MUG CCArly: A Novel Autologous 3D Cholangiocarcinoma Model Presents an Increased Angiogenic Potential
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient History
2.2. Cell Culture
2.3. 3D Cell Culture
2.4. Immunohistochemistry
2.5. Cell Line Identification by Short Tandem Repeat (STR) Analysis
2.6. Telomerase Activity
2.7. Tumorigenicity
2.8. Ultrasound Imaging
2.9. Protein Isolation of 3D Cultures
2.10. Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
2.11. Protein Data Evaluation
2.12. Luminex xMAP Technology
2.13. Statistical Analysis
3. Results
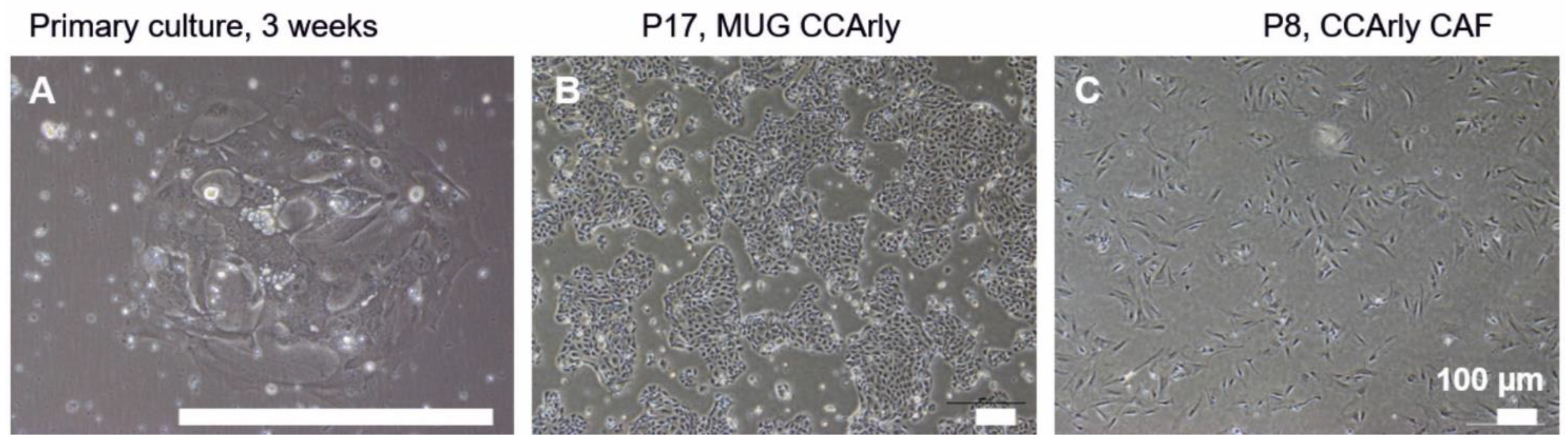
3.1. Establishment and Characterization of the New CCA Cell Line MUG CCArly and Its Autologous hTERT Immortalized CCArly CAF
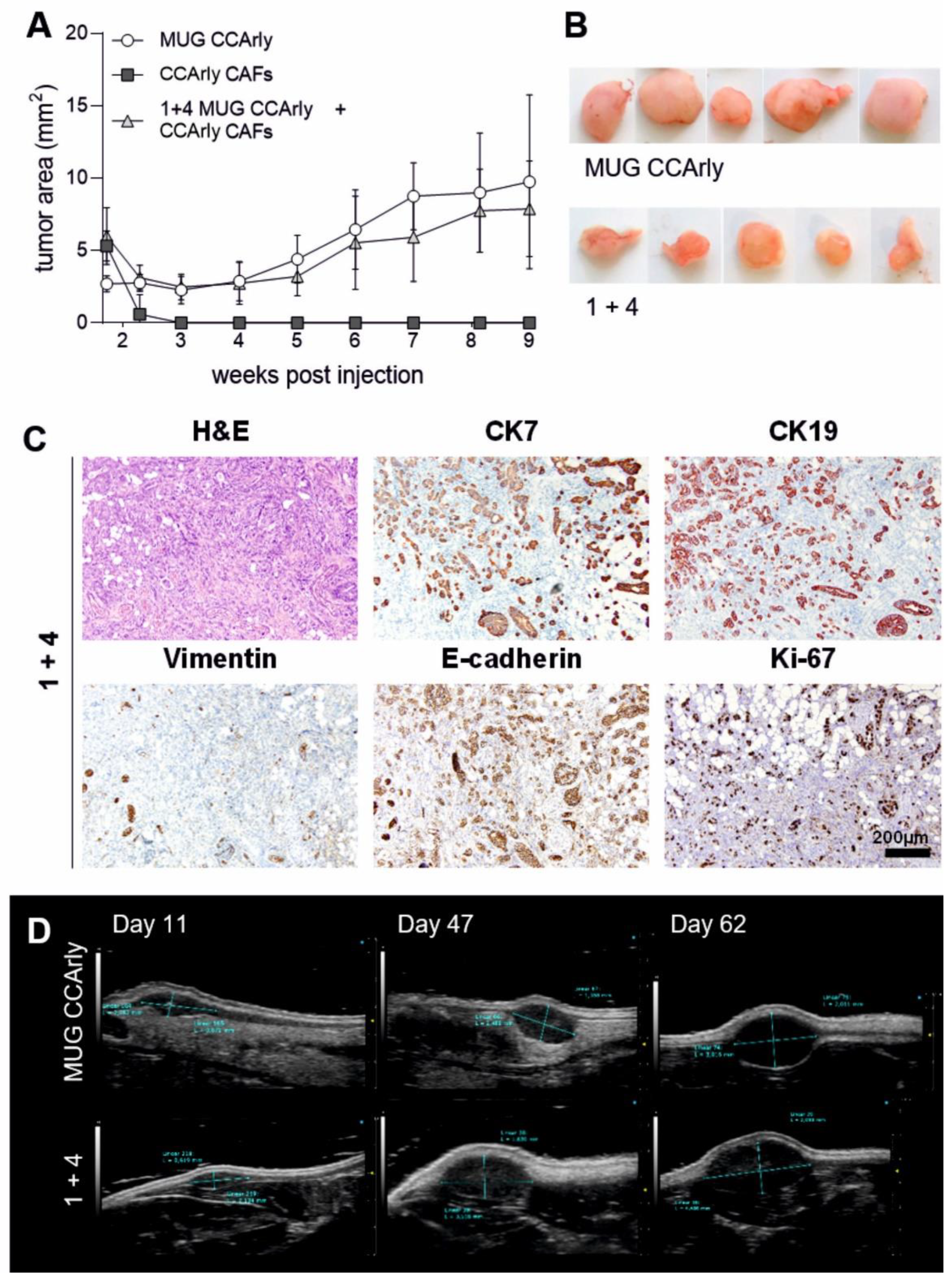
3.2. In Vivo Studies Confirm the Tumorigenic Potential of MUG CCArly
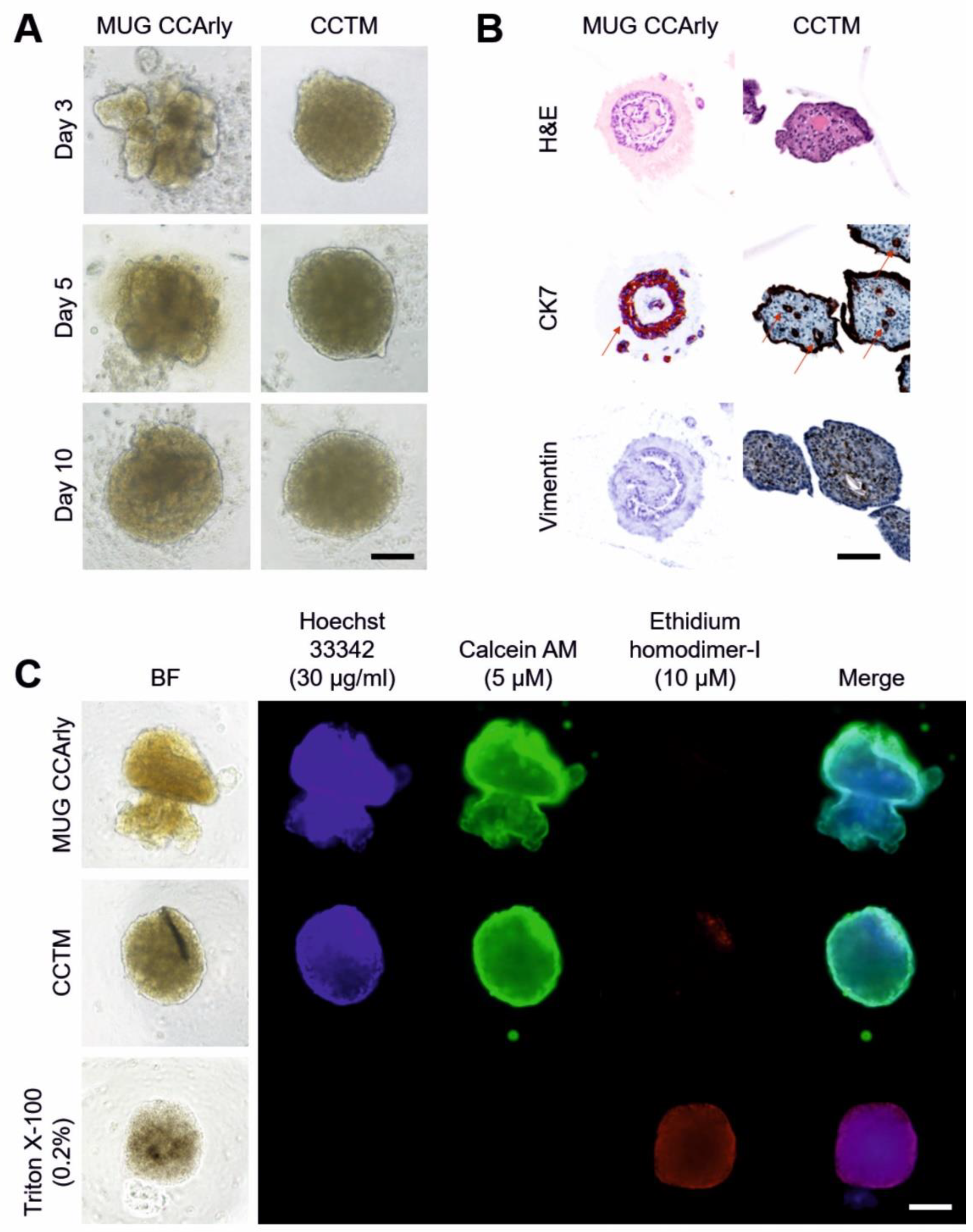
3.3. Quality Assessment of the 3D In Vitro Co-Culture Tumor Model
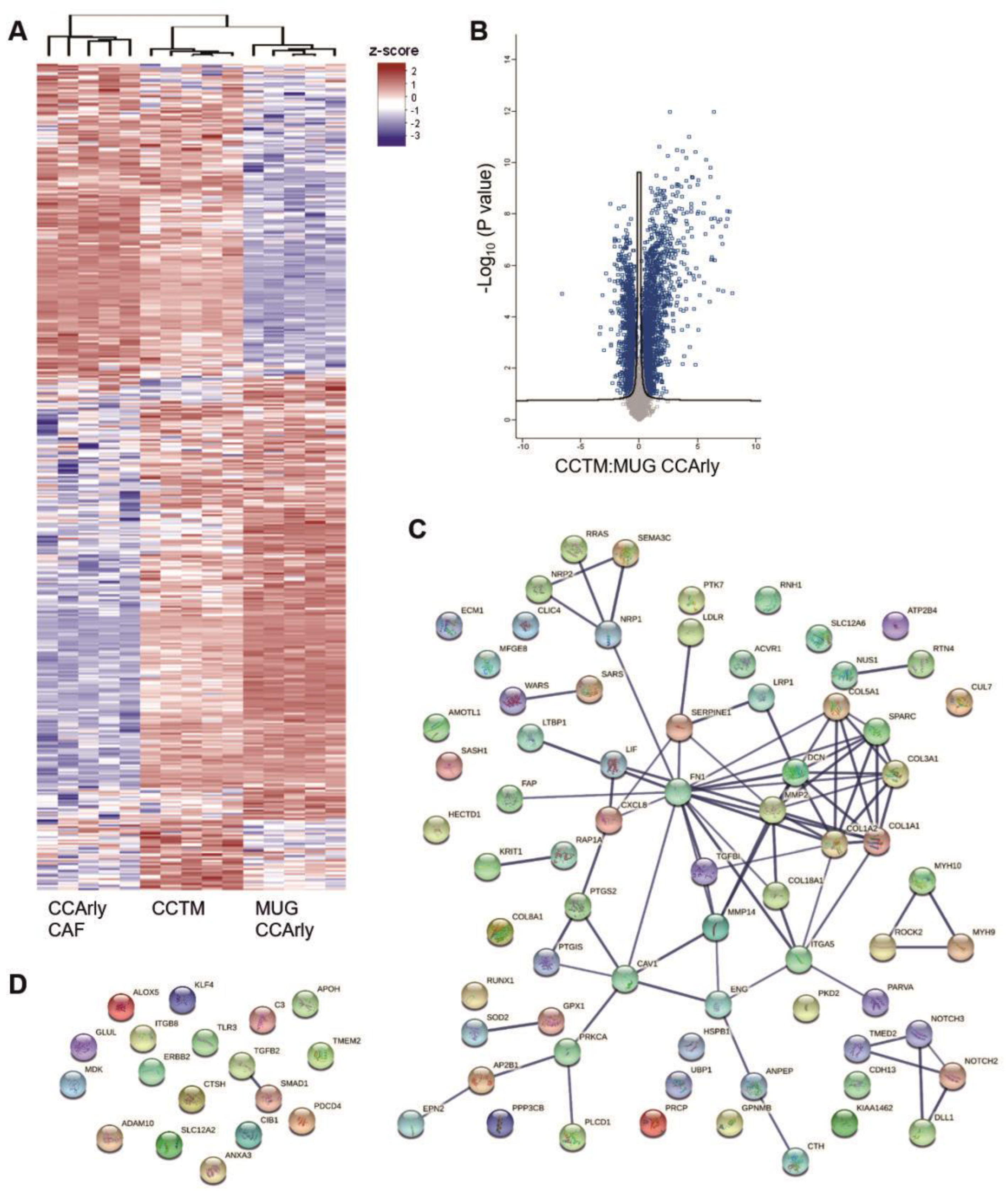
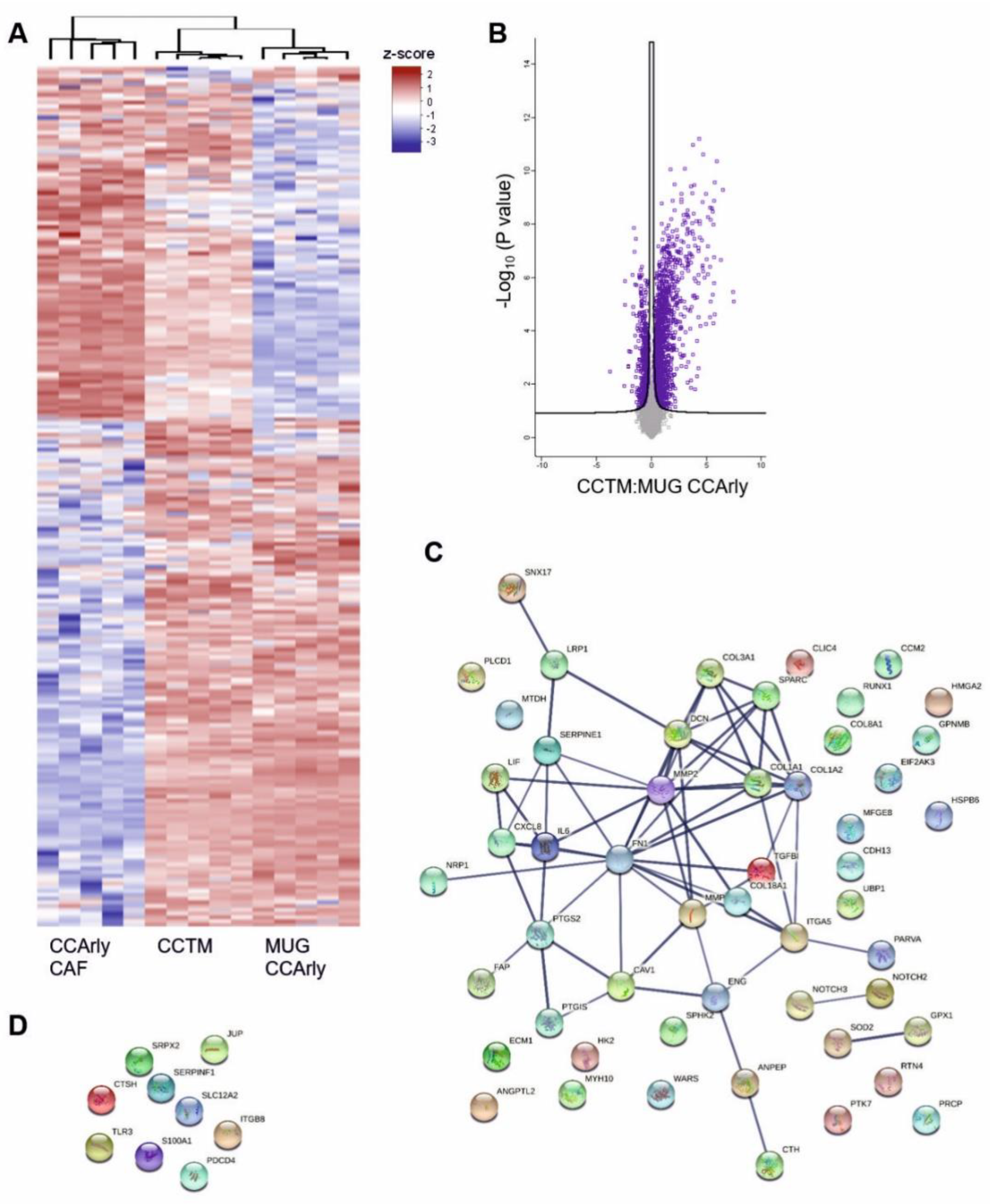
3.4. Mass Spectrometric Analysis of 3D Cultures
3.5. The Angiogenic Proteome Is Enriched Particularly in the CCTM
3.6. CCArly CAF Contribute to An Angiogenic TME
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BF | Bright field |
| BMWFW | Austrian Federal Ministry of Science and Research |
| CA19.9 | Carbohydrate antigen 19.9 |
| CAF | Cancer-associated fibroblasts |
| CCA/iCCA/pCCA/dCCA | Cholangiocarcinoma/intrahepatic/perihilar/distal CCA |
| CCTM | Co-culture tumor model |
| CK | Cytokeratin |
| DC | Detergent compatible |
| FDR | False discovery rate |
| FFPE | Fixed in 4% formalin and embedded in paraffin |
| FN1 | Fibronectin |
| HF-US | High-frequency ultrasound imaging |
| hTERT | Human telomerase reverse transcriptase |
| LIF | Leukemia inhibitory factor |
| PBMC | Peripheral blood mononuclear cells |
| PCA | Principal component analysis |
| PIC | Protease-inhibitor cocktail |
| STR | Short tandem repeat analysis |
| TAM | Tumor-associated macrophage |
| TME | Tumor microenvironment |
| TNC | Tenascin C |
| TRAP | Telomerase-repeated amplification protocol |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Wiggers, J.K.; Ruys, A.T.; Groot Koerkamp, B.; Beuers, U.; ten Kate, F.J.; van Gulik, T.M. Differences in immunohistochemical biomarkers between intra- and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 2014, 29, 1582–1594. [Google Scholar] [CrossRef] [PubMed]
- Reicher, A.; Harris, A.L.; Prinz, F.; Kiesslich, T.; Wei, M.; Öllinger, R.; Rad, R.; Pichler, M.; Kwong, L.N. Generation of An Endogenous FGFR2-BICC1 Gene Fusion/58 Megabase Inversion Using Single-Plasmid CRISPR/Cas9 Editing in Biliary Cells. Int. J. Mol. Sci. 2020, 21, 2460. [Google Scholar] [CrossRef] [PubMed]
- Massa, A.; Varamo, C.; Vita, F.; Tavolari, S.; Peraldo-Neia, C.; Brandi, G.; Rizzo, A.; Cavalloni, G.; Aglietta, M. Evolution of the Experimental Models of Cholangiocarcinoma. Cancers 2020, 12, 2308. [Google Scholar] [CrossRef]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef]
- Zabron, A.; Edwards, R.J.; Khan, S.A. The challenge of cholangiocarcinoma: Dissecting the molecular mechanisms of an insidious cancer. Dis. Model. Mech. 2013, 6, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.W.; Borbath, I.; Khan, S.A.; Huguet, F.; Gruenberger, T.; Arnold, D. Biliary cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, v28–v37. [Google Scholar] [CrossRef]
- Oh, D.Y.; Lee, K.H.; Lee, D.W.; Yoon, J.; Kim, T.Y.; Bang, J.H.; Nam, A.R.; Oh, K.S.; Kim, J.M.; Lee, Y.; et al. Gemcitabine and cisplatin plus durvalumab with or without tremelimumab in chemotherapy-naive patients with advanced biliary tract cancer: An open-label, single-centre, phase 2 study. Lancet Gastroenterol. Hepatol. 2022, 7, 522–532. [Google Scholar] [CrossRef]
- Wu, H.J.; Chu, P.Y. Role of Cancer Stem Cells in Cholangiocarcinoma and Therapeutic Implications. Int. J. Mol. Sci. 2019, 20, 4154. [Google Scholar] [CrossRef]
- Sha, M.; Jeong, S.; Qiu, B.J.; Tong, Y.; Xia, L.; Xu, N.; Zhang, J.J.; Xia, Q. Isolation of cancer-associated fibroblasts and its promotion to the progression of intrahepatic cholangiocarcinoma. Cancer Med. 2018, 7, 4665–4677. [Google Scholar] [CrossRef]
- Vaquero, J.; Guedj, N.; Clapéron, A.; Nguyen Ho-Bouldoires, T.H.; Paradis, V.; Fouassier, L. Epithelial-mesenchymal transition in cholangiocarcinoma: From clinical evidence to regulatory networks. J. Hepatol. 2017, 66, 424–441. [Google Scholar] [CrossRef] [PubMed]
- Affo, S.; Yu, L.X.; Schwabe, R.F. The Role of Cancer-Associated Fibroblasts and Fibrosis in Liver Cancer. Annu. Rev. Pathol. 2017, 12, 153–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhou, L.; Li, D.; Andl, T.; Zhang, Y. Cancer-Associated Fibroblasts Build and Secure the Tumor Microenvironment. Front. Cell Dev. Biol. 2019, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Posch, F.; Prinz, F.; Balihodzic, A.; Mayr, C.; Kiesslich, T.; Klec, C.; Jonas, K.; Barth, D.A.; Riedl, J.M.; Gerger, A.; et al. MiR-200c-3p Modulates Cisplatin Resistance in Biliary Tract Cancer by ZEB1-Independent Mechanisms. Cancers 2021, 13, 3996. [Google Scholar] [CrossRef]
- Muciño-Olmos, E.A.; Vázquez-Jiménez, A.; Avila-Ponce de León, U.; Matadamas-Guzman, M.; Maldonado, V.; López-Santaella, T.; Hernández-Hernández, A.; Resendis-Antonio, O. Unveiling functional heterogeneity in breast cancer multicellular tumor spheroids through single-cell RNA-seq. Sci. Rep. 2020, 10, 12728. [Google Scholar] [CrossRef] [PubMed]
- Novotný, J.; Strnadová, K.; Dvořánková, B.; Kocourková, Š.; Jakša, R.; Dundr, P.; Pačes, V.; Smetana, K., Jr.; Kolář, M.; Lacina, L. Single-Cell RNA Sequencing Unravels Heterogeneity of the Stromal Niche in Cutaneous Melanoma Heterogeneous Spheroids. Cancers 2020, 12, 3324. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.S.; Barros, A.S.; Costa, E.C.; Moreira, A.F.; Correia, I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019, 116, 206–226. [Google Scholar] [CrossRef]
- Maia, A.; Wiemann, S. Cancer-Associated Fibroblasts: Implications for Cancer Therapy. Cancers 2021, 13, 3526. [Google Scholar] [CrossRef]
- Ludlow, A.T.; Shelton, D.; Wright, W.E.; Shay, J.W. ddTRAP: A Method for Sensitive and Precise Quantification of Telomerase Activity. Methods Mol. Biol. 2018, 1768, 513–529. [Google Scholar] [CrossRef]
- Mender, I.; Shay, J.W. Telomerase Repeated Amplification Protocol (TRAP). Bio-Protocol 2015, 5, e1657. [Google Scholar] [CrossRef]
- Meier, F.; Brunner, A.D.; Frank, M.; Ha, A.; Bludau, I.; Voytik, E.; Kaspar-Schoenefeld, S.; Lubeck, M.; Raether, O.; Bache, N.; et al. diaPASEF: Parallel accumulation-serial fragmentation combined with data-independent acquisition. Nat. Methods 2020, 17, 1229–1236. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Demichev, V.; Yu, F.; Teo, G.C.; Szyrwiel, L.; Rosenberger, G.A.; Decker, J.; Kaspar-Schoenefeld, S.; Lilley, K.S.; Mülleder, M.; Nesvizhskii, A.I.; et al. High sensitivity dia-PASEF proteomics with DIA-NN and FragPipe. bioRxiv 2021. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Ross, J.S.; Linette, G.P.; Stec, J.; Ross, M.S.; Anwar, S.; Boguniewicz, A. DNA ploidy and cell cycle analysis in breast cancer. Am. J. Clin. Pathol. 2003, 120, S72–S84. [Google Scholar] [CrossRef]
- Malenica, I.; Donadon, M.; Lleo, A. Molecular and Immunological Characterization of Biliary Tract Cancers: A Paradigm Shift Towards a Personalized Medicine. Cancers 2020, 12, 2190. [Google Scholar] [CrossRef]
- Zach, S.; Birgin, E.; Rückert, F. Primary cholangiocellular carcinoma cell lines. J. Stem Cell Res. Transplant. 2015, 2, 1013. [Google Scholar]
- Sirica, A.E. The role of cancer-associated myofibroblasts in intrahepatic cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011, 9, 44–54. [Google Scholar] [CrossRef]
- Fiori, M.E.; Di Franco, S.; Villanova, L.; Bianca, P.; Stassi, G.; De Maria, R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol. Cancer 2019, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Thongchot, S.; Ferraresi, A.; Vidoni, C.; Loilome, W.; Yongvanit, P.; Namwat, N.; Isidoro, C. Resveratrol interrupts the pro-invasive communication between cancer associated fibroblasts and cholangiocarcinoma cells. Cancer Lett. 2018, 430, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, J.; Aoudjehane, L.; Fouassier, L. Cancer-associated fibroblasts in cholangiocarcinoma. Curr. Opin. Gastroenterol. 2020, 36, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, P.; Wang, L.; Li, M.; Ge, Z.; Noordam, L.; Lieshout, R.; Verstegen, M.M.A.; Ma, B.; Su, J.; et al. Cancer-Associated Fibroblasts Provide a Stromal Niche for Liver Cancer Organoids That Confers Trophic Effects and Therapy Resistance. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 407–431. [Google Scholar] [CrossRef]
- Nii, T.; Makino, K.; Tabata, Y. A Cancer Invasion Model Combined with Cancer-Associated Fibroblasts Aggregates Incorporating Gelatin Hydrogel Microspheres Containing a p53 Inhibitor. Tissue Eng. Part C Methods 2019, 25, 711–720. [Google Scholar] [CrossRef]
- Arora, L.; Kalia, M.; Dasgupta, S.; Singh, N.; Verma, A.K.; Pal, D. Development of a Multicellular 3D Tumor Model to Study Cellular Heterogeneity and Plasticity in NSCLC Tumor Microenvironment. Front. Oncol. 2022, 12, 881207. [Google Scholar] [CrossRef]
- Alzeeb, G.; Dubreuil, M.; Arzur, D.; Rivet, S.; Corcos, L.; Grand, Y.L.; Le Jossic-Corcos, C. Gastric cancer multicellular spheroid analysis by two-photon microscopy. Biomed. Opt. Express 2022, 13, 3120–3130. [Google Scholar] [CrossRef]
- Gao, Q.; Yang, Z.; Xu, S.; Li, X.; Yang, X.; Jin, P.; Liu, Y.; Zhou, X.; Zhang, T.; Gong, C.; et al. Heterotypic CAF-tumor spheroids promote early peritoneal metastasis of ovarian cancer. J. Exp. Med. 2019, 216, 688–703. [Google Scholar] [CrossRef]
- Leung, B.M.; Lesher-Perez, S.C.; Matsuoka, T.; Moraes, C.; Takayama, S. Media additives to promote spheroid circularity and compactness in hanging drop platform. Biomater. Sci. 2015, 3, 336–344. [Google Scholar] [CrossRef]
- Affo, S.; Nair, A.; Brundu, F.; Ravichandra, A.; Bhattacharjee, S.; Matsuda, M.; Chin, L.; Filliol, A.; Wen, W.; Song, X.; et al. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell 2021, 39, 866–882.e811. [Google Scholar] [CrossRef]
- Campbell, D.J.; Dumur, C.I.; Lamour, N.F.; Dewitt, J.L.; Sirica, A.E. Novel organotypic culture model of cholangiocarcinoma progression. Hepatol. Res. Off. J. Jpn. Soc. Hepatol. 2012, 42, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.A.; Pedram, K.; Malaker, S.A.; Batista, P.J.; Smith, B.A.H.; Johnson, A.G.; George, B.M.; Majzoub, K.; Villalta, P.W.; Carette, J.E.; et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 2021, 184, 3109–3124.e3122. [Google Scholar] [CrossRef] [PubMed]
- Nachtergaele, S.; Krishnan, Y. New Vistas for Cell-Surface GlycoRNAs. N. Engl. J. Med. 2021, 385, 658–660. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, G.; Correia, V.G.; Palma, A.S.; Chai, W.; Li, C.; Feizi, T.; Martin, E.; Laux, B.; Franz, A.; Fuchs, K.; et al. Siglec-15 recognition of sialoglycans on tumor cell lines can occur independently of sialyl Tn antigen expression. Glycobiology 2021, 31, 44–54. [Google Scholar] [CrossRef]
- Roy, S.; Glaser, S.; Chakraborty, S. Inflammation and Progression of Cholangiocarcinoma: Role of Angiogenic and Lymphangiogenic Mechanisms. Front. Med. 2019, 6, 293. [Google Scholar] [CrossRef]
- Mancinelli, R.; Mammola, C.L.; Sferra, R.; Pompili, S.; Vetuschi, A.; Pannarale, L. Role of the angiogenic factors in cholangiocarcinoma. Appl. Sci. 2019, 9, 1393. [Google Scholar] [CrossRef]
- Rizvi, S.; Gores, G.J. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology 2013, 145, 1215–1229. [Google Scholar] [CrossRef]
- Labib, P.L.; Goodchild, G.; Pereira, S.P. Molecular Pathogenesis of Cholangiocarcinoma. BMC Cancer 2019, 19, 185. [Google Scholar] [CrossRef]
- Brindley, P.J.; Bachini, M.; Ilyas, S.I.; Khan, S.A.; Loukas, A.; Sirica, A.E.; Teh, B.T.; Wongkham, S.; Gores, G.J. Cholangiocarcinoma. Nat. Rev. Dis. Prim. 2021, 7, 65. [Google Scholar] [CrossRef]
- Kleinegger, F.; Hofer, E.; Wodlej, C.; Golob-Schwarzl, N.; Birkl-Toeglhofer, A.M.; Stallinger, A.; Petzold, J.; Orlova, A.; Krassnig, S.; Reihs, R.; et al. Pharmacologic IL-6Rα inhibition in cholangiocarcinoma promotes cancer cell growth and survival. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 308–321. [Google Scholar] [CrossRef]
- Sun, Q.; Li, F.; Sun, F.; Niu, J. Interleukin-8 is a prognostic indicator in human hilar cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 8376–8384. [Google Scholar]
- Bakouny, Z.; Choueiri, T.K. IL-8 and cancer prognosis on immunotherapy. Nat. Med. 2020, 26, 650–651. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, C.; Sanmamed, M.F.; Rodríguez-Ruiz, M.E.; Teijeira, Á.; Oñate, C.; González, Á.; Ponz, M.; Schalper, K.A.; Pérez-Gracia, J.L.; Melero, I. Interleukin-8 in cancer pathogenesis, treatment and follow-up. Cancer Treat. Rev. 2017, 60, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Chen, J.J.; Yao, P.L.; Yang, P.C. The role of interleukin-8 in cancer cells and microenvironment interaction. Front. Biosci. A J. Virtual Libr. 2005, 10, 853–865. [Google Scholar] [CrossRef]
- Thongchot, S.; Jamjuntra, P.; Therasakvichya, S.; Warnnissorn, M.; Ferraresi, A.; Thuwajit, P.; Isidoro, C.; Thuwajit, C. Interleukin-8 released by cancer-associated fibroblasts attenuates the autophagy and promotes the migration of ovarian cancer cells. Int. J. Oncol. 2021, 58, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Shen, J.; Xie, G.; Wu, J.; He, M.; Gao, L.; Zhang, Y.; Yao, X.; Shen, L. Cancer-associated fibroblasts-derived IL-8 mediates resistance to cisplatin in human gastric cancer. Cancer Lett. 2019, 454, 37–43. [Google Scholar] [CrossRef]
- You, Z.; Bei, L.; Cheng, L.P.; Cheng, N.S. Expression of COX-2 and VEGF-C in cholangiocarcinomas at different clinical and pathological stages. Genet. Mol. Res. GMR 2015, 14, 6239–6246. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, C.; Lu, S.; Xu, Y.; Li, Z.; Jiang, H.; Ma, Y. Tumor-associated macrophages in cholangiocarcinoma: Complex interplay and potential therapeutic target. EBioMedicine 2021, 67, 103375. [Google Scholar] [CrossRef]
- Subimerb, C.; Pinlaor, S.; Khuntikeo, N.; Leelayuwat, C.; Morris, A.; McGrath, M.S.; Wongkham, S. Tissue invasive macrophage density is correlated with prognosis in cholangiocarcinoma. Mol. Med. Rep. 2010, 3, 597–605. [Google Scholar] [CrossRef]
- Lowy, C.M.; Oskarsson, T. Tenascin C in metastasis: A view from the invasive front. Cell Adhes. Migr. 2015, 9, 112–124. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, H.; Feng, Y.; Ye, Q.; Hu, J.; Guo, Y.; Feng, Y. Pan-Cancer Analysis of the Associations of TGFBI Expression With Prognosis and Immune Characteristics. Front. Mol. Biosci. 2021, 8, 745649. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, J.; Sim, W.; Kim, J.H. Soluble TGFBI aggravates the malignancy of cholangiocarcinoma through activation of the ITGB1 dependent PPARγ signalling pathway. Cell. Oncol. 2022, 45, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.P.; Zhang, J.X.; Gu, S.P.; Wu, Y.B.; Liu, J.F. Overexpression of ECM1 contributes to migration and invasion in cholangiocarcinoma cell. Neoplasma 2012, 59, 409–415. [Google Scholar] [CrossRef]
- Binder, B.R.; Mihaly, J. The plasminogen activator inhibitor “paradox” in cancer. Immunol. Lett. 2008, 118, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Lv, Q.; Zhao, Y.; Liu, C.; Sun, Y.; Xi, K.; Xiao, J.; Li, C. Wnt/β-catenin pathway is required for epithelial to mesenchymal transition in CXCL12 over expressed breast cancer cells. Int. J. Clin. Exp. Pathol. 2015, 8, 12357–12367. [Google Scholar]
- Leelawat, K.; Leelawat, S.; Tepaksorn, P.; Rattanasinganchan, P.; Leungchaweng, A.; Tohtong, R.; Sobhon, P. Involvement of c-Met/hepatocyte growth factor pathway in cholangiocarcinoma cell invasion and its therapeutic inhibition with small interfering RNA specific for c-Met. J. Surg. Res. 2006, 136, 78–84. [Google Scholar] [CrossRef]
- Naka, D.; Shimomura, T.; Yoshiyama, Y.; Sato, M.; Sato, M.; Ishii, T.; Hara, H. Internalization and degradation of hepatocyte growth factor in hepatocytes with down-regulation of the receptor/c-Met. FEBS Lett. 1993, 329, 147–152. [Google Scholar] [CrossRef]
- Viticchiè, G.; Muller, P.A.J. c-Met and Other Cell Surface Molecules: Interaction, Activation and Functional Consequences. Biomedicines 2015, 3, 46–70. [Google Scholar] [CrossRef]
- Kimura, T.; Sakisaka, T.; Baba, T.; Yamada, T.; Takai, Y. Involvement of the Ras-Ras-activated Rab5 guanine nucleotide exchange factor RIN2-Rab5 pathway in the hepatocyte growth factor-induced endocytosis of E-cadherin. J. Biol. Chem. 2006, 281, 10598–10609. [Google Scholar] [CrossRef]
- Chatterjee, M.; Seizer, P.; Borst, O.; Schönberger, T.; Mack, A.; Geisler, T.; Langer, H.F.; May, A.E.; Vogel, S.; Lang, F.; et al. SDF-1α induces differential trafficking of CXCR4-CXCR7 involving cyclophilin A, CXCR7 ubiquitination and promotes platelet survival. FASEB J. 2014, 28, 2864–2878. [Google Scholar] [CrossRef]
- Boldajipour, B.; Mahabaleshwar, H.; Kardash, E.; Reichman-Fried, M.; Blaser, H.; Minina, S.; Wilson, D.; Xu, Q.; Raz, E. Control of chemokine-guided cell migration by ligand sequestration. Cell 2008, 132, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Yang, S.; Ingle, G.; Zlot, C.; Rangell, L.; Kowalski, J.; Schwall, R.; Ferrara, N.; Gerritsen, M.E. Hepatocyte growth factor enhances vascular endothelial growth factor-induced angiogenesis in vitro and in vivo. Am. J. Pathol. 2001, 158, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Nagy, J.A.; Vasile, E.; Feng, D.; Sundberg, C.; Brown, L.F.; Detmar, M.J.; Lawitts, J.A.; Benjamin, L.; Tan, X.; Manseau, E.J.; et al. Vascular permeability factor/vascular endothelial growth factor induces lymphangiogenesis as well as angiogenesis. J. Exp. Med. 2002, 196, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Bag, A.K.; Singh, R.K.; Talmadge, J.E.; Batra, S.K.; Datta, K. Multifaceted Role of Neuropilins in the Immune System: Potential Targets for Immunotherapy. Front. Immunol. 2017, 8, 1228. [Google Scholar] [CrossRef] [PubMed]
- Morton, S.D.; Cadamuro, M.; Brivio, S.; Vismara, M.; Stecca, T.; Massani, M.; Bassi, N.; Furlanetto, A.; Joplin, R.E.; Floreani, A.; et al. Leukemia inhibitory factor protects cholangiocarcinoma cells from drug-induced apoptosis via a PI3K/AKT-dependent Mcl-1 activation. Oncotarget 2015, 6, 26052–26064. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.C.; Silva, D.N.; Fortuna, V.; Silveira, B.M.; Orge, I.D.; de Santana, T.A.; Sampaio, G.L.; Paredes, B.D.; Ribeiro-Dos-Santos, R.; Soares, M.B.P. Leukemia Inhibitory Factor (LIF) Overexpression Increases the Angiogenic Potential of Bone Marrow Mesenchymal Stem/Stromal Cells. Front. Cell Dev. Biol. 2020, 8, 778. [Google Scholar] [CrossRef]
- Zoetemelk, M.; Rausch, M.; Colin, D.J.; Dormond, O.; Nowak-Sliwinska, P. Short-term 3D culture systems of various complexity for treatment optimization of colorectal carcinoma. Sci. Rep. 2019, 9, 7103. [Google Scholar] [CrossRef]
- Kuo, C.T.; Wang, J.Y.; Lin, Y.F.; Wo, A.M.; Chen, B.P.C.; Lee, H. Three-dimensional spheroid culture targeting versatile tissue bioassays using a PDMS-based hanging drop array. Sci. Rep. 2017, 7, 4363. [Google Scholar] [CrossRef]
- Gopal, S.; Kwon, S.J.; Ku, B.; Lee, D.W.; Kim, J.; Dordick, J.S. 3D tumor spheroid microarray for high-throughput, high-content natural killer cell-mediated cytotoxicity. Commun. Biol. 2021, 4, 893. [Google Scholar] [CrossRef]


| Day Three | Day Seven | |||||
|---|---|---|---|---|---|---|
| Fold Change | ||||||
| Protein ID | Protein Name | Gene | CCTM: MUG CCArly | CCTM: CCArly CAF | CCTM: MUG CCArly | CCTM: CCArly CAF |
| P05121 | PAI1 | SERPINE1 | 187.4341 | 2.9192 | 30.8141 | 2.2039 |
| P35354 | COX-2 | PTGS2 | 172.0948 | 5.7055 | 42.3465 | 5.7281 |
| P10145 | IL8 | CXCL8 | 109.7289 | 26.4974 | 30.8723 | 18.0813 |
| P15018 | LIF | LIF | 71.3025 | 41.38 | 19.1683 | 12.0751 |
| Q15582 | BGH3 | TGFBI | 22.3808 | 3.1987 | 19.4534 | 2.7792 |
| Q16610 | ECM1 | ECM1 | 9.7985 | 2.6402 | 2.8068 | 2.6888 |
| O60462 | NRP2 | NRP2 | 2.179 | 2.2517 | <2 | 2.5282 |
| P05231 | IL6 | IL6 | - | - | 47.3197 | 25.7217 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schrom, S.; Kleinegger, F.; Anders, I.; Hebesberger, T.; Karner, C.; Liesinger, L.; Birner-Gruenberger, R.; Renner, W.; Pichler, M.; Grillari, R.; et al. MUG CCArly: A Novel Autologous 3D Cholangiocarcinoma Model Presents an Increased Angiogenic Potential. Cancers 2023, 15, 1757. https://doi.org/10.3390/cancers15061757
Schrom S, Kleinegger F, Anders I, Hebesberger T, Karner C, Liesinger L, Birner-Gruenberger R, Renner W, Pichler M, Grillari R, et al. MUG CCArly: A Novel Autologous 3D Cholangiocarcinoma Model Presents an Increased Angiogenic Potential. Cancers. 2023; 15(6):1757. https://doi.org/10.3390/cancers15061757
Chicago/Turabian StyleSchrom, Silke, Florian Kleinegger, Ines Anders, Thomas Hebesberger, Christina Karner, Laura Liesinger, Ruth Birner-Gruenberger, Wilfried Renner, Martin Pichler, Regina Grillari, and et al. 2023. "MUG CCArly: A Novel Autologous 3D Cholangiocarcinoma Model Presents an Increased Angiogenic Potential" Cancers 15, no. 6: 1757. https://doi.org/10.3390/cancers15061757
APA StyleSchrom, S., Kleinegger, F., Anders, I., Hebesberger, T., Karner, C., Liesinger, L., Birner-Gruenberger, R., Renner, W., Pichler, M., Grillari, R., Aigelsreiter, A., & Rinner, B. (2023). MUG CCArly: A Novel Autologous 3D Cholangiocarcinoma Model Presents an Increased Angiogenic Potential. Cancers, 15(6), 1757. https://doi.org/10.3390/cancers15061757







