Soluble Guanylate Cyclase β1 Subunit Represses Human Glioblastoma Growth
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and Cell Culture
2.2. Cell Viability Assay
2.3. Colony Formation Assay
2.4. Orthotopic Xenograft Models
2.5. Assay of cGMP in Intact Cells
2.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.7. Western Blot Analysis
2.8. Flow Cytometry
2.9. Confocal Microscopy
2.10. Chromatin Immunoprecipitation (ChIP) Assay
2.11. Dual Luciferase Assay
2.12. Statistical Analysis
3. Results
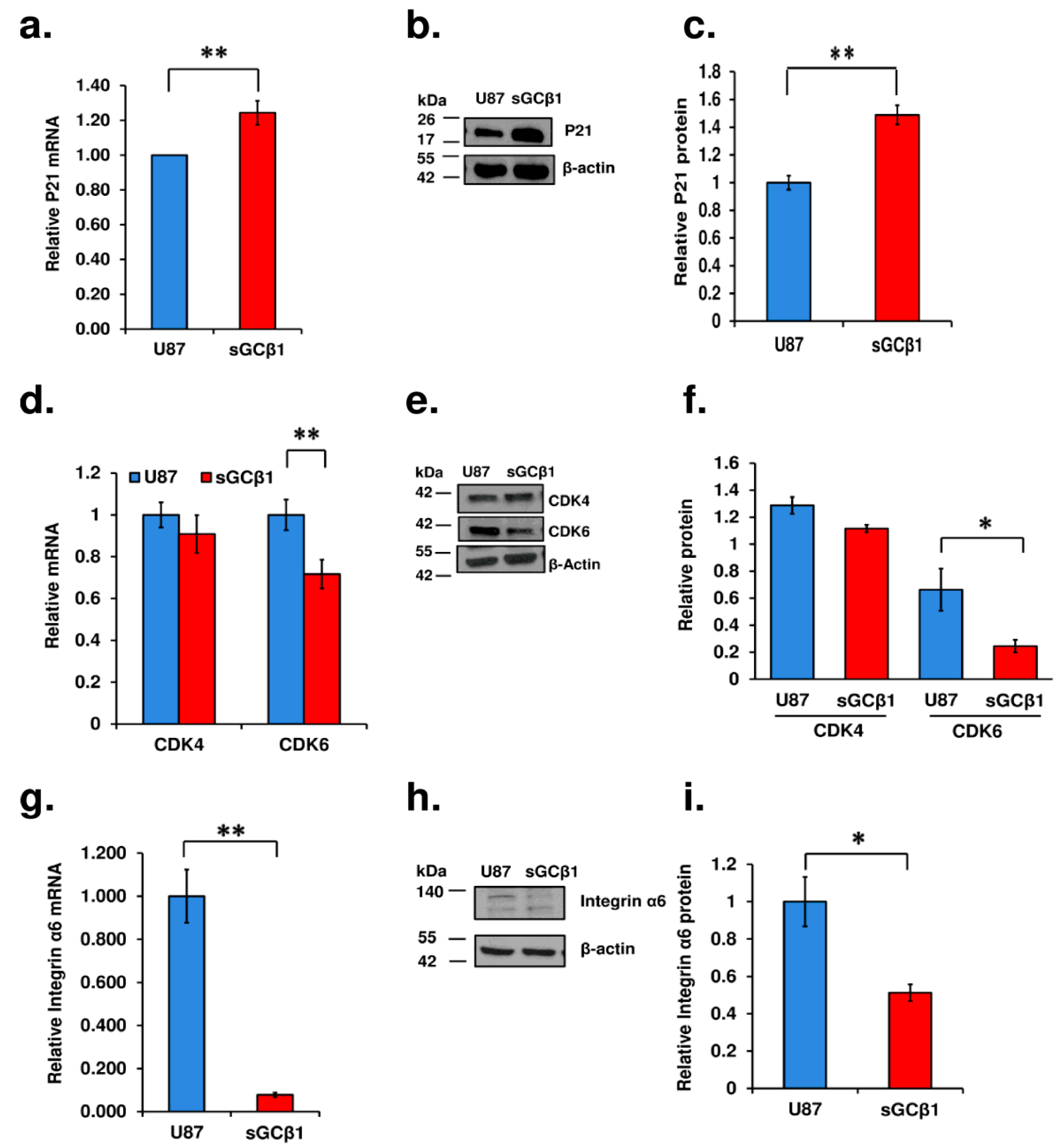
3.1. sGCβ1 Overexpression Represses the Growth of Human Glioblastoma
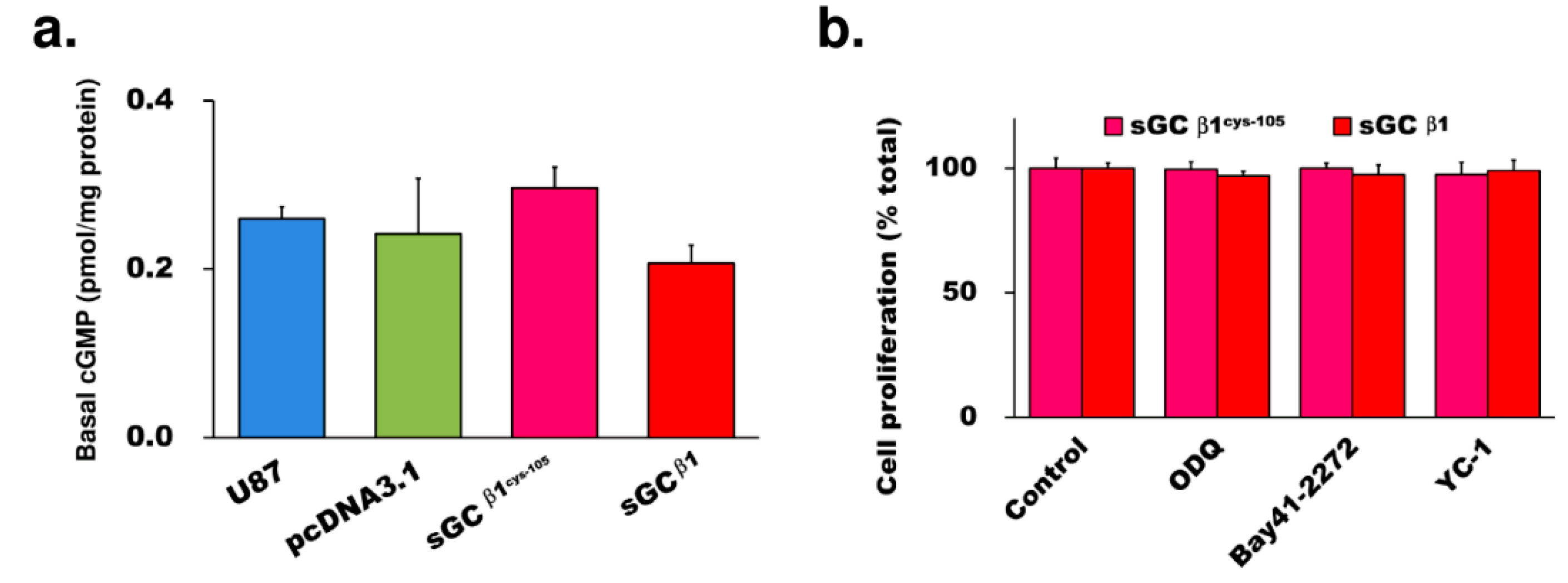
3.2. The Growth Repression by sGCβ1 Overexpression Is cGMP-Independent
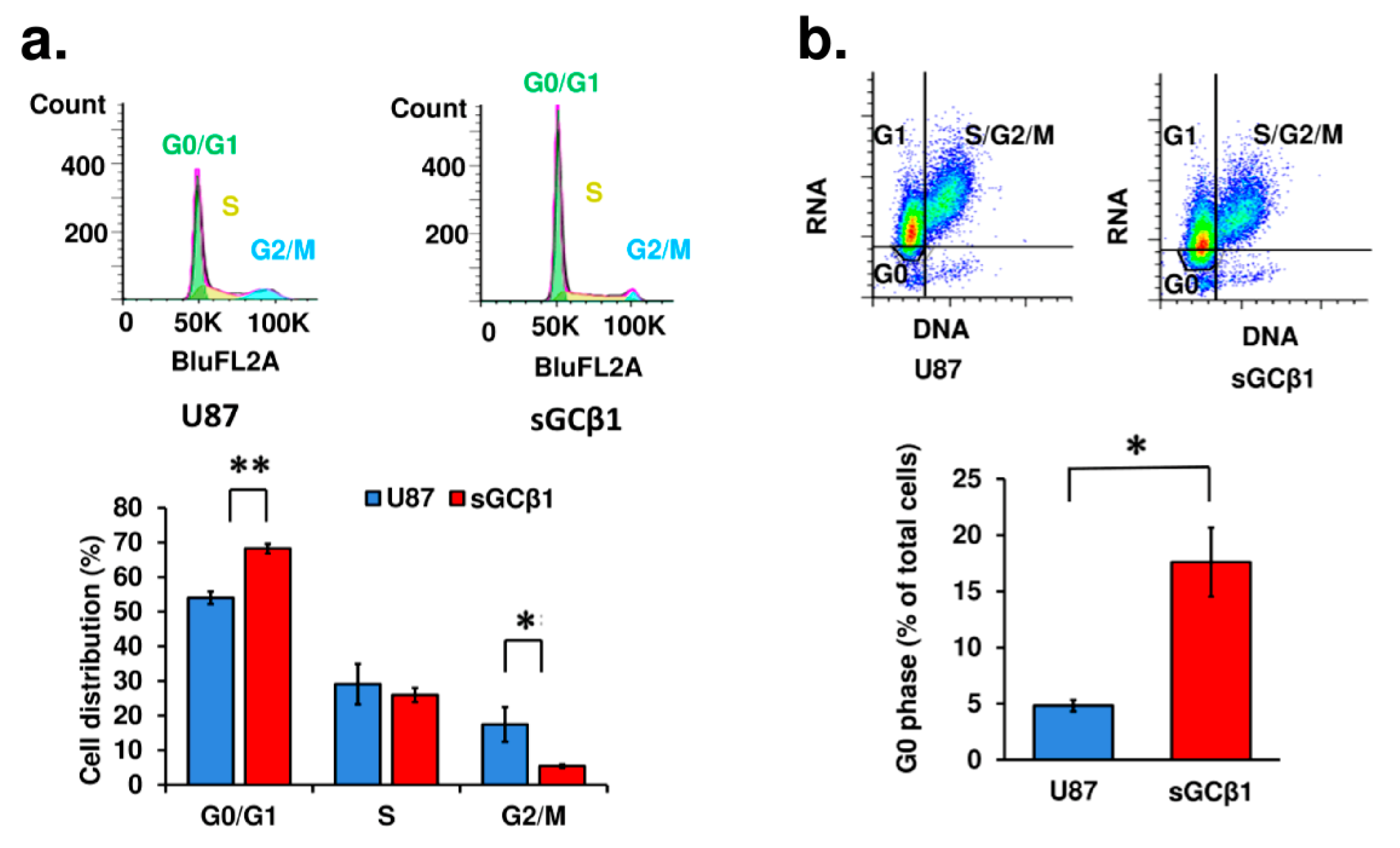
3.3. sGCβ1 Overexpression Induces G0 Phase Arrest of Human Glioblastoma Cells
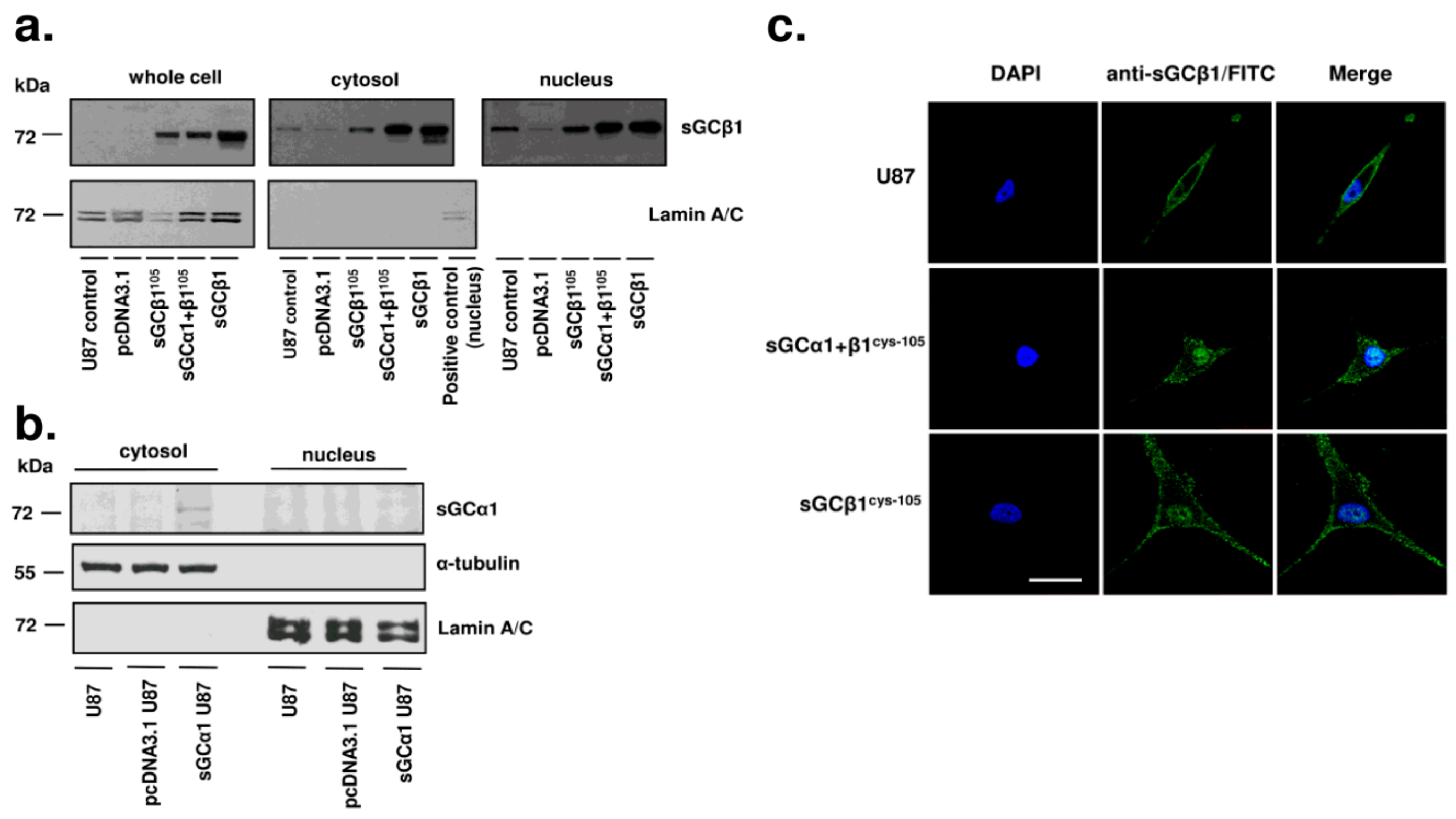
3.4. sGCβ1 Is Localized in the Nucleus in Human Glioblastoma Cells
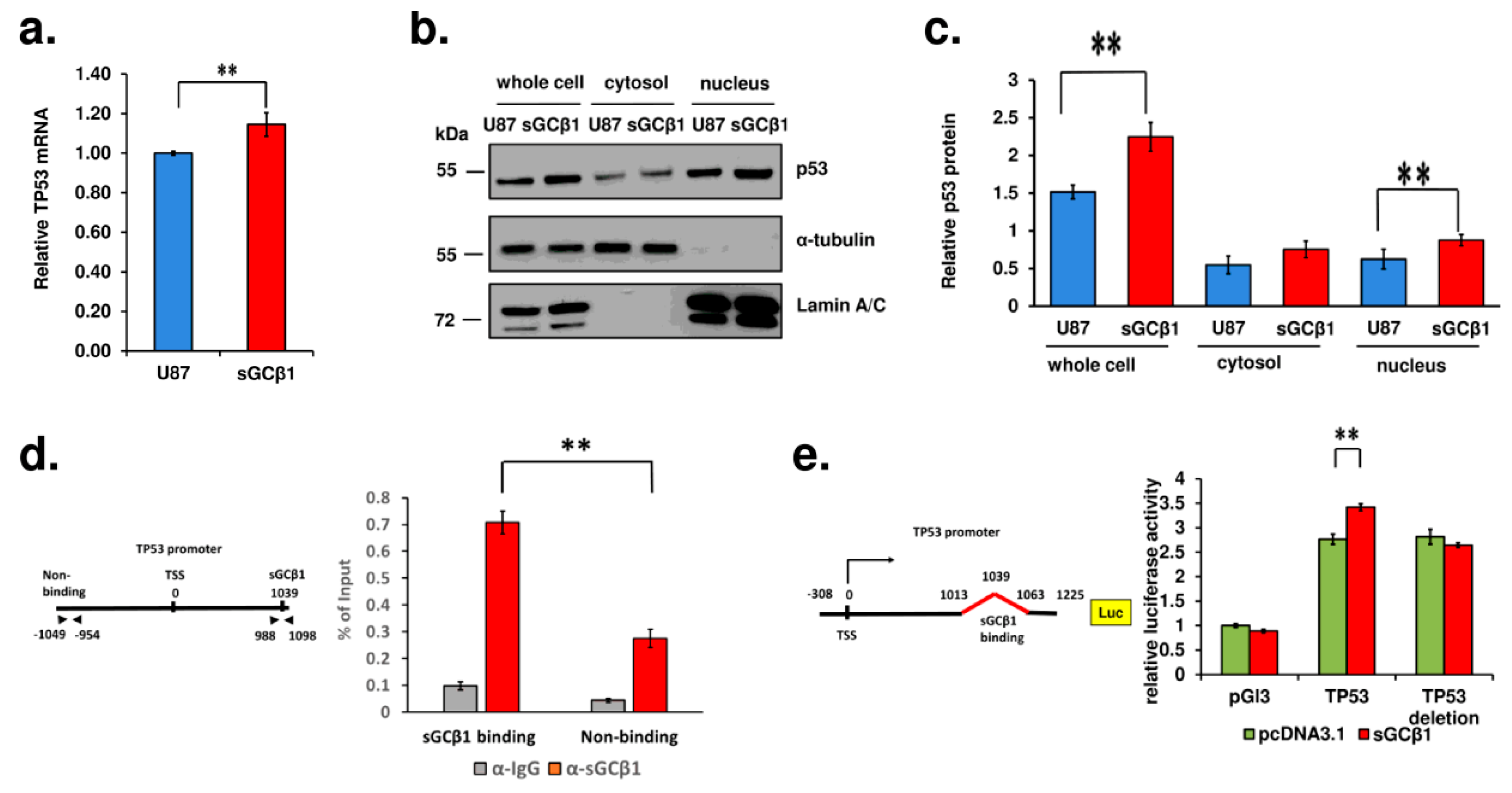
3.5. A Link between sGCβ1 and p53
3.6. sGCβ1 Overexpression Represses Glioblastoma Multiforme Signaling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.K.; Dursa, E.K.; Bossarte, R.M.; Schneiderman, A.I. Trends in brain cancer mortality among U.S. Gulf War veterans: 21 year follow-up. Cancer Epidemiol. 2017, 50, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Bullman, T.A.; Mahan, C.M.; Kang, H.K.; Page, W.F. Mortality in US Army Gulf War veterans exposed to 1991 Khamisiyah chemical munitions destruction. Am. J. Public Health 2005, 95, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.K.; Kang, H.K.; Bullman, T.A.; Wallin, M.T. Neurological mortality among U.S. veterans of the Persian Gulf War: 13-year follow-up. Am. J. Ind. Med. 2009, 52, 663–670. [Google Scholar] [CrossRef]
- Fallahi, P.; Elia, G.; Foddis, R.; Cristaudo, A.; Antonelli, A. High risk of brain tumors in military personnel: A case control study. Clin. Ter. 2017, 168, e376–e379. [Google Scholar] [CrossRef]
- Zhu, H.; Li, J.T.; Zheng, F.; Martin, E.; Kots, A.Y.; Krumenacker, J.S.; Choi, B.K.; McCutcheon, I.E.; Weisbrodt, N.; Bogler, O.; et al. Restoring soluble guanylyl cyclase expression and function blocks the aggressive course of glioma. Mol. Pharm. 2011, 80, 1076–1084. [Google Scholar] [CrossRef]
- Kamisaki, Y.; Saheki, S.; Nakane, M.; Palmieri, J.A.; Kuno, T.; Chang, B.Y.; Waldman, S.A.; Murad, F. Soluble guanylate cyclase from rat lung exists as a heterodimer. J. Biol. Chem. 1986, 261, 7236–7241. [Google Scholar] [CrossRef]
- Nakane, M.; Murad, F. Cloning of guanylyl cyclase isoforms. Adv. Pharm. 1994, 26, 7–18. [Google Scholar] [CrossRef]
- Budworth, J.; Meillerais, S.; Charles, I.; Powell, K. Tissue distribution of the human soluble guanylate cyclases. Biochem. Biophys. Res. Commun. 1999, 263, 696–701. [Google Scholar] [CrossRef]
- Armstrong, P.W.; Pieske, B.; Anstrom, K.J.; Ezekowitz, J.; Hernandez, A.F.; Butler, J.; Lam, C.S.P.; Ponikowski, P.; Voors, A.A.; Jia, G.; et al. Vericiguat in Patients with Heart Failure and Reduced Ejection Fraction. N. Engl. J. Med. 2020, 382, 1883–1893. [Google Scholar] [CrossRef]
- Sandner, P.; Follmann, M.; Becker-Pelster, E.; Hahn, M.G.; Meier, C.; Freitas, C.; Roessig, L.; Stasch, J.P. Soluble GC stimulators and activators: Past, present and future. Br. J. Pharm. 2021, 10, 1–22. [Google Scholar] [CrossRef]
- Sotolongo, A.; Monica, F.Z.; Kots, A.; Xiao, H.; Liu, J.; Seto, E.; Bian, K.; Murad, F. Epigenetic regulation of soluble guanylate cyclase (sGC) beta1 in breast cancer cells. FASEB J. 2016, 30, 3171–3180. [Google Scholar] [CrossRef]
- Kots, A.Y.; Choi, B.K.; Estrella-Jimenez, M.E.; Warren, C.A.; Gilbertson, S.R.; Guerrant, R.L.; Murad, F. Pyridopyrimidine derivatives as inhibitors of cyclic nucleotide synthesis: Application for treatment of diarrhea. Proc. Natl. Acad. Sci. USA 2008, 105, 8440–8445. [Google Scholar] [CrossRef]
- Bian, K.; Harari, Y.; Zhong, M.; Lai, M.; Castro, G.; Weisbrodt, N.; Murad, F. Down-regulation of inducible nitric-oxide synthase (NOS-2) during parasite-induced gut inflammation: A path to identify a selective NOS-2 inhibitor. Mol. Pharm. 2001, 59, 939–947. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Hu, M.; Polyak, K. Serial analysis of gene expression. Nat. Protoc. 2006, 1, 1743–1760. [Google Scholar] [CrossRef]
- Martin, E.; Sharina, I.; Kots, A.; Murad, F. A constitutively activated mutant of human soluble guanylyl cyclase (sGC): Implication for the mechanism of sGC activation. Proc. Natl. Acad. Sci. USA 2003, 100, 9208–9213. [Google Scholar] [CrossRef]
- Fischbach, C.; Chen, R.; Matsumoto, T.; Schmelzle, T.; Brugge, J.S.; Polverini, P.J.; Mooney, D.J. Engineering tumors with 3D scaffolds. Nat. Methods 2007, 4, 855–860. [Google Scholar] [CrossRef]
- Sharina, I.G.; Jelen, F.; Bogatenkova, E.P.; Thomas, A.; Martin, E.; Murad, F. Alpha1 soluble guanylyl cyclase (sGC) splice forms as potential regulators of human sGC activity. J. Biol. Chem. 2008, 283, 15104–15113. [Google Scholar] [CrossRef]
- Corbalan, R.; Chatauret, N.; Behrends, S.; Butterworth, R.F.; Felipo, V. Region selective alterations of soluble guanylate cyclase content and modulation in brain of cirrhotic patients. Hepatology 2002, 36, 1155–1162. [Google Scholar] [CrossRef]
- Bonkale, W.L.; Winblad, B.; Ravid, R.; Cowburn, R.F. Reduced nitric oxide responsive soluble guanylyl cyclase activity in the superior temporal cortex of patients with Alzheimer’s disease. Neurosci. Lett. 1995, 187, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Haase, N.; Haase, T.; Seeanner, M.; Behrends, S. Nitric oxide sensitive guanylyl cyclase activity decreases during cerebral postnatal development because of a reduction in heterodimerization. J. Neurochem. 2010, 112, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Nedvetsky, P.I.; Kleinschnitz, C.; Schmidt, H.H. Regional distribution of protein and activity of the nitric oxide receptor, soluble guanylyl cyclase, in rat brain suggests multiple mechanisms of regulation. Brain Res. 2002, 950, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, R.; Montoliu, C.; Chatauret, N.; Butterworth, R.; Behrends, S.; Del Olmo, J.A.; Serra, M.A.; Rodrigo, J.M.; Erceg, S.; Felipo, V. Alterations in soluble guanylate cyclase content and modulation by nitric oxide in liver disease. Neurochem. Int. 2004, 45, 947–953. [Google Scholar] [CrossRef]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, A.C.; Hiller, D.J.; Chen, A.J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 2008, 455, 1129–1133. [Google Scholar] [CrossRef]
- Sembritzki, O.; Hagel, C.; Lamszus, K.; Deppert, W.; Bohn, W. Cytoplasmic localization of wild-type p53 in glioblastomas correlates with expression of vimentin and glial fibrillary acidic protein. Neuro Oncol. 2002, 4, 171–178. [Google Scholar] [CrossRef]
- Shaulsky, G.; Goldfinger, N.; Ben-Ze’ev, A.; Rotter, V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol. Cell Biol. 1990, 10, 6565–6577. [Google Scholar] [CrossRef]
- O’Brate, A.; Giannakakou, P. The importance of p53 location: Nuclear or cytoplasmic zip code? Drug Resist. Updat. 2003, 6, 313–322. [Google Scholar] [CrossRef]
- Green, D.R.; Kroemer, G. Cytoplasmic functions of the tumour suppressor p53. Nature 2009, 458, 1127–1130. [Google Scholar] [CrossRef]
- Harms, K.; Nozell, S.; Chen, X. The common and distinct target genes of the p53 family transcription factors. Cell Mol. Life Sci. 2004, 61, 822–842. [Google Scholar] [CrossRef]
- Gomez-Manzano, C.; Fueyo, J.; Kyritsis, A.P.; McDonnell, T.J.; Steck, P.A.; Levin, V.A.; Yung, W.K. Characterization of p53 and p21 functional interactions in glioma cells en route to apoptosis. J. Natl. Cancer Inst. 1997, 89, 1036–1044. [Google Scholar] [CrossRef]
- Canhoto, A.J.; Chestukhin, A.; Litovchick, L.; DeCaprio, J.A. Phosphorylation of the retinoblastoma-related protein p130 in growth-arrested cells. Oncogene 2000, 19, 5116–5122. [Google Scholar] [CrossRef]
- Li, M.; Xiao, A.; Floyd, D.; Olmez, I.; Lee, J.; Godlewski, J.; Bronisz, A.; Bhat, K.P.L.; Sulman, E.P.; Nakano, I.; et al. CDK4/6 inhibition is more active against the glioblastoma proneural subtype. Oncotarget 2017, 8, 55319–55331. [Google Scholar] [CrossRef]
- Goel, S.; DeCristo, M.J.; McAllister, S.S.; Zhao, J.J. CDK4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol. 2018, 28, 911–925. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Alvarado, A.G.; Thiagarajan, P.S.; Mulkearns-Hubert, E.E.; Silver, D.J.; Hale, J.S.; Alban, T.J.; Turaga, S.M.; Jarrar, A.; Reizes, O.; Longworth, M.S.; et al. Glioblastoma Cancer Stem Cells Evade Innate Immune Suppression of Self-Renewal through Reduced TLR4 Expression. Cell Stem Cell 2017, 20, 450–461.e4. [Google Scholar] [CrossRef]
- Stanzani, E.; Pedrosa, L.; Bourmeau, G.; Anezo, O.; Noguera-Castells, A.; Esteve-Codina, A.; Passoni, L.; Matteoli, M.; De la Iglesia, N.; Seano, G.; et al. Dual Role of Integrin Alpha-6 in Glioblastoma: Supporting Stemness in Proneural Stem-Like Cells While Inducing Radioresistance in Mesenchymal Stem-Like Cells. Cancers 2021, 13, 3055. [Google Scholar] [CrossRef]
- Ying, M.; Tilghman, J.; Wei, Y.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Ji, H.; Laterra, J. Kruppel-like factor-9 (KLF9) inhibits glioblastoma stemness through global transcription repression and integrin alpha6 inhibition. J. Biol. Chem. 2014, 289, 32742–32756. [Google Scholar] [CrossRef]
- Krumenacker, J.S.; Kots, A.; Murad, F. Effects of the JNK inhibitor anthra[1,9-cd]pyrazol-6(2H)-one (SP-600125) on soluble guanylyl cyclase alpha1 gene regulation and cGMP synthesis. Am. J. Physiol. Cell Physiol. 2005, 289, C778–C784. [Google Scholar] [CrossRef]
- Sharina, I.G.; Cote, G.J.; Martin, E.; Doursout, M.F.; Murad, F. RNA splicing in regulation of nitric oxide receptor soluble guanylyl cyclase. Nitric Oxide 2011, 25, 265–274. [Google Scholar] [CrossRef]
- Sharin, V.G.; Mujoo, K.; Kots, A.Y.; Martin, E.; Murad, F.; Sharina, I.G. Nitric oxide receptor soluble guanylyl cyclase undergoes splicing regulation in differentiating human embryonic cells. Stem Cells Dev. 2011, 20, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Sharina, I.; Lezgyieva, K.; Krutsenko, Y.; Martin, E. Higher susceptibility to heme oxidation and lower protein stability of the rare alpha(1)C517Ybeta(1) sGC variant associated with moyamoya syndrome. Biochem. Pharm. 2021, 186, 114459. [Google Scholar] [CrossRef] [PubMed]
- Sharina, I.G.; Martin, E. The Role of Reactive Oxygen and Nitrogen Species in the Expression and Splicing of Nitric Oxide Receptor. Antioxid. Redox Signal. 2017, 26, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Bian, K.; Ghassemi, F.; Sotolongo, A.; Siu, A.; Shauger, L.; Kots, A.; Murad, F. NOS-2 signaling and cancer therapy. IUBMB Life 2012, 64, 676–683. [Google Scholar] [CrossRef]
- Pifarre, P.; Baltrons, M.A.; Foldi, I.; Garcia, A. NO-sensitive guanylyl cyclase beta1 subunit is peripherally associated to chromosomes during mitosis. Novel role in chromatin condensation and cell cycle progression. Int. J. Biochem. Cell Biol. 2009, 41, 1719–1730. [Google Scholar] [CrossRef]
- Djuzenova, C.S.; Fiedler, V.; Memmel, S.; Katzer, A.; Hartmann, S.; Krohne, G.; Zimmermann, H.; Scholz, C.J.; Polat, B.; Flentje, M.; et al. Actin cytoskeleton organization, cell surface modification and invasion rate of 5 glioblastoma cell lines differing in PTEN and p53 status. Exp. Cell Res. 2015, 330, 346–357. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Haronikova, L.; Olivares-Illana, V.; Wang, L.; Karakostis, K.; Chen, S.; Fahraeus, R. The p53 mRNA: An integral part of the cellular stress response. Nucleic Acids Res. 2019, 47, 3257–3271. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef]
- Yount, G.L.; Haas-Kogan, D.A.; Vidair, C.A.; Haas, M.; Dewey, W.C.; Israel, M.A. Cell cycle synchrony unmasks the influence of p53 function on radiosensitivity of human glioblastoma cells. Cancer Res. 1996, 56, 500–506. [Google Scholar]
- Cerrato, J.A.; Yung, W.K.; Liu, T.J. Introduction of mutant p53 into a wild-type p53-expressing glioma cell line confers sensitivity to Ad-p53-induced apoptosis. Neuro Oncol. 2001, 3, 113–122. [Google Scholar] [CrossRef]
- Benson, E.K.; Zhao, B.; Sassoon, D.A.; Lee, S.W.; Aaronson, S.A. Effects of p21 deletion in mouse models of premature aging. Cell Cycle 2009, 8, 2002–2004. [Google Scholar] [CrossRef]
- Gartel, A.L.; Tyner, A.L. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol. Cancer Ther. 2002, 1, 639–649. [Google Scholar]
- Mlcochova, P.; Winstone, H.; Zuliani-Alvarez, L.; Gupta, R.K. TLR4-Mediated Pathway Triggers Interferon-Independent G0 Arrest and Antiviral SAMHD1 Activity in Macrophages. Cell Rep. 2020, 30, 3972–3980.e5. [Google Scholar] [CrossRef]
- Speidel, D. Transcription-independent p53 apoptosis: An alternative route to death. Trends Cell Biol. 2010, 20, 14–24. [Google Scholar] [CrossRef]
- Marchenko, N.D.; Moll, U.M. Mitochondrial death functions of p53. Mol. Cell Oncol. 2014, 1, e955995. [Google Scholar] [CrossRef]
- Cai, C.; Chen, S.Y.; Zheng, Z.; Omwancha, J.; Lin, M.F.; Balk, S.P.; Shemshedini, L. Androgen regulation of soluble guanylyl cyclasealpha1 mediates prostate cancer cell proliferation. Oncogene 2007, 26, 1606–1615. [Google Scholar] [CrossRef]
- Cai, C.; Hsieh, C.L.; Gao, S.; Kannan, A.; Bhansali, M.; Govardhan, K.; Dutta, R.; Shemshedini, L. Soluble guanylyl cyclase alpha1 and p53 cytoplasmic sequestration and down-regulation in prostate cancer. Mol. Endocrinol. 2012, 26, 292–307. [Google Scholar] [CrossRef]
- Frock, R.L.; Sadeghi, C.; Meng, J.; Wang, J.L. DNA End Joining: G0-ing to the Core. Biomolecules 2021, 11, 1487. [Google Scholar] [CrossRef]
- Sagot, I.; Laporte, D. Quiescence, an individual journey. Curr. Genet. 2019, 65, 695–699. [Google Scholar] [CrossRef]
- Frade, J.M.; Ovejero-Benito, M.C. Neuronal cell cycle: The neuron itself and its circumstances. Cell Cycle 2015, 14, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Patel, A. Benign vs Malignant Tumors. JAMA Oncol. 2020, 6, 1488. [Google Scholar] [CrossRef] [PubMed]
- De The, H. Differentiation therapy revisited. Nat. Rev. Cancer 2018, 18, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Krumenacker, J.S.; Katsuki, S.; Kots, A.; Murad, F. Differential expression of genes involved in cGMP-dependent nitric oxide signaling in murine embryonic stem (ES) cells and ES cell-derived cardiomyocytes. Nitric Oxide 2006, 14, 1–11. [Google Scholar] [CrossRef]
- Mujoo, K.; Krumenacker, J.S.; Wada, Y.; Murad, F. Differential expression of nitric oxide signaling components in undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2006, 15, 779–787. [Google Scholar] [CrossRef]
- Rossi, F.; Noren, H.; Jove, R.; Beljanski, V.; Grinnemo, K.H. Differences and similarities between cancer and somatic stem cells: Therapeutic implications. Stem Cell Res. Ther. 2020, 11, 489. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Bellutti, F.; Tigan, A.S.; Nebenfuehr, S.; Dolezal, M.; Zojer, M.; Grausenburger, R.; Hartenberger, S.; Kollmann, S.; Doma, E.; Prchal-Murphy, M.; et al. CDK6 Antagonizes p53-Induced Responses during Tumorigenesis. Cancer Discov. 2018, 8, 884–897. [Google Scholar] [CrossRef]
- Nebenfuehr, S.; Bellutti, F.; Sexl, V. Cdk6: At the interface of Rb and p53. Mol. Cell Oncol. 2018, 5, e1511206. [Google Scholar] [CrossRef]
- Kollmann, K.; Heller, G.; Schneckenleithner, C.; Warsch, W.; Scheicher, R.; Ott, R.G.; Schafer, M.; Fajmann, S.; Schlederer, M.; Schiefer, A.I.; et al. A Kinase-Independent Function of CDK6 Links the Cell Cycle to Tumor Angiogenesis. Cancer Cell 2016, 30, 359–360. [Google Scholar] [CrossRef]
- Hydbring, P.; Malumbres, M.; Sicinski, P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat. Rev. Mol. Cell Biol. 2016, 17, 280–292. [Google Scholar] [CrossRef]
- Chamberlain, M.C.; Cloughsey, T.; Reardon, D.A.; Wen, P.Y. A novel treatment for glioblastoma: Integrin inhibition. Expert Rev. Neurother. 2012, 12, 421–435. [Google Scholar] [CrossRef]
- Colin, C.; Baeza, N.; Bartoli, C.; Fina, F.; Eudes, N.; Nanni, I.; Martin, P.M.; Ouafik, L.; Figarella-Branger, D. Identification of genes differentially expressed in glioblastoma versus pilocytic astrocytoma using Suppression Subtractive Hybridization. Oncogene 2006, 25, 2818–2826. [Google Scholar] [CrossRef]
- Ramalho-Santos, M.; Yoon, S.; Matsuzaki, Y.; Mulligan, R.C.; Melton, D.A. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science 2002, 298, 597–600. [Google Scholar] [CrossRef]
- Lathia, J.D.; Gallagher, J.; Heddleston, J.M.; Wang, J.; Eyler, C.E.; Macswords, J.; Wu, Q.; Vasanji, A.; McLendon, R.E.; Hjelmeland, A.B.; et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell 2010, 6, 421–432. [Google Scholar] [CrossRef]
- Hoey, T.; Yen, W.C.; Axelrod, F.; Basi, J.; Donigian, L.; Dylla, S.; Fitch-Bruhns, M.; Lazetic, S.; Park, I.K.; Sato, A.; et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell 2009, 5, 168–177. [Google Scholar] [CrossRef]
- Levine, A.J. Targeting the P53 Protein for Cancer Therapies: The Translational Impact of P53 Research. Cancer Res. 2022, 82, 362–364. [Google Scholar] [CrossRef]
- Rampioni Vinciguerra, G.L.; Sonego, M.; Segatto, I.; Dall’Acqua, A.; Vecchione, A.; Baldassarre, G.; Belletti, B. CDK4/6 Inhibitors in Combination Therapies: Better in Company Than Alone: A Mini Review. Front. Oncol. 2022, 12, 891580. [Google Scholar] [CrossRef]
- Alday-Parejo, B.; Stupp, R.; Ruegg, C. Are Integrins Still Practicable Targets for Anti-Cancer Therapy? Cancers 2019, 11, 978. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, H.; Zhu, H.; Bögler, O.; Mónica, F.Z.; Kots, A.Y.; Murad, F.; Bian, K. Soluble Guanylate Cyclase β1 Subunit Represses Human Glioblastoma Growth. Cancers 2023, 15, 1567. https://doi.org/10.3390/cancers15051567
Xiao H, Zhu H, Bögler O, Mónica FZ, Kots AY, Murad F, Bian K. Soluble Guanylate Cyclase β1 Subunit Represses Human Glioblastoma Growth. Cancers. 2023; 15(5):1567. https://doi.org/10.3390/cancers15051567
Chicago/Turabian StyleXiao, Haijie, Haifeng Zhu, Oliver Bögler, Fabiola Zakia Mónica, Alexander Y. Kots, Ferid Murad, and Ka Bian. 2023. "Soluble Guanylate Cyclase β1 Subunit Represses Human Glioblastoma Growth" Cancers 15, no. 5: 1567. https://doi.org/10.3390/cancers15051567
APA StyleXiao, H., Zhu, H., Bögler, O., Mónica, F. Z., Kots, A. Y., Murad, F., & Bian, K. (2023). Soluble Guanylate Cyclase β1 Subunit Represses Human Glioblastoma Growth. Cancers, 15(5), 1567. https://doi.org/10.3390/cancers15051567








