Insights into the Link Between DNA Damage and Apoptosis. Antioxid. Redox Signal. 2013, 19, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Drögemöller, B.I.; Brooks, B.; Critchley, C.; Monzon, J.G.; Wright, G.E.B.; Liu, G.; Renouf, D.J.; Kollmannsberger, C.K.; Bedard, P.L.; Hayden, M.R.; et al. Further Investigation of the Role of ACYP2 and WFS1 Pharmacogenomic Variants in the Development of Cisplatin-Induced Ototoxicity in Testicular Cancer Patients. Clin. Cancer Res. 2018, 24, 1866. [Google Scholar] [CrossRef] [PubMed]
- Driessen, C.M.; Ham, J.C.; te Loo, M.; van Meerten, E.; van Lamoen, M.; Hakobjan, M.H.; Takes, R.P.; van der Graaf, W.T.; Kaanders, J.H.; Coenen, M.J.H.; et al. Genetic Variants as Predictive Markers for Ototoxicity and Nephrotoxicity in Patients with Locally Advanced Head and Neck Cancer Treated with Cisplatin-Containing Chemoradiotherapy (The PRONE Study). Cancers 2019, 11, 551. [Google Scholar] [CrossRef] [PubMed]
- Tserga, E.; Nandwani, T.; Edvall, N.K.; Bulla, J.; Patel, P.; Canlon, B.; Cederroth, C.R.; Baguley, D.M. The Genetic Vulnerability to Cisplatin Ototoxicity: A Systematic Review. Sci. Rep. 2019, 9, 3455. [Google Scholar] [CrossRef]
- Wheeler, H.E.; Gamazon, E.R.; Frisina, R.D.; Perez-Cervantes, C.; El Charif, O.; Mapes, B.; Fossa, S.D.; Feldman, D.R.; Hamilton, R.J.; Vaughn, D.J.; et al. Variants in WFS1 and Other Mendelian Deafness Genes Are Associated with Cisplatin-Associated Ototoxicity. Clin. Cancer Res. 2017, 23, 3325. [Google Scholar] [CrossRef]
- Trendowski, M.R.; El Charif, O.; Dinh, P.C.; Travis, L.B.; Dolan, M.E. Genetic and Modifiable Risk Factors Contributing to Cisplatin-Induced Toxicities. Clin. Cancer Res. 2019, 25, 1147–1155. [Google Scholar] [CrossRef]
- Peters, U.; Preisler-Adams, S.; Hebeisen, A.; Hahn, M.; Seifert, E.; Lanvers, C.; Heinecke, A.; Horst, J.; Jürgens, H.; Lamprecht-Dinnesen, A. Glutathione S-Transferase Genetic Polymorphisms and Individual Sensitivity to the Ototoxic Effect of Cisplatin. Anticancer Drugs 2000, 11, 639–643. [Google Scholar] [CrossRef]
- Oldenburg, J.; Kraggerud, S.M.; Cvancarova, M.; Lothe, R.A.; Fossa, S.D. Cisplatin-Induced Long-Term Hearing Impairment Is Associated With Specific Glutathione S-Transferase Genotypes in Testicular Cancer Survivors. J. Clin. Oncol. 2007, 25, 708–714. [Google Scholar] [CrossRef]
- Choeyprasert, W.; Sawangpanich, R.; Lertsukprasert, K.; Udomsubpayakul, U.; Songdej, D.; Unurathapan, U.; Pakakasama, S.; Hongeng, S. Cisplatin-Induced Ototoxicity in Pediatric Solid Tumors: The Role of Glutathione S-Transferases and Megalin Genetic Polymorphisms. J. Pediatr. Hematol. Oncol. 2013, 35, e138–e143. [Google Scholar] [CrossRef]
- Lui, G.; Bouazza, N.; Denoyelle, F.; Moine, M.; Chastagner, P.; Corradini, N.; Entz-Werle, N.; Vérité, C.; Landmanparker, J.; Sudour-Bonnange, H.; et al. Association between Genetic Polymorphisms and Platinum-Induced Ototoxicity in Children. Oncotarget 2018, 9, 30883–30893. [Google Scholar] [CrossRef]
- Budai, B.; Prekopp, P.; Noszek, L.; Kovács, E.; Szőnyi, M.; Erdélyi, D.; Biró, K.; Geczi, L. GSTM1 Null and GSTT1 Null: Predictors of Cisplatin-Caused Acute Ototoxicity Measured by DPOAEs. J. Mol. Med. 2020, 98, 963–971. [Google Scholar] [CrossRef]
- Sherief, L.M.; Rifky, E.; Attia, M.; Ahmed, R.; Kamal, N.M.; Oshi, M.A.M.; Hanna, D. Platinum-Induced Ototoxicity in Pediatric Cancer Survivors: GSTP1 c.313A>G Variant Association. Medicine 2022, 101, e31627. [Google Scholar] [CrossRef] [PubMed]
- Caronia, D.; Patiño-García, A.; Milne, R.L.; Zalacain-Díez, M.; Pita, G.; Alonso, M.R.; Moreno, L.T.; Sierrasesumaga-Ariznabarreta, L.; Benítez, J.; González-Neira, A. Common Variations in ERCC2 Are Associated with Response to Cisplatin Chemotherapy and Clinical Outcome in Osteosarcoma Patients. Pharm. J. 2009, 9, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Pincinato, E.C.; Costa, E.F.D.; Lopes-Aguiar, L.; Nogueira, G.A.S.; Lima, T.R.P.; Visacri, M.B.; Costa, A.P.L.; Lourenço, G.J.; Calonga, L.; Mariano, F.V.; et al. GSTM1, GSTT1 and GSTP1 Ile105Val Polymorphisms in Outcomes of Head and Neck Squamous Cell Carcinoma Patients Treated with Cisplatin Chemoradiation. Sci. Rep. 2019, 9, 9312. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Aguiar, L.; Costa, E.F.D.; Nogueira, G.A.S.; Lima, T.R.P.; Visacri, M.B.; Pincinato, E.C.; Calonga, L.; Mariano, F.V.; de Almeida Milani Altemani, A.M.; Altemani, J.M.C.; et al. XPD c.934G>A Polymorphism of Nucleotide Excision Repair Pathway in Outcome of Head and Neck Squamous Cell Carcinoma Patients Treated with Cisplatin Chemoradiation. Oncotarget 2017, 8, 16190–16201. [Google Scholar] [CrossRef]
- Costa, E.F.D.; Lima, T.R.P.; Lopes-Aguiar, L.; Nogueira, G.A.S.; Visacri, M.B.; Quintanilha, J.C.F.; Pincinato, E.C.; Calonga, L.; Mariano, F.V.; de Almeida Milani Altemani, A.M.; et al. FAS and FASL Variations in Outcomes of Tobacco- and Alcohol-Related Head and Neck Squamous Cell Carcinoma Patients. Tumor Biol. 2020, 42, 1010428320938494. [Google Scholar] [CrossRef]
- Oken, M.; Creech, R.; Tormey, D.; Horton, J.; Davis, T.; McFadden, E.; Carbone, P. Toxicity and Response Criteria of the Eastern Cooperative Oncology Group. Am. J. Clin. Oncol. 1982, 6, 649–655. [Google Scholar] [CrossRef]
- Forastiere, A.A.; Zhang, Q.; Weber, R.S.; Maor, M.H.; Goepfert, H.; Pajak, T.F.; Morrison, W.; Glisson, B.; Trotti, A.; Ridge, J.A.; et al. Long-Term Results of RTOG 91-11: A Comparison of Three Nonsurgical Treatment Strategies to Preserve the Larynx in Patients with Locally Advanced Larynx Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 845–852. [Google Scholar] [CrossRef]
- Common Terminology Criteria for Adverse Events (CTCAE) v5.0. Published November, 27, 2017. Available online: https://ctep.cancer.gov/protocoldevelopment/electronic_applications/ctc.htm#ctc_50 (accessed on 15 July 2022).
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for Reporting Observational Studies. PLoS Med. 2007, 4, e296. [Google Scholar] [CrossRef]
- Hayes, R.B.; Bravo-Otero, E.; Kleinman, D.V.; Brown, L.M.; Fraumeni, J.F.; Harty, L.C.; Winn, D.M. Tobacco and Alcohol Use and Oral Cancer in Puerto Rico. Cancer Causes Control 1999, 10, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Bray, G.A.; Heisel, W.E.; Afshin, A.; Jensen, M.D.; Dietz, W.H.; Long, M.; Kushner, R.F.; Daniels, S.R.; Wadden, T.A.; Tsai, A.G.; et al. The Science of Obesity Management: An Endocrine Society Scientific Statement. Endocr. Rev. 2018, 39, 79–132. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association Professional Practice Committee 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2022. Diabetes Care 2021, 45, S17–S38. [Google Scholar] [CrossRef]
- Toto, R.D. Defining Hypertension: Role of New Trials and Guidelines. Clin. J. Am. Soc. Nephrol. CJASN 2018, 13, 1578–1580. [Google Scholar] [CrossRef] [PubMed]
- Lydiatt, W.M.; Patel, S.G.; O’Sullivan, B.; Brandwein, M.S.; Ridge, J.A.; Migliacci, J.C.; Loomis, A.M.; Shah, J.P. Head and Neck Cancers—Major Changes in the American Joint Committee on Cancer Eighth Edition Cancer Staging Manual. CA Cancer J. Clin. 2017, 67, 122–137. [Google Scholar] [CrossRef]
- Clark, J. Uses and Abuses of Hearing Loss Classification. ASHA 1981, 23, 493–500. [Google Scholar]
- American Speech-Language-Hearing Association Determining Threshold Level for Speech [Guidelines]. 1988. Available online: https://www.asha.org/policy/GL1988-00008/ (accessed on 15 July 2022).
- Jerger, J.; Speaks, C.; Trammell, J.L. A New Approach to Speech Audiometry. J. Speech Hear. Disord. 1968, 33, 318–328. [Google Scholar] [CrossRef]
- Informal Working Group on Prevention of Deafness and Hearing Impairment Programme Planning (1991: Geneva, Switzerland); World Health Organization. Programme for the Prevention of Deafness and Hearing Impairment. In Report of the Informal Working Group on Prevention of Deafness and Hearing Impairment Programme Planning, Geneva, 18–21 June 1991; World Health Organization: Geneva, Switzerland, 1991. [Google Scholar]
- Stevens, G.; Flaxman, S.; Brunskill, E.; Mascarenhas, M.; Mathers, C.D.; Finucane, M. On behalf of the Global Burden of Disease Hearing Loss Expert Group Global and Regional Hearing Impairment Prevalence: An Analysis of 42 Studies in 29 Countries. Eur. J. Public Health 2013, 23, 146–152. [Google Scholar] [CrossRef]
- Moyer, A.M.; Salavaggione, O.E.; Hebbring, S.J.; Moon, I.; Hildebrandt, M.A.T.; Eckloff, B.W.; Schaid, D.J.; Wieben, E.D.; Weinshilboum, R.M. Glutathione S-Transferase T1 and M1: Gene Sequence Variation and Functional Genomics. Clin. Cancer Res. 2007, 13, 7207–7216. [Google Scholar] [CrossRef]
- Terrier, P.; Townsend, A.J.; Coindre, J.M.; Triche, T.J.; Cowan, K.H. An Immunohistochemical Study of Pi Class Glutathione S-Transferase Expression in Normal Human Tissue. Am. J. Pathol. 1990, 137, 845–853. [Google Scholar]
- Zhu, Y.; Yang, H.; Chen, Q.; Lin, J.; Grossman, H.B.; Dinney, C.P.; Wu, X.; Gu, J. Modulation of DNA Damage/DNA Repair Capacity by XPC Polymorphisms. DNA Repair 2008, 7, 141–148. [Google Scholar] [CrossRef]
- Spitz, M.R.; Wu, X.; Wang, Y.; Wang, L.-E.; Shete, S.; Amos, C.I.; Guo, Z.; Lei, L.; Mohrenweiser, H.; Wei, Q. Modulation of Nucleotide Excision Repair Capacity by XPD Polymorphisms in Lung Cancer Patients. Cancer Res. 2001, 61, 1354. [Google Scholar] [PubMed]
- Vaezi, A.; Wang, X.; Buch, S.; Gooding, W.; Wang, L.; Seethala, R.R.; Weaver, D.T.; D’Andrea, A.D.; Argiris, A.; Romkes, M.; et al. XPF Expression Correlates with Clinical Outcome in Squamous Cell Carcinoma of the Head and Neck. Clin. Cancer Res. 2011, 17, 5513–5522. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.J.; Lee, K.B.; Mu, C.; Li, Q.; Abernathy, T.V.; Bostick-Bruton, F.; Reed, E. Comparison of Two Human Ovarian Carcinoma Cell Lines (A2780/CP70 and MCAS) That Are Equally Resistant to Platinum, but Differ at Codon 118 of the ERCC1 Gene. Int. J. Oncol. 2000, 16, 555–615. [Google Scholar] [CrossRef] [PubMed]
- Perera, S.; Mrkonjic, M.; Rawson, J.B.; Bapat, B. Functional Effects of the MLH1-93G>A Polymorphism on MLH1/EPM2AIP1 Promoter Activity. Oncol. Rep. 2011, 25, 809–815. [Google Scholar] [CrossRef]
- Marra, G.; D’Atri, S.; Corti, C.; Bonmassar, L.; Cattaruzza, M.S.; Schweizer, P.; Heinimann, K.; Bartosova, Z.; Nyström-Lahti, M.; Jiricny, J. Tolerance of Human MSH2+/− Lymphoblastoid Cells to the Methylating Agent Temozolomide. Proc. Natl. Acad. Sci. USA 2001, 98, 7164–7169. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, G.A.S.; Lourenço, G.J.; Oliveira, C.B.M.; Marson, F.A.L.; Lopes-Aguiar, L.; Costa, E.F.D.; Lima, T.R.P.; Liutti, V.T.; Leal, F.; Santos, V.C.A.; et al. Association between Genetic Polymorphisms in DNA Mismatch Repair-Related Genes with Risk and Prognosis of Head and Neck Squamous Cell Carcinoma. Int. J. Cancer 2015, 137, 810–818. [Google Scholar] [CrossRef]
- Wei, K.; Clark, A.B.; Wong, E.; Kane, M.F.; Mazur, D.J.; Parris, T.; Kolas, N.K.; Russell, R.; Hou, H.; Kneitz, B.; et al. Inactivation of Exonuclease 1 in Mice Results in DNA Mismatch Repair Defects, Increased Cancer Susceptibility, and Male and Female Sterility. Genes Dev. 2003, 17, 603–614. [Google Scholar] [CrossRef]
- Dumont, P.; Leu, J.I.-J.; Della Pietra, A.C.; George, D.L.; Murphy, M. The Codon 72 Polymorphic Variants of P53 Have Markedly Different Apoptotic Potential. Nat. Genet. 2003, 33, 357–365. [Google Scholar] [CrossRef]
- Sibley, K.; Rollinson, S.; Allan, J.M.; Smith, A.G.; Law, G.R.; Roddam, P.L.; Skibola, C.F.; Smith, M.T.; Morgan, G.J. Functional FAS Promoter Polymorphisms Are Associated with Increased Risk of Acute Myeloid Leukemia1. Cancer Res. 2003, 63, 4327–4330. [Google Scholar]
- Wu, J.; Metz, C.; Xu, X.; Abe, R.; Gibson, A.W.; Edberg, J.C.; Cooke, J.; Xie, F.; Cooper, G.S.; Kimberly, R.P. A Novel Polymorphic CAAT/Enhancer-Binding Protein Beta Element in the FasL Gene Promoter Alters Fas Ligand Expression: A Candidate Background Gene in African American Systemic Lupus Erythematosus Patients. J. Immunol. 2003, 170, 132–138. [Google Scholar] [CrossRef]
- Jang, J.S.; Kim, K.M.; Choi, J.E.; Cha, S.I.; Kim, C.H.; Lee, W.K.; Kam, S.; Jung, T.H.; Park, J.Y. Identification of Polymorphisms in the Caspase-3 Gene and Their Association with Lung Cancer Risk. Mol. Carcinog. 2008, 47, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhao, H.; Hu, Z.; Wang, L.-E.; Zhang, W.; Sturgis, E.M.; Wei, Q. CASP3 Polymorphisms and Risk of Squamous Cell Carcinoma of the Head and Neck. Clin. Cancer Res. 2008, 14, 6343. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Levine, M.; Ensom, M.H.H. Post Hoc Power Analysis: An Idea Whose Time Has Passed? Pharm. J. Hum. Pharmacol. Drug Ther. 2001, 21, 405–409. [Google Scholar] [CrossRef]
- Wang, M.; Xu, S. Statistical Power in Genome-Wide Association Studies and Quantitative Trait Locus Mapping. Heredity 2019, 123, 287–306. [Google Scholar] [CrossRef]
- Efron, B. Bootstrap Methods: Another Look at the Jackknife. Ann. Stat. 1979, 7, 1–26. [Google Scholar] [CrossRef]
- Kopke, R.; Liu, W.; Gabaizadeh, R.; Jacono, A.; Feghali, J.; Spray, D.; Garcia, P.; Steinman, H.; Malgrange, B.; Ruben, R.; et al. Use of Organotypic Cultures of Corti’s Organ to Study the Protective Effects of Antioxidant Molecules on Cisplatin-Induced Damage of Auditory Hair Cells. Am. J. Otol. 1997, 18, 559–571. [Google Scholar]
- Breglio, A.M.; Rusheen, A.E.; Shide, E.D.; Fernandez, K.A.; Spielbauer, K.K.; McLachlin, K.M.; Hall, M.D.; Amable, L.; Cunningham, L.L. Cisplatin Is Retained in the Cochlea Indefinitely Following Chemotherapy. Nat. Commun. 2017, 8, 1654. [Google Scholar] [CrossRef]
- Nomura, K.; Nakao, M.; Morimoto, T. Effect of Smoking on Hearing Loss: Quality Assessment and Meta-Analysis. Prev. Med. 2005, 40, 138–144. [Google Scholar] [CrossRef]
- Capra Marques de Oliveira, D.C.; de Melo Tavares de Lima, M.A. Low and High Frequency Tonal Threshold Audiometry: Comparing Hearing Thresholds between Smokers and Non-Smokers. Braz. J. Otorhinolaryngol. 2009, 75, 738–744. [Google Scholar] [CrossRef]
- Le, T.N.; Straatman, L.V.; Lea, J.; Westerberg, B. Current Insights in Noise-Induced Hearing Loss: A Literature Review of the Underlying Mechanism, Pathophysiology, Asymmetry, and Management Options. J. Otolaryngol.—Head Neck Surg. 2017, 46, 41. [Google Scholar] [CrossRef] [PubMed]
- Berg, R.; Pickett, W.; Linneman, J.; Wood, D.; Marlenga, B. Asymmetry in Noise-Induced Hearing Loss: Evaluation of Two Competing Theories. Noise Health 2014, 16, 102–107. [Google Scholar] [PubMed]
- Schmidt, C.-M.; Knief, A.; Lagosch, A.K.; Deuster, D.; am Zehnhoff-Dinnesen, A. Left-Right Asymmetry in Hearing Loss Following Cisplatin Therapy in Children—The Left Ear Is Slightly but Significantly More Affected. Ear Hear. 2008, 29, 830–837. [Google Scholar] [CrossRef] [PubMed]
- GBD 2019 Hearing Loss Collaborators. Hearing Loss Prevalence and Years Lived with Disability, 1990–2019: Findings from the Global Burden of Disease Study 2019. Lancet 2021, 397, 996–1009. [Google Scholar] [CrossRef]
- Dillard, L.K.; Lopez-Perez, L.; Martinez, R.X.; Fullerton, A.M.; Chadha, S.; McMahon, C.M. Global Burden of Ototoxic Hearing Loss Associated with Platinum-Based Cancer Treatment: A Systematic Review and Meta-Analysis. Cancer Epidemiol. 2022, 79, 102203. [Google Scholar] [CrossRef]
- Hornsby, B.W.Y.; Ricketts, T.A. The Effects of Hearing Loss on the Contribution of High- and Low-Frequency Speech Information to Speech Understanding. II. Sloping Hearing Loss. J. Acoust. Soc. Am. 2006, 119, 1752–1763. [Google Scholar] [CrossRef]
- Shahbazi, M.; Zhang, X.; Dinh, P.C.; Sanchez, V.A.; Trendowski, M.R.; Shuey, M.M.; Nguyen, T.; Regeneron Genetics Center; Feldman, D.R.; Vaughn, D.J.; et al. Comprehensive Association Analysis of Speech Recognition Thresholds after Cisplatin-Based Chemotherapy in Survivors of Adult-Onset Cancer. Cancer Med. 2022, 12, 2999–3012. [Google Scholar] [CrossRef]
- Eckerling, A.; Ricon-Becker, I.; Sorski, L.; Sandbank, E.; Ben-Eliyahu, S. Stress and Cancer: Mechanisms, Significance and Future Directions. Nat. Rev. Cancer 2021, 21, 767–785. [Google Scholar] [CrossRef]
- Miaskowski, C.; Paul, S.M.; Mastick, J.; Abrams, G.; Topp, K.; Smoot, B.; Kober, K.M.; Chesney, M.; Mazor, M.; Mausisa, G.; et al. Associations Between Perceived Stress and Chemotherapy-Induced Peripheral Neuropathy and Otoxicity in Adult Cancer Survivors. J. Pain Symptom Manag. 2018, 56, 88–97. [Google Scholar] [CrossRef]
- Watson, M.A.; Stewart, R.K.; Smith, G.B.; Massey, T.E.; Bell, D.A. Human Glutathione S-Transferase P1 Polymorphisms: Relationship to Lung Tissue Enzyme Activity and Population Frequency Distribution. Carcinogenesis 1998, 19, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How Does P53 Induce Apoptosis and How Does This Relate to P53-Mediated Tumour Suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Aasland, D.; Götzinger, L.; Hauck, L.; Berte, N.; Meyer, J.; Effenberger, M.; Schneider, S.; Reuber, E.E.; Roos, W.P.; Tomicic, M.T.; et al. Temozolomide Induces Senescence and Repression of DNA Repair Pathways in Glioblastoma Cells via Activation of ATR–CHK1, P21, and NF-ΚB. Cancer Res. 2019, 79, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Allmann, S.; Mayer, L.; Olma, J.; Kaina, B.; Hofmann, T.G.; Tomicic, M.T.; Christmann, M. Benzo[a]Pyrene Represses DNA Repair through Altered E2F1/E2F4 Function Marking an Early Event in DNA Damage-Induced Cellular Senescence. Nucleic Acids Res. 2020, 48, 12085–12101. [Google Scholar] [CrossRef] [PubMed]
- Benkafadar, N.; François, F.; Affortit, C.; Casas, F.; Ceccato, J.-C.; Menardo, J.; Venail, F.; Malfroy-Camine, B.; Puel, J.-L.; Wang, J. ROS-Induced Activation of DNA Damage Responses Drives Senescence-Like State in Postmitotic Cochlear Cells: Implication for Hearing Preservation. Mol. Neurobiol. 2019, 56, 5950–5969. [Google Scholar] [CrossRef]
- Acklin, S.; Zhang, M.; Du, W.; Zhao, X.; Plotkin, M.; Chang, J.; Campisi, J.; Zhou, D.; Xia, F. Depletion of Senescent-like Neuronal Cells Alleviates Cisplatin-Induced Peripheral Neuropathy in Mice. Sci. Rep. 2020, 10, 14170. [Google Scholar] [CrossRef]

| Variable | Median (IQR) or N (%) |
|---|---|
| Age (years) | 56 (37–69) |
| Sex | |
| Male | 82 (92.1) |
| Female | 7 (7.9) |
| Race (non-white) | 9 (10.1) |
| Tobacco consumption | |
| Smokers | 87 (97.7) |
| Non-smokers | 2 (2.3) |
| Alcohol consumption | |
| Active | 81 (91) |
| Abstainers | 8 (9) |
| ECOG performance status | |
| 0 | 58 (63.5) |
| 1 | 31 (36.5) |
| Comorbidities | |
| BMI | 19.4 (13.7–27.5) |
| Diabetes | 9 (10.1) |
| Hypertension | 24 (26.9) |
| Tumor location | |
| Oral cavity or oropharynx | 55 (61.8) |
| Hypopharynx or larynx | 34 (38.2) |
| Tumor side | |
| Right | 37 (42.0) |
| Left | 42 (47.7) |
| Bilateral/medial | 9 (10.2) |
| Histological grade | |
| Well or moderately differentiated | 73 (82.0) |
| Poorly or undifferentiated | 16 (18.0) |
| Tumor stage | |
| I or II | 6 (6.7) |
| III or IV | 83 (93.3) |
| Cumulative cisplatin dose (mg/m2) | 260 (160–300) |
| Radiotherapy technique (2D) | 88 (98.8) |
| Radiotherapy dose (Gy) | |
| Supraclavicular fossa | 50 (44–50) |
| Facial (right and left) | 44 (44–44) |
| Boost (right and left) | 20 (20–20) |
| Frequency (kHz) | Right Ear | Left Ear | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre-Treatment | Post-Treatment | Difference | p-Value | BH p-Value | Pre-Treatment | Post-Treatment | Difference | p-Value | BH p-Value | |
| Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | |||||||
| Air conduction | ||||||||||
| 0.25 | 15 (10–20) | 15 (10–20) | 0 (−5–+5) | 0.97 | 0.97 | 15 (5–20) | 15 (10–20) | 0 (−5–+5) | 0.10 | 0.11 |
| 0.5 | 10 (5–15) | 10 (5–15) | 0 (−5–+5) | 0.40 | 0.45 | 10 (5–15) | 15 (10–20) | 0 (−5–+5) | 0.06 | 0.09 |
| 1 | 10 (5–15) | 10 (5–15) | 0 (−5–+5) | 0.05 | 0.07 | 10 (5–20) | 10 (5–20) | 0 (−5–+5) | 0.11 | 0.11 |
| 2 | 10 (5–20) | 15 (10–35) | 5 (0–15) | <0.001 | <0.001 | 15 (5–20) | 20 (10–35) | 5 (0–20) | <0.001 | <0.001 |
| 3 | 20 (10–35) | 45 (20–60) | 10 (0–25) | <0.001 | <0.001 | 25 (10–35) | 45 (20–60) | 10 (0–30) | <0.001 | <0.001 |
| 4 | 35 (20–50) | 55 (35–65) | 15 (0–25) | <0.001 | <0.001 | 35 (20–45) | 55 (45–65) | 10 (5–30) | <0.001 | <0.001 |
| 6 | 40 (20–55) | 65 (45–75) | 15 (5–35) | <0.001 | <0.001 | 35 (20–55) | 65 (50–75) | 20 (5–35) | <0.001 | <0.001 |
| 8 | 35 (15–55) | 65 (50–75) | 20 (10–35) | <0.001 | <0.001 | 30 (15–55) | 65 (55–75) | 30 (10–40) | <0.001 | <0.001 |
| Bone conduction | ||||||||||
| 0.25 | 15 (10–20) | 15 (10–20) | 0 (−5–+5) | 0.90 | 0.99 | 15 (5–15) | 15 (10–20) | 0 (−5–+5) | 0.03 | 0.05 |
| 0.5 | 10 (5–15) | 10 (5–15) | 0 (−5–+5) | 0.99 | 0.99 | 10 (5–15) | 10 (5–15) | 0 (−5–+5) | 0.13 | 0.15 |
| 1 | 10 (5–15) | 10 (5–15) | 0 (−5–+5) | 0.15 | 0.23 | 10 (5–15) | 10 (5–20) | 0 (−5–+5) | 0.16 | 0.15 |
| 2 | 10 (5–20) | 15 (10–35) | 5 (0–10) | <0.001 | <0.001 | 10 (5–20) | 20 (10–35) | 5 (0–20) | <0.001 | <0.001 |
| 3 | 20 (10–30) | 40 (20–55) | 10 (0–25) | <0.001 | <0.001 | 20 (10–35) | 40 (20–55) | 10 (0–25) | <0.001 | <0.001 |
| 4 | 30 (15–45) | 50 (30–60) | 10 (0–30) | <0.001 | <0.001 | 30 (20–45) | 50 (40–60) | 10 (0–25) | <0.001 | <0.001 |
| Variable | Pre-Treatment | Post-Treatment | Difference | p-Value |
|---|---|---|---|---|
| Median (IQR) or N (%) | Median (IQR) or N (%) | Median (IQR) or N (%) | ||
| Pure tone averages | ||||
| ISO average (0.5, 1, 2, and 4 kHz) | ||||
| Air conduction (dB) | ||||
| Right ear | 17.5 (8–83) | 22.5 (8.7–48.7) | 5 (−6.25–22.5) | <0.001 |
| Left ear | 18.75 (5–43.7) | 25 (6.2–75) | 6.25 (−5–30) | <0.001 |
| Bone conduction (dB) | ||||
| Right ear | 16.25 (6.2–37.5) | 22.5 (10–45) | 6.25 (−7.5–20) | <0.001 |
| Left ear | 16.25 (5–37.5) | 22.5 (6.2–52.5) | 6.25 (−5–20) | <0.001 |
| High-frequency averages (3, 4, 6, and 8 kHz) | ||||
| Air conduction (dB) | ||||
| Right ear | 35 (0.25–8) | 54 (0.25–8) | 18 (−51–8) | <0.001 |
| Left ear | 34 (0.25–8) | 55 (0.25–8) | 19 (−54–4) | <0.001 |
| Speech audiometry | ||||
| SRT (dB) | ||||
| Right ear | 10 (5–30) | 15 (5–35) | 0 (−10–15) | 0.12 |
| Left ear | 15 (5–35) | 15 (5–45) | 0 (−10–25) | 0.30 |
| SDS (%) | ||||
| Right ear | 96 (88–100) | 92 (80–100) | −4 (−12–4) | 0.001 |
| Left ear | 96 (72–100) | 92 (72–100) | 0 (−16–12) | 0.06 |
| GBD hearing loss classification | ||||
| Air conduction | ||||
| No loss | 57 (64) | 32 (35.9) | <0.001 | |
| Unilateral | 1 (1.1) | 4 (4.5) | ||
| Mild | 28 (31.4) | 37 (41.5) | ||
| Moderate | 3 (3.4) | 16 (17.9) | ||
| Bone conduction | ||||
| No loss | 62 (69.6) | 39 (43.8) | <0.001 | |
| Unilateral | 1 (1.1) | 2 (2.2) | ||
| Mild | 23 (25.8) | 38 (42.7) | ||
| Moderate | 3 (3.4) | 10 (11.2) | ||
| Variable | N | Right Ear | Left Ear | ||||
|---|---|---|---|---|---|---|---|
| Pre-Treatment | Post-Treatment | Difference | Pre-Treatment | Post-Treatment | Difference | ||
| Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | ||||
| XPC c.2815A>C | |||||||
| AA | 34 | 26.3 (16.9–42.2) | 60.0 (43.4–67.8) | 23.8 (10.0–39.4) | 25.6 (18.8–46.3) | 57.5 (40.0–68.8) | 27.5 (8.8–40.3) |
| AC or CC | 55 | 35.0 (20.0–51.3) | 58.8 (32.5–68.8) | 17.5 (5.0–22.5) | 35.0 (20.0–48.8) | 55.0 (41.3–65.0) | 16.3 (6.3–26.3) |
| p-value | 0.008 | 0.04 | |||||
| PA | 60.5 | 49.6 | |||||
| GSTP1 c.313A>G + XPC c.2815A>C | |||||||
| AA + AC or CC | 25 | 28.8 (18.8–46.9) | 50.0 (30.0–65.6) | 17.5 (7.5–23.8) | 33.8 (17.5–42.5) | 52.5 (38.1–62.5) | 16.3 (8.8–23.8) |
| AG or GG + AA | 19 | 20.0 (15.0–35.0) | 61.3 (50.0–67.5) | 30.0 (16.3–48.8) | 23.8 (13.8–41.3) | 62.5 (42.5–68.8) | 38.8 (16.3–52.5) |
| p-value | 0.005 | 0.01 | |||||
| PA | 88.0 | 76.0 | |||||
| GSTM1 + EXO1 c.1762G>A | |||||||
| Present + GG | 13 | 21.3 (18.8–48.1) | 31.3 (25.6–63.8) | 5.0 (2.5–18.8) | 23.8 (17.5–48.1) | 38.8 (28.8–60.0) | 8.8 (5.0–15.0) |
| Null + GA or AA | 27 | 37.5 (21.3–50.0) | 62.5 (57.5–68.8) | 21.3 (16.3–30.0) | 40.0 (25.0–48.8) | 61.3 (52.5–68.8) | 22.5 (11.3–28.8) |
| p-value | 0.008 | 0.005 | |||||
| PA | 75.0 | 85.0 |
| Variable | N | Ototoxicity | ||||
|---|---|---|---|---|---|---|
| G0–G2 | G3–G4 | OR (95% CI) | p-Value | PA (%) | ||
| GSTP1 c.313A>G | ||||||
| AA | 40 | 33 (52.4) | 7 (26.9) | Reference | 0.01 1 | 65 |
| AG or GG | 49 | 30 (47.6) | 19 (73.1) | 4.20 (1.34–13.16) | ||
| XPC c.2815A>C | ||||||
| AC or CC | 55 | 45 (71.4) | 10 (38.5) | Reference | 0.01 2 | 65 |
| AA | 34 | 18 (28.6) | 16 (61.5) | 3.13 (1.27–7.70) | ||
| GSTM1 + XPC c.2815A>C | ||||||
| Present + AC or CC | 22 | 19 (70.4) | 3 (27.3) | Reference | 0.02 3 | 99 |
| Null + AA | 16 | 8 (29.6) | 8 (72.7) | 8.19 (1.28–52.20) | ||
| GSTP1 c.313A>G + XPC 2815A>C | ||||||
| AA + AC or CC | 25 | 24 (72.7) | 1 (9.1) | Reference | 0.004 4 | 97 |
| AG or GG + AA | 19 | 9 (27.3) | 10 (90.9) | 32.22 (3.09–335.52) | ||
| GSTP1 c.313A>G + XPD c.934G>A | ||||||
| AA + GG or GA | 37 | 30 (96.8) | 7 (63.6) | Reference | 0.02 5 | 92 |
| AG or GG + AA | 5 | 1 (3.2) | 4 (36.4) | 19.44 (1.59–237.72) | ||
| GSTP1 c.313A>G + EXO1 c.1762G>A | ||||||
| AA + GG or GA | 37 | 31 (93.9) | 6 (54.5) | Reference | 0.01 6 | 81 |
| AG or GG + AA | 7 | 2 (6.1) | 5 (45.5) | 12.08 (1.60–91.01) | ||
| XPC c.2815A>C + MSH3 c.3133A>G | ||||||
| AC or CC + AG or GG | 23 | 21 (65.6) | 2 (22.2) | Reference | 0.009 7 | 88 |
| AA + AA | 18 | 11 (34.4) | 7 (77.8) | 17.09 (2.02–144.32) | ||
| XPC c.2815A>C + FASL c.-844C>T | ||||||
| AC or CC + TT | 12 | 11 (50.0) | 1 (7.7) | Reference | 0.01 8 | 82 |
| AA + CC or CT | 23 | 11 (50.0) | 12 (92.3) | 22.29 (1.79–276.99) | ||
| EXO1 c.1762G>A + P53 c.215G>C | ||||||
| GG + GG or GC | 31 | 24 (96.0) | 7 (58.3) | Reference | 0.01 9 | 85 |
| GA or AA + CC | 6 | 1 (4.0) | 5 (41.7) | 20.97 (1.66–264.08) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macedo, L.T.; Costa, E.F.D.; Carvalho, B.F.; Lourenço, G.J.; Calonga, L.; Castilho, A.M.; Chone, C.T.; Lima, C.S.P. Association of Clinical Aspects and Genetic Variants with the Severity of Cisplatin-Induced Ototoxicity in Head and Neck Squamous Cell Carcinoma: A Prospective Cohort Study. Cancers 2023, 15, 1759. https://doi.org/10.3390/cancers15061759
Macedo LT, Costa EFD, Carvalho BF, Lourenço GJ, Calonga L, Castilho AM, Chone CT, Lima CSP. Association of Clinical Aspects and Genetic Variants with the Severity of Cisplatin-Induced Ototoxicity in Head and Neck Squamous Cell Carcinoma: A Prospective Cohort Study. Cancers. 2023; 15(6):1759. https://doi.org/10.3390/cancers15061759
Chicago/Turabian StyleMacedo, Ligia Traldi, Ericka Francislaine Dias Costa, Bruna Fernandes Carvalho, Gustavo Jacob Lourenço, Luciane Calonga, Arthur Menino Castilho, Carlos Takahiro Chone, and Carmen Silvia Passos Lima. 2023. "Association of Clinical Aspects and Genetic Variants with the Severity of Cisplatin-Induced Ototoxicity in Head and Neck Squamous Cell Carcinoma: A Prospective Cohort Study" Cancers 15, no. 6: 1759. https://doi.org/10.3390/cancers15061759
APA StyleMacedo, L. T., Costa, E. F. D., Carvalho, B. F., Lourenço, G. J., Calonga, L., Castilho, A. M., Chone, C. T., & Lima, C. S. P. (2023). Association of Clinical Aspects and Genetic Variants with the Severity of Cisplatin-Induced Ototoxicity in Head and Neck Squamous Cell Carcinoma: A Prospective Cohort Study. Cancers, 15(6), 1759. https://doi.org/10.3390/cancers15061759