Insight on Non-Coding RNAs from Biofluids in Ovarian Tumors
Abstract
:Simple Summary
Abstract
1. Introduction
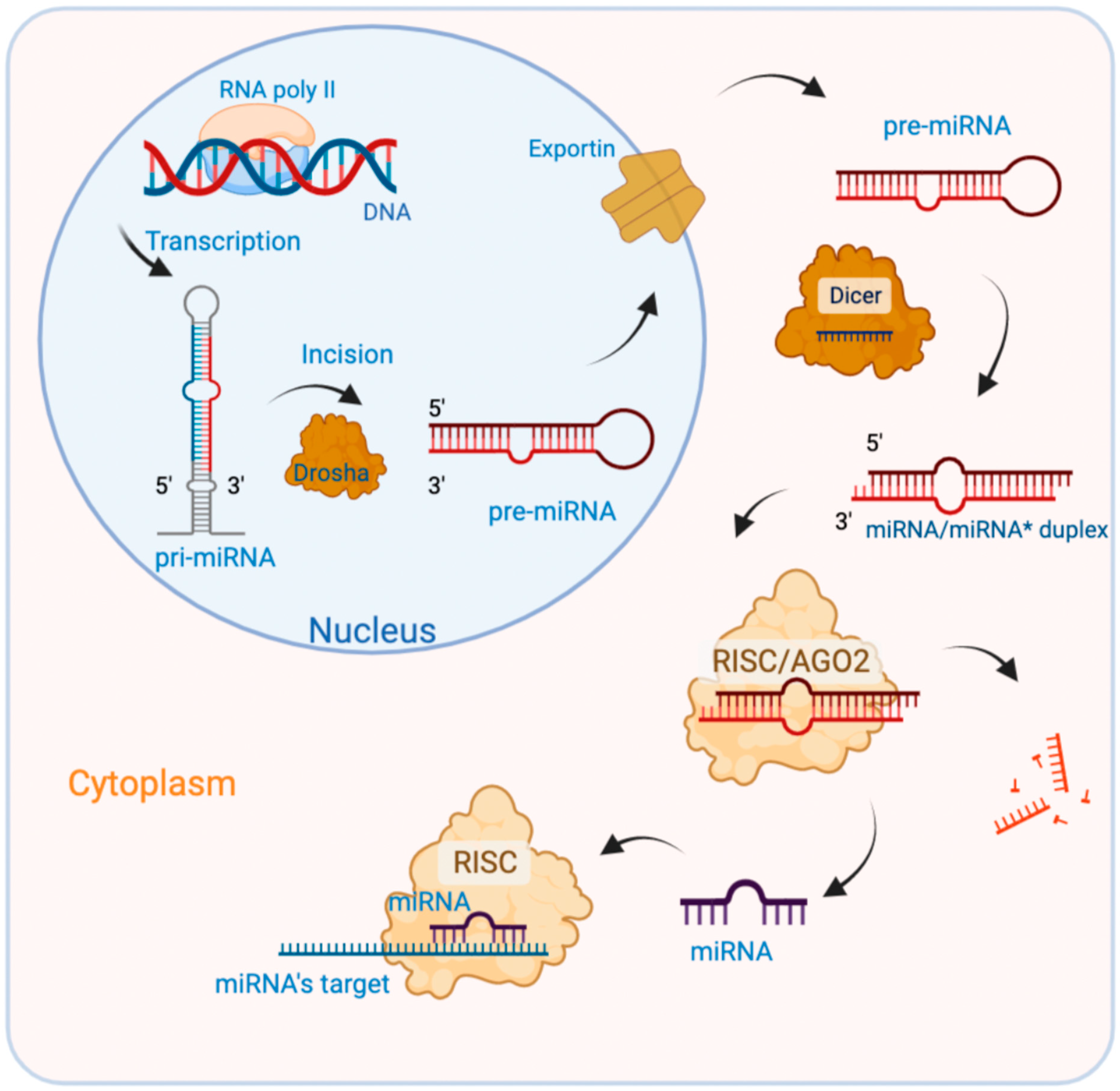
2. miRNAs
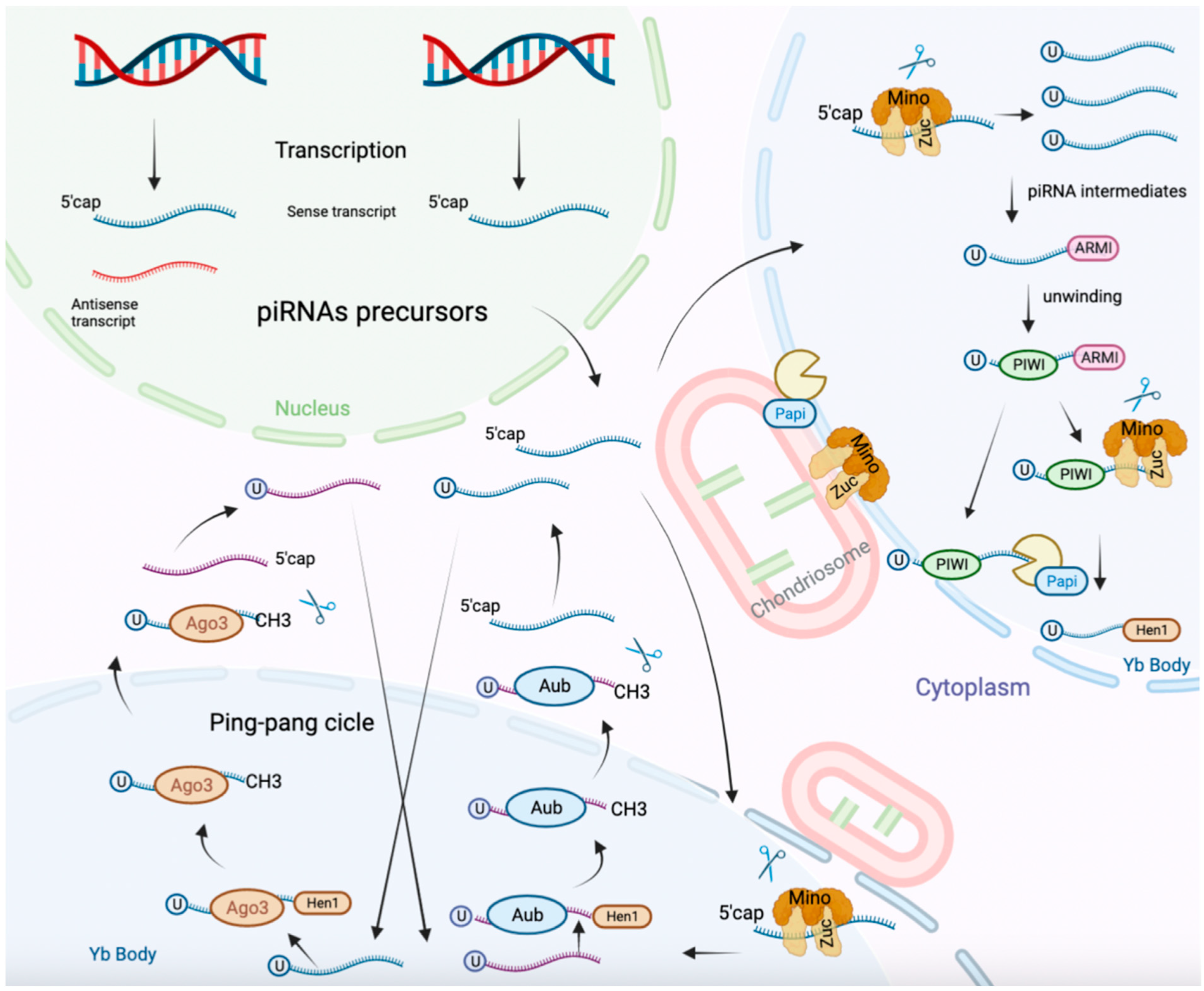
3. PiRNAs
4. Transfer-RNAs (tRNAs)
5. Circular RNAs (CircRNAs)
6. Small Nucleolar RNAs (snoRNAs)
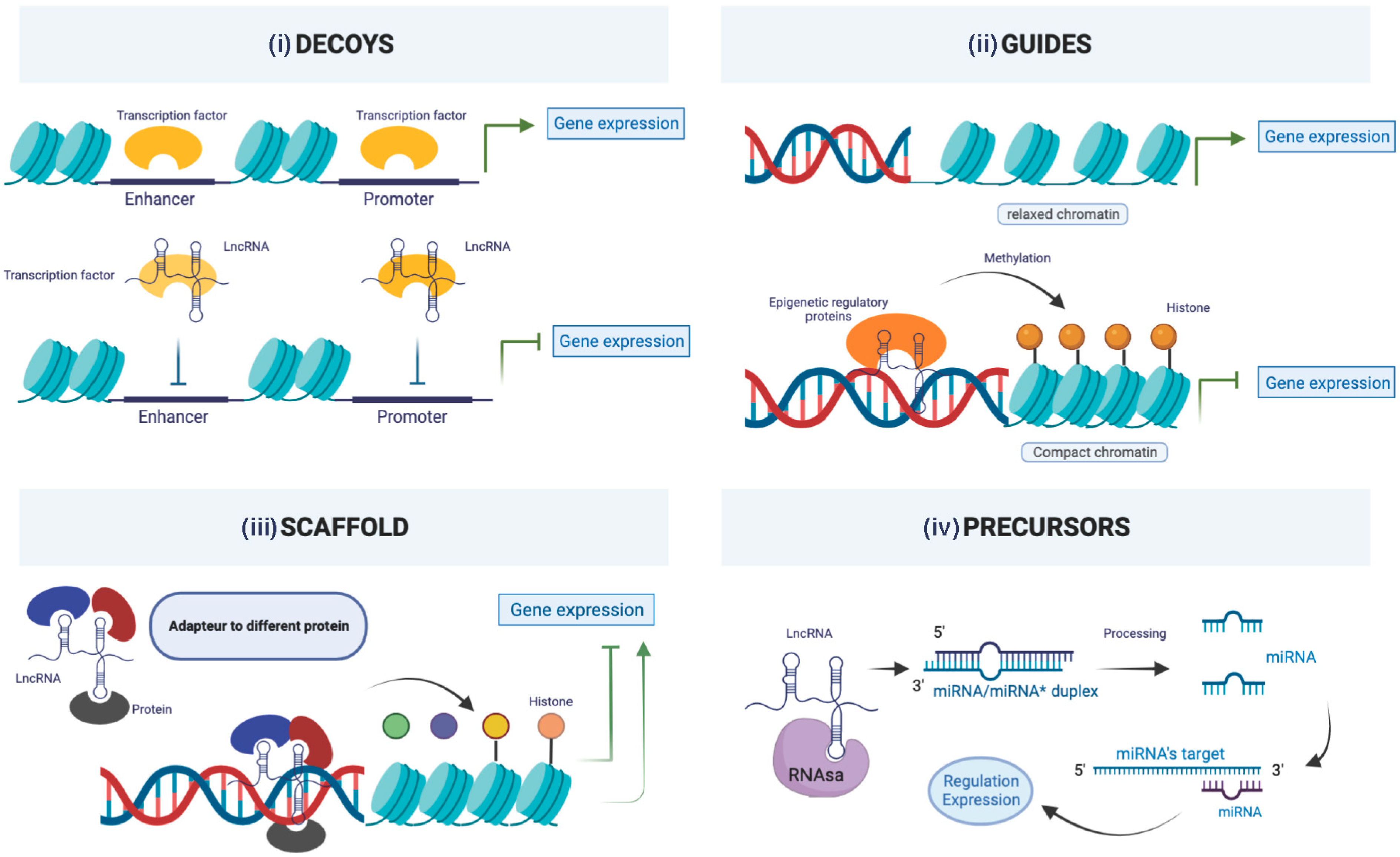
7. Long Non-Coding RNAs (lncRNAs)
8. Perspectives and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jung, A.; Reheis, L.; Host, A.; Hummel, M.; Billing, M.; Garbin, O. Retrospective evaluation of relevance of care in the management of presumed benign ovarian tumors. Gynecol. Obstet. Fertil. Senol. 2020, 48, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Bullock, W.K.; Houts, R.E.; Gilrane, J.J. Ovarian Tumors; a Survey of All Surgically Treated Ovarian Tumors in a Large General Hospital over a Ten-Year Period. AMA Arch. Surg. 1955, 71, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Guidelines Detail. Available online: https://www.nccn.org/guidelines/guidelines-detail (accessed on 1 October 2022).
- SFOG. fr—Société Française d’Oncologie Gynécologie—Un Site Utilisant. Available online: https://sfog.fr/ (accessed on 14 January 2023).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef]
- Lavoué, V.; Huchon, C.; Daraï, E. Management of Epithelial Ovarian Cancer: French joint recommendations of FRANCOGYN, CNGOF, SFOG, GINECO-ARCAGY and endorsed by INCa. Introduction. Gynecol. Obstet. Fertil. Senol. 2019, 47, 93–94. [Google Scholar] [CrossRef] [PubMed]
- Reiser, E.; Pils, D.; Grimm, C.; Hoffmann, I.; Polterauer, S.; Kranawetter, M.; Aust, S. Defining Models to Classify between Benign and Malignant Adnexal Masses Using Routine Laboratory Parameters. Cancers 2022, 14, 3210. [Google Scholar] [CrossRef]
- Froyman, W.; Landolfo, C.; De Cock, B.; Wynants, L.; Sladkevicius, P.; Testa, A.C.; Van Holsbeke, C.; Domali, E.; Fruscio, R.; Epstein, E.; et al. Risk of Complications in Patients with Conservatively Managed Ovarian Tumours (IOTA5): A 2-Year Interim Analysis of a Multicentre, Prospective, Cohort Study. Lancet Oncol. 2019, 20, 448–458. [Google Scholar] [CrossRef]
- Van Calster, B.; Valentin, L.; Froyman, W.; Landolfo, C.; Ceusters, J.; Testa, A.C.; Wynants, L.; Sladkevicius, P.; Van Holsbeke, C.; Domali, E.; et al. Validation of Models to Diagnose Ovarian Cancer in Patients Managed Surgically or Conservatively: Multicentre Cohort Study. BMJ 2020, 370, m2614. [Google Scholar] [CrossRef]
- Meys, E.M.J.; Kaijser, J.; Kruitwagen, R.F.P.M.; Slangen, B.F.M.; Van Calster, B.; Aertgeerts, B.; Verbakel, J.Y.; Timmerman, D.; Van Gorp, T. Subjective Assessment versus Ultrasound Models to Diagnose Ovarian Cancer: A Systematic Review and Meta-Analysis. Eur. J. Cancer Oxf. Engl. 1990 2016, 58, 17–29. [Google Scholar] [CrossRef]
- Thomassin-Naggara, I.; Poncelet, E.; Jalaguier-Coudray, A.; Guerra, A.; Fournier, L.S.; Stojanovic, S.; Millet, I.; Bharwani, N.; Juhan, V.; Cunha, T.M.; et al. Ovarian-Adnexal Reporting Data System Magnetic Resonance Imaging (O-RADS MRI) Score for Risk Stratification of Sonographically Indeterminate Adnexal Masses. JAMA Netw. Open 2020, 3, e1919896. [Google Scholar] [CrossRef]
- Thomassin-Naggara, I.; Aubert, E.; Rockall, A.; Jalaguier-Coudray, A.; Rouzier, R.; Daraï, E.; Bazot, M. Adnexal Masses: Development and Preliminary Validation of an MR Imaging Scoring System. Radiology 2013, 267, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Maggino, T.; Gadducci, A.; D’Addario, V.; Pecorelli, S.; Lissoni, A.; Stella, M.; Romagnolo, C.; Federghini, M.; Zucca, S.; Trio, D. Prospective Multicenter Study on CA 125 in Postmenopausal Pelvic Masses. Gynecol. Oncol. 1994, 54, 117–123. [Google Scholar] [CrossRef]
- Choi, H.-J.; Lee, Y.-Y.; Sohn, I.; Kim, Y.-M.; Kim, J.-W.; Kang, S.; Kim, B.-G. Comparison of CA 125 Alone and Risk of Ovarian Malignancy Algorithm (ROMA) in Patients with Adnexal Mass: A Multicenter Study. Curr. Probl. Cancer 2020, 44, 100508. [Google Scholar] [CrossRef]
- Sevinc, A.; Adli, M.; Kalender, M.E.; Camci, C. Benign Causes of Increased Serum CA-125 Concentration. Lancet Oncol. 2007, 8, 1054–1055. [Google Scholar] [CrossRef] [PubMed]
- Høgdall, E.V.S.; Christensen, L.; Kjaer, S.K.; Blaakaer, J.; Kjaerbye-Thygesen, A.; Gayther, S.; Jacobs, I.J.; Høgdall, C.K. CA125 Expression Pattern, Prognosis and Correlation with Serum CA125 in Ovarian Tumor Patients. From The Danish “MALOVA” Ovarian Cancer Study. Gynecol. Oncol. 2007, 104, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Dunton, C.J.; Hutchcraft, M.L.; Bullock, R.G.; Northrop, L.E.; Ueland, F.R. Salvaging Detection of Early-Stage Ovarian Malignancies When CA125 Is Not Informative. Diagnostics 2021, 11, 1440. [Google Scholar] [CrossRef]
- Davenport, C.; Rai, N.; Sharma, P.; Deeks, J.J.; Berhane, S.; Mallett, S.; Saha, P.; Champaneria, R.; Bayliss, S.E.; Snell, K.I.; et al. Menopausal Status, Ultrasound and Biomarker Tests in Combination for the Diagnosis of Ovarian Cancer in Symptomatic Women. Cochrane Database Syst. Rev. 2022, 7, CD011964. [Google Scholar] [CrossRef] [PubMed]
- Funston, G.; Van Melle, M.; Baun, M.-L.L.; Jensen, H.; Helsper, C.; Emery, J.; Crosbie, E.J.; Thompson, M.; Hamilton, W.; Walter, F.M. Variation in the Initial Assessment and Investigation for Ovarian Cancer in Symptomatic Women: A Systematic Review of International Guidelines. BMC Cancer 2019, 19, 1028. [Google Scholar] [CrossRef]
- Shahrouki, P.; Larsson, E. The Non-Coding Oncogene: A Case of Missing DNA Evidence? Front. Genet. 2012, 3, 170. [Google Scholar] [CrossRef]
- Green, D.; Fraser, W.D.; Dalmay, T. Transfer RNA-Derived Small RNAs in the Cancer Transcriptome. Pflugers Arch. 2016, 468, 1041–1047. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The Functional Role of Long Non-Coding RNA in Human Carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Jiang, Z.; Li, T.; Hu, Y.; Guo, J. Circular RNAs in Hepatocellular Carcinoma: Functions and Implications. Cancer Med. 2018, 7, 3101–3109. [Google Scholar] [CrossRef]
- Yang, L.; Froberg, J.E.; Lee, J.T. Long Noncoding RNAs: Fresh Perspectives into the RNA World. Trends Biochem. Sci. 2014, 39, 35–43. [Google Scholar] [CrossRef]
- Shen, Y.; Yu, X.; Zhu, L.; Li, T.; Yan, Z.; Guo, J. Transfer RNA-Derived Fragments and TRNA Halves: Biogenesis, Biological Functions and Their Roles in Diseases. J. Mol. Med. 2018, 96, 1167–1176. [Google Scholar] [CrossRef]
- Ambros, V. The Functions of Animal MicroRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wu, S.; Ding, J.; Lin, J.; Wei, L.; Gu, J.; He, X. MicroRNA-181a Modulates Gene Expression of Zinc Finger Family Members by Directly Targeting Their Coding Regions. Nucleic Acids Res. 2010, 38, 7211–7218. [Google Scholar] [CrossRef]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a Binds the 5′UTR of Ribosomal Protein MRNAs and Enhances Their Translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.; Patel, T.; et al. Plasma Extracellular RNA Profiles in Healthy and Cancer Patients. Sci. Rep. 2016, 6, 19413. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.-G.; Alder, H.; et al. MicroRNA Signatures in Human Ovarian Cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef]
- Dwivedi, S.K.D.; Mustafi, S.B.; Mangala, L.S.; Jiang, D.; Pradeep, S.; Rodriguez-Aguayo, C.; Ling, H.; Ivan, C.; Mukherjee, P.; Calin, G.A.; et al. Therapeutic Evaluation of MicroRNA-15a and MicroRNA-16 in Ovarian Cancer. Oncotarget 2016, 7, 15093–15104. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Nicoloso, M.; Arvizo, R.; Wang, E.; Cortez, A.; Rossi, S.; Calin, G.A.; Mukherjee, P. MiR-15a and MiR-16 Control Bmi-1 Expression in Ovarian Cancer. Cancer Res. 2009, 69, 9090–9095. [Google Scholar] [CrossRef] [PubMed]
- Vang, S.; Wu, H.-T.; Fischer, A.; Miller, D.H.; MacLaughlan, S.; Douglass, E.; Comisar, L.; Steinhoff, M.; Collins, C.; Smith, P.J.S.; et al. Identification of Ovarian Cancer Metastatic MiRNAs. PLoS ONE 2013, 8, e58226. [Google Scholar] [CrossRef]
- Dwivedi, S.K.D.; Rao, G.; Dey, A.; Mukherjee, P.; Wren, J.D.; Bhattacharya, R. Small Non-Coding-RNA in Gynecological Malignancies. Cancers 2021, 13, 1085. [Google Scholar] [CrossRef]
- Shu, C.; Wang, W.; Wu, L.; Qi, C.; Yan, W.; Lu, W.; Tian, J.; Shang, A. LINC00936/MicroRNA-221-3p Regulates Tumor Progression in Ovarian Cancer by Interacting with LAMA3. Recent Pat. Anticancer Drug Discov. 2023, 18, 66–79. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, Z.; Wang, M.; Zhang, M.; Chen, Y.; Yang, X.; Zhou, C.; Liu, Y.; Hong, L.; Zhang, L. Detection of Plasma Exosomal MiRNA-205 as a Biomarker for Early Diagnosis and an Adjuvant Indicator of Ovarian Cancer Staging. J. Ovarian Res. 2022, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.Y.; Sun, L.; Guo, Z.R.; Li, J.C.; Bai, T.T.; Cai, X.X.; Li, W.H.; Zhu, Y.F. Upregulated Expression of Serum Exosomal MiR-375 and MiR-1307 Enhance the Diagnostic Power of CA125 for Ovarian Cancer. J. Ovarian Res. 2019, 12, 6. [Google Scholar] [CrossRef]
- Shah, J.S.; Gard, G.B.; Yang, J.; Maidens, J.; Valmadre, S.; Soon, P.S.; Marsh, D.J. Combining Serum MicroRNA and CA-125 as Prognostic Indicators of Preoperative Surgical Outcome in Women with High-Grade Serous Ovarian Cancer. Gynecol. Oncol. 2018, 148, 181–188. [Google Scholar] [CrossRef]
- Robelin, P.; Tod, M.; Colomban, O.; Lachuer, J.; Ray-Coquard, I.; Rauglaudre, G.D.; Joly, F.; Chevalier-Place, A.; Combe, P.; Lortholary, A.; et al. Comparative Analysis of Predictive Values of the Kinetics of 11 Circulating MiRNAs and of CA125 in Ovarian Cancer during First Line Treatment (a GINECO Study). Gynecol. Oncol. 2020, 159, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Tian, J.; Lin, Y.; Jin, Y.; Wang, L.; Cui, M. Serum MicroRNA-92 Expression in Patients with Ovarian Epithelial Carcinoma. J. Int. Med. Res. 2013, 41, 1456–1461. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, P.D.R.; Margiotti, K.; Fabiani, M.; Barros-Filho, M.C.; Sparacino, D.; Cima, A.; Longo, S.A.; Cupellaro, M.; Mesoraca, A.; Giorlandino, C. Multi-Analytical Test Based on Serum MiRNAs and Proteins Quantification for Ovarian Cancer Early Detection. PLoS ONE 2021, 16, e0255804. [Google Scholar] [CrossRef]
- Meng, X.; Joosse, S.A.; Müller, V.; Trillsch, F.; Milde-Langosch, K.; Mahner, S.; Geffken, M.; Pantel, K.; Schwarzenbach, H. Diagnostic and Prognostic Potential of Serum MiR-7, MiR-16, MiR-25, MiR-93, MiR-182, MiR-376a and MiR-429 in Ovarian Cancer Patients. Br. J. Cancer 2015, 113, 1358–1366. [Google Scholar] [CrossRef]
- Cui, Y.; Hong, S.; Zhu, X. The Accuracy of Single MicroRNAs in Peripheral Blood to Diagnose Ovarian Cancer: An Updated Meta-Analysis. Dis. Markers 2020, 2020, 1075942. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, L.; Zhao, Y.; Yang, D.; Song, F.; Wen, Y.; Hao, Q.; Hu, Z.; Zhang, W.; Chen, K. Plasma MiRNAs as Diagnostic and Prognostic Biomarkers for Ovarian Cancer. PLoS ONE 2013, 8, e77853. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, Y.; Lyu, T.; Liu, H.; Shen, L.; Zhou, T.; Feng, W. Identification and Functional Validation of Differentially Expressed MicroRNAs in Ascites-Derived Ovarian Cancer Cells Compared with Primary Tumour Tissue. Cancer Manag. Res. 2021, 13, 6585–6597. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Wang, C.; Gao, Y.; Wang, J. Decreased Expression of MicroRNA-148a Predicts Poor Prognosis in Ovarian Cancer and Associates with Tumor Growth and Metastasis. Biomed. Pharmacother. Biomed. Pharmacother. 2016, 83, 58–63. [Google Scholar] [CrossRef]
- Oliveira, D.N.P.; Carlsen, A.L.; Heegaard, N.H.H.; Prahm, K.P.; Christensen, I.J.; Høgdall, C.K.; Høgdall, E.V. Diagnostic Plasma MiRNA-Profiles for Ovarian Cancer in Patients with Pelvic Mass. PLoS ONE 2019, 14, e0225249. [Google Scholar] [CrossRef]
- Savolainen, K.; Scaravilli, M.; Ilvesmäki, A.; Staff, S.; Tolonen, T.; Mäenpää, J.U.; Visakorpi, T.; Auranen, A. Expression of the MiR-200 Family in Tumor Tissue, Plasma and Urine of Epithelial Ovarian Cancer Patients in Comparison to Benign Counterparts. BMC Res. Notes 2020, 13, 311. [Google Scholar] [CrossRef]
- Salem, M.; O’Brien, J.A.; Bernaudo, S.; Shawer, H.; Ye, G.; Brkić, J.; Amleh, A.; Vanderhyden, B.C.; Refky, B.; Yang, B.B.; et al. MiR-590-3p Promotes Ovarian Cancer Growth and Metastasis via a Novel FOXA2-Versican Pathway. Cancer Res. 2018, 78, 4175–4190. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.K.; Li, X.; Mu, N.; Hrydziuszko, O.; Garcia-Majano, B.; Larsson, C.; Lui, W.-O. MicroRNA Expression Profiles in Non-epithelial Ovarian Tumors. Int. J. Oncol. 2018, 52, 55–66. [Google Scholar] [CrossRef]
- Yokoi, A.; Yoshioka, Y.; Hirakawa, A.; Yamamoto, Y.; Ishikawa, M.; Ikeda, S.-I.; Kato, T.; Niimi, K.; Kajiyama, H.; Kikkawa, F.; et al. A Combination of Circulating MiRNAs for the Early Detection of Ovarian Cancer. Oncotarget 2017, 8, 89811–89823. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Gao, W.; Chen, X.; Zhang, Y.; Wu, M.; Zhang, P.; Wang, S. A Pilot Study of Circulating MicroRNA-125b as a Diagnostic and Prognostic Biomarker for Epithelial Ovarian Cancer. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2017, 27, 3–10. [Google Scholar] [CrossRef]
- Meng, X.; Müller, V.; Milde-Langosch, K.; Trillsch, F.; Pantel, K.; Schwarzenbach, H. Diagnostic and Prognostic Relevance of Circulating Exosomal MiR-373, MiR-200a, MiR-200b and MiR-200c in Patients with Epithelial Ovarian Cancer. Oncotarget 2016, 7, 16923–16935. [Google Scholar] [CrossRef] [PubMed]
- Resnick, K.E.; Alder, H.; Hagan, J.P.; Richardson, D.L.; Croce, C.M.; Cohn, D.E. The Detection of Differentially Expressed MicroRNAs from the Serum of Ovarian Cancer Patients Using a Novel Real-Time PCR Platform. Gynecol. Oncol. 2009, 112, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Weng, G. The Diagnostic Value of Serum MiR-21 in Patients with Ovarian Cancer: A Systematic Review and Meta-Analysis. J. Ovarian Res. 2022, 15, 51. [Google Scholar] [CrossRef]
- Wang, W.; Jo, H.; Park, S.; Kim, H.; Kim, S.I.; Han, Y.; Lee, J.; Seol, A.; Kim, J.; Lee, M.; et al. Integrated Analysis of Ascites and Plasma Extracellular Vesicles Identifies a MiRNA-Based Diagnostic Signature in Ovarian Cancer. Cancer Lett. 2022, 542, 215735. [Google Scholar] [CrossRef]
- Berner, K.; Hirschfeld, M.; Weiß, D.; Rücker, G.; Asberger, J.; Ritter, A.; Nöthling, C.; Jäger, M.; Juhasz-Böss, I.; Erbes, T. Evaluation of Circulating MicroRNAs as Non-Invasive Biomarkers in the Diagnosis of Ovarian Cancer: A Case-Control Study. Arch. Gynecol. Obstet. 2022, 306, 151–163. [Google Scholar] [CrossRef]
- Gao, Y.-C.; Wu, J. MicroRNA-200c and MicroRNA-141 as Potential Diagnostic and Prognostic Biomarkers for Ovarian Cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental. Biol. Med. 2015, 36, 4843–4850. [Google Scholar] [CrossRef]
- Zuberi, M.; Mir, R.; Khan, I.; Javid, J.; Guru, S.A.; Bhat, M.; Sumi, M.P.; Ahmad, I.; Masroor, M.; Yadav, P.; et al. The Promising Signatures of Circulating MicroRNA-145 in Epithelial Ovarian Cancer Patients. MicroRNA Shariqah United Arab. Emir. 2020, 9, 49–57. [Google Scholar] [CrossRef]
- Zuberi, M.; Khan, I.; Mir, R.; Gandhi, G.; Ray, P.C.; Saxena, A. Utility of Serum MiR-125b as a Diagnostic and Prognostic Indicator and Its Alliance with a Panel of Tumor Suppressor Genes in Epithelial Ovarian Cancer. PLoS ONE 2016, 11, e0153902. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, D.; Jopek, K.; Sterzynska, K.; Nowicki, M.; Rucinski, M.; Januchowski, R. The Profile of MicroRNA Expression and Potential Role in the Regulation of Drug-Resistant Genes in Cisplatin- and Paclitaxel-Resistant Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2022, 23, 526. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, P.; Kaźmierczak, D.; Jopek, K.; Nowicki, M.; Rucinski, M.; Januchowski, R. The Profile of MicroRNA Expression and Potential Role in the Regulation of Drug-Resistant Genes in Doxorubicin and Topotecan Resistant Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2022, 23, 5846. [Google Scholar] [CrossRef]
- Yang, H.; Kong, W.; He, L.; Zhao, J.-J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA Expression Profiling in Human Ovarian Cancer: MiR-214 Induces Cell Survival and Cisplatin Resistance by Targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef]
- Echevarría-Vargas, I.M.; Valiyeva, F.; Vivas-Mejía, P.E. Upregulation of MiR-21 in Cisplatin Resistant Ovarian Cancer via JNK-1/c-Jun Pathway. PLoS ONE 2014, 9, e97094. [Google Scholar] [CrossRef]
- Lu, L.; Schwartz, P.; Scarampi, L.; Rutherford, T.; Canuto, E.M.; Yu, H.; Katsaros, D. MicroRNA Let-7a: A Potential Marker for Selection of Paclitaxel in Ovarian Cancer Management. Gynecol. Oncol. 2011, 122, 366–371. [Google Scholar] [CrossRef]
- Langhe, R.; Norris, L.; Saadeh, F.A.; Blackshields, G.; Varley, R.; Harrison, A.; Gleeson, N.; Spillane, C.; Martin, C.; O’Donnell, D.M.; et al. A Novel Serum MicroRNA Panel to Discriminate Benign from Malignant Ovarian Disease. Cancer Lett. 2015, 356, 628–636. [Google Scholar] [CrossRef]
- Saburi, A.; Kahrizi, M.S.; Naghsh, N.; Etemadi, H.; İlhan, A.; Adili, A.; Ghoreishizadeh, S.; Tamjidifar, R.; Akbari, M.; Ercan, G. A Comprehensive Survey into the Role of MicroRNAs in Ovarian Cancer Chemoresistance; an Updated Overview. J. Ovarian Res. 2022, 15, 81. [Google Scholar] [CrossRef]
- Bendifallah, S.; Dabi, Y.; Suisse, S.; Jornea, L.; Bouteiller, D.; Touboul, C.; Puchar, A.; Daraï, E. A Bioinformatics Approach to MicroRNA-Sequencing Analysis Based on Human Saliva Samples of Patients with Endometriosis. Int. J. Mol. Sci. 2022, 23, 8045. [Google Scholar] [CrossRef]
- Ishizu, H.; Siomi, H.; Siomi, M.C. Biology of PIWI-Interacting RNAs: New Insights into Biogenesis and Function inside and Outside of Germlines. Genes Dev. 2012, 26, 2361–2373. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, S.; Matushansky, I. Piwis and Piwi-Interacting RNAs in the Epigenetics of Cancer. J. Cell Biochem. 2012, 113, 373–380. [Google Scholar] [CrossRef]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z.; Guo, J. PiR-823, a Novel Non-Coding Small RNA, Demonstrates in Vitro and in Vivo Tumor Suppressive Activity in Human Gastric Cancer Cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, G.; Cordero, F.; Tarallo, S.; Arigoni, M.; Riccardo, F.; Gallo, G.; Ronco, G.; Allasia, M.; Kulkarni, N.; Matullo, G.; et al. Small Non-Coding RNA Profiling in Human Biofluids and Surrogate Tissues from Healthy Individuals: Description of the Diverse and Most Represented Species. Oncotarget 2017, 9, 3097. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Roy, J.; Rout, P.; Mallick, B. Genome-Wide Profiling of the PIWI-Interacting RNA-MRNA Regulatory Networks in Epithelial Ovarian Cancers. PLoS ONE 2018, 13, e0190485. [Google Scholar] [CrossRef]
- Hulstaert, E.; Morlion, A.; Levanon, K.; Vandesompele, J.; Mestdagh, P. Candidate RNA Biomarkers in Biofluids for Early Diagnosis of Ovarian Cancer: A Systematic Review. Gynecol. Oncol. 2021, 160, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, Z.; Sheng, J. TRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes 2018, 9, 246. [Google Scholar] [CrossRef]
- Balatti, V.; Pekarsky, Y.; Croce, C.M. Role of the TRNA-Derived Small RNAs in Cancer: New Potential Biomarkers and Target for Therapy. Adv. Cancer Res. 2017, 135, 173–187. [Google Scholar] [CrossRef]
- Telonis, A.G.; Loher, P.; Honda, S.; Jing, Y.; Palazzo, J.; Kirino, Y.; Rigoutsos, I. Dissecting TRNA-Derived Fragment Complexities Using Personalized Transcriptomes Reveals Novel Fragment Classes and Unexpected Dependencies. Oncotarget 2015, 6, 24797–24822. [Google Scholar] [CrossRef]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human TRNA-Derived Small RNAs in the Global Regulation of RNA Silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.B.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous TRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Rounge, T.B.; Furu, K.; Skotheim, R.I.; Haugen, T.B.; Grotmol, T.; Enerly, E. Profiling of the Small RNA Populations in Human Testicular Germ Cell Tumors Shows Global Loss of PiRNAs. Mol. Cancer 2015, 14, 153. [Google Scholar] [CrossRef]
- Sobala, A.; Hutvagner, G. Small RNAs Derived from the 5′ End of TRNA Can Inhibit Protein Translation in Human Cells. RNA Biol. 2013, 10, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Telonis, A.G.; Loher, P.; Magee, R.; Pliatsika, V.; Londin, E.; Kirino, Y.; Rigoutsos, I. TRNA Fragments Show Intertwining with MRNAs of Specific Repeat Content and Have Links to Disparities. Cancer Res. 2019, 79, 3034–3049. [Google Scholar] [CrossRef]
- Gebetsberger, J.; Wyss, L.; Mleczko, A.M.; Reuther, J.; Polacek, N. A TRNA-Derived Fragment Competes with MRNA for Ribosome Binding and Regulates Translation during Stress. RNA Biol. 2017, 14, 1364–1373. [Google Scholar] [CrossRef]
- Balatti, V.; Nigita, G.; Veneziano, D.; Drusco, A.; Stein, G.S.; Messier, T.L.; Farina, N.H.; Lian, J.B.; Tomasello, L.; Liu, C.-G.; et al. TsRNA Signatures in Cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 8071–8076. [Google Scholar] [CrossRef]
- Peng, E.Y.; Shu, Y.; Wu, Y.; Zeng, F.; Tan, S.; Deng, Y.; Deng, Y.; Chen, H.; Zhu, L.; Xu, H. Presence and Diagnostic Value of Circulating TsncRNA for Ovarian Tumor. Mol. Cancer 2018, 17, 163. [Google Scholar] [CrossRef]
- Zhang, M.; Li, F.; Wang, J.; He, W.; Li, Y.; Li, H.; Wei, Z.; Cao, Y. TRNA-Derived Fragment TRF-03357 Promotes Cell Proliferation, Migration and Invasion in High-Grade Serous Ovarian Cancer. OncoTargets Ther. 2019, 12, 6371–6383. [Google Scholar] [CrossRef]
- Jia, Y.; Tan, W.; Zhou, Y. Transfer RNA-Derived Small RNAs: Potential Applications as Novel Biomarkers for Disease Diagnosis and Prognosis. Ann. Transl. Med. 2020, 8, 1092. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-like Structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Hsu, M.T.; Coca-Prados, M. Electron Microscopic Evidence for the Circular Form of RNA in the Cytoplasm of Eukaryotic Cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Han, P.; Zhou, T.; Guo, X.; Song, X.; Li, Y. CircRNADb: A Comprehensive Database for Human Circular RNAs with Protein-Coding Annotations. Sci. Rep. 2016, 6, 34985. [Google Scholar] [CrossRef]
- Lai, H.; Li, Y.; Zhang, H.; Hu, J.; Liao, J.; Su, Y.; Li, Q.; Chen, B.; Li, C.; Wang, Z.; et al. ExoRBase 2.0: An Atlas of MRNA, LncRNA and CircRNA in Extracellular Vesicles from Human Biofluids. Nucleic Acids Res. 2022, 50, D118–D128. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded Identification and Characterization of Mammalian Circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef]
- Kelly, S.; Greenman, C.; Cook, P.R.; Papantonis, A. Exon Skipping Is Correlated with Exon Circularization. J. Mol. Biol. 2015, 427, 2414–2417. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, S.; Chen, X.; Li, N.; Li, J.; Jia, R.; Pan, Y.; Liang, H. EIF4A3-Induced Circular RNA MMP9 (CircMMP9) Acts as a Sponge of MiR-124 and Promotes Glioblastoma Multiforme Cell Tumorigenesis. Mol. Cancer 2018, 17, 166. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, L.; Wu, J.; Khadka, B.; Fang, Z.; Gu, J.; Tang, B.; Xiao, R.; Pan, G.; Liu, J. CircRNA Circ_0000190 Inhibits the Progression of Multiple Myeloma through Modulating MiR-767-5p/MAPK4 Pathway. J. Exp. Clin. Cancer Res. CR 2019, 38, 54. [Google Scholar] [CrossRef]
- Lu, Q.; Liu, T.; Feng, H.; Yang, R.; Zhao, X.; Chen, W.; Jiang, B.; Qin, H.; Guo, X.; Liu, M.; et al. Circular RNA CircSLC8A1 Acts as a Sponge of MiR-130b/MiR-494 in Suppressing Bladder Cancer Progression via Regulating PTEN. Mol. Cancer 2019, 18, 111. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Ahmed, I.; Karedath, T.; Al-Dasim, F.M.; Malek, J.A. Identification of Human Genetic Variants Controlling Circular RNA Expression. RNA 2019, 25, 1765–1778. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Wang, P.L.; Bao, Y.; Yee, M.-C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA Is Expressed across the Eukaryotic Tree of Life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef]
- Wang, J.; Wu, A.; Yang, B.; Zhu, X.; Teng, Y.; Ai, Z. Profiling and Bioinformatics Analyses Reveal Differential Circular RNA Expression in Ovarian Cancer. Gene 2020, 724, 144150. [Google Scholar] [CrossRef]
- Gan, X.; Zhu, H.; Jiang, X.; Obiegbusi, S.C.; Yong, M.; Long, X.; Hu, J. CircMUC16 Promotes Autophagy of Epithelial Ovarian Cancer via Interaction with ATG13 and MiR-199a. Mol. Cancer 2020, 19, 45. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Zhao, W.; Liu, G.; Yang, Q. Circ-PGAM1 Promotes Malignant Progression of Epithelial Ovarian Cancer through Regulation of the MiR-542-3p/CDC5L/PEAK1 Pathway. Cancer Med. 2020, 9, 3500–3521. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, W.; Li, Q.-H.; Xie, B.-M.; Shen, F.; Du, Y.-P.; Zong, Z.-H.; Wang, L.-L.; Wei, X.-Q.; Zhao, Y. Circ-NOLC1 Promotes Epithelial Ovarian Cancer Tumorigenesis and Progression by Binding ESRP1 and Modulating CDK1 and RhoA Expression. Cell Death Discov. 2021, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Liu, J.; Zhou, L. Upregulation of Circular RNA Circ-FAM53B Predicts Adverse Prognosis and Accelerates the Progression of Ovarian Cancer via the MiR-646/VAMP2 and MiR-647/MDM2 Signaling Pathways. Oncol. Rep. 2019, 42, 2728–2737. [Google Scholar] [CrossRef]
- Ma, R.; Ye, X.; Cheng, H.; Cui, H.; Chang, X. Tumor-Derived Exosomal CircRNA051239 Promotes Proliferation and Migration of Epithelial Ovarian Cancer. Am. J. Transl. Res. 2021, 13, 1125–1139. [Google Scholar]
- Ding, J.; Wang, Q.; Guo, N.; Wang, H.; Chen, H.; Ni, G.; Li, P. CircRNA Circ_0072995 Promotes the Progression of Epithelial Ovarian Cancer by Modulating MiR-147a/CDK6 Axis. Aging 2020, 12, 17209–17223. [Google Scholar] [CrossRef]
- Yong, M.; Hu, J.; Zhu, H.; Jiang, X.; Gan, X.; Hu, L. Erratum: Circ-EEF2 Facilitated Autophagy via Interaction with Mir-6881-3p and ANXA2 in EOC. Am. J. Cancer Res. 2021, 11, 1795–1799. [Google Scholar]
- Zhang, Z.; Zhu, H.; Hu, J. CircRAB11FIP1 Promoted Autophagy Flux of Ovarian Cancer through DSC1 and MiR-129. Cell Death Dis. 2021, 12, 219. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Dong, Z.-N.; Qiu, B.-Q.; Hu, M.; Liang, X.-Q.; Dai, X.; Hong, D.; Sun, Y.-F. CircRNA FGFR3 Induces Epithelial-Mesenchymal Transition of Ovarian Cancer by Regulating MiR-29a-3p/E2F1 Axis. Aging 2020, 12, 14080–14091. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Wang, L.; Xia, Y. Circ_0015756 Promotes the Progression of Ovarian Cancer by Regulating MiR-942-5p/CUL4B Pathway. Cancer Cell Int. 2020, 20, 572. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Liu, L.U.; He, C.; Zhao, M.; Ni, R.; Zhang, Z.; Shui, C. Circ_0002711 Knockdown Suppresses Cell Growth and Aerobic Glycolysis by Modulating MiR-1244/ROCK1 Axis in Ovarian Cancer. J. Biosci. 2021, 46, 21. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yao, Y.; Jin, M. Circ-0001068 Is a Novel Biomarker for Ovarian Cancer and Inducer of PD1 Expression in T Cells. Aging 2020, 12, 19095–19106. [Google Scholar] [CrossRef]
- Hou, W.; Zhang, Y. Circ_0025033 Promotes the Progression of Ovarian Cancer by Activating the Expression of LSM4 via Targeting MiR-184. Pathol. Res. Pract. 2021, 217, 153275. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Zhang, X.; Liu, G. Serum CircSETDB1 Is a Promising Biomarker for Predicting Response to Platinum-Taxane-Combined Chemotherapy and Relapse in High-Grade Serous Ovarian Cancer. OncoTargets Ther. 2019, 12, 7451–7457. [Google Scholar] [CrossRef]
- Guo, M.; Li, S.; Zhao, X.; Yuan, Y.; Zhang, B.; Guan, Y. Knockdown of Circular RNA Hsa_circ_0000714 Can Regulate RAB17 by Sponging MiR-370-3p to Reduce Paclitaxel Resistance of Ovarian Cancer Through CDK6/RB Pathway. OncoTargets Ther. 2020, 13, 13211–13224. [Google Scholar] [CrossRef]
- Xia, B.; Zhao, Z.; Wu, Y.; Wang, Y.; Zhao, Y.; Wang, J. Circular RNA CircTNPO3 Regulates Paclitaxel Resistance of Ovarian Cancer Cells by MiR-1299/NEK2 Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 21, 780–791. [Google Scholar] [CrossRef]
- Li, M.; Cai, J.; Han, X.; Ren, Y. Downregulation of CircNRIP1 Suppresses the Paclitaxel Resistance of Ovarian Cancer via Regulating the MiR-211-5p/HOXC8 Axis. Cancer Manag. Res. 2020, 12, 9159–9171. [Google Scholar] [CrossRef]
- You, J.; Han, Y.; Qiao, H.; Han, Y.; Lu, X.; Lu, Y.; Wang, X.; Kai, H.; Zheng, Y. Hsa_circ_0063804 Enhances Ovarian Cancer Cells Proliferation and Resistance to Cisplatin by Targeting MiR-1276/CLU Axis. Aging 2022, 14, 4699–4713. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, H.; Li, Y.; Wang, S. Noncoding RNAs Interplay in Ovarian Cancer Therapy and Drug Resistance. Cancer Biother. Radiopharm. 2022, 37, 186–198. [Google Scholar] [CrossRef]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.-P.; Zhang, B.-X.; Chu, L. Small Nucleolar RNAs: Insight into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef]
- Liu, Y.; Ruan, H.; Li, S.; Ye, Y.; Hong, W.; Gong, J.; Zhang, Z.; Jing, Y.; Zhang, X.; Diao, L.; et al. The Genetic and Pharmacogenomic Landscape of SnoRNAs in Human Cancer. Mol. Cancer 2020, 19, 108. [Google Scholar] [CrossRef]
- Lemus-Diaz, N.; Ferreira, R.R.; Bohnsack, K.E.; Gruber, J.; Bohnsack, M.T. The Human Box C/D SnoRNA U3 Is a MiRNA Source and MiR-U3 Regulates Expression of Sortin Nexin 27. Nucleic Acids Res. 2020, 48, 8074–8089. [Google Scholar] [CrossRef]
- Zhou, F.; Liu, Y.; Rohde, C.; Pauli, C.; Gerloff, D.; Köhn, M.; Misiak, D.; Bäumer, N.; Cui, C.; Göllner, S.; et al. AML1-ETO Requires Enhanced C/D Box SnoRNA/RNP Formation to Induce Self-Renewal and Leukaemia. Nat. Cell Biol. 2017, 19, 844–855. [Google Scholar] [CrossRef]
- McCann, K.L.; Kavari, S.L.; Burkholder, A.B.; Phillips, B.T.; Hall, T.M.T. H/ACA SnoRNA Levels Are Regulated during Stem Cell Differentiation. Nucleic Acids Res. 2020, 48, 8686–8703. [Google Scholar] [CrossRef]
- Oliveira, D.V.N.P.; Prahm, K.P.; Christensen, I.J.; Hansen, A.; Høgdall, C.K.; Høgdall, E.V. Noncoding RNA (NcRNA) Profile Association with Patient Outcome in Epithelial Ovarian Cancer Cases. Reprod. Sci. 2021, 28, 757–765. [Google Scholar] [CrossRef]
- Lin, H.; Shen, L.; Lin, Q.; Dong, C.; Maswela, B.; Illahi, G.S.; Wu, X. SNHG5 Enhances Paclitaxel Sensitivity of Ovarian Cancer Cells through Sponging MiR-23a. Biomed. Pharmacother. 2020, 123, 109711. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, T.; Luan, S.; Kong, Q.; Hu, W.; Zou, X.; Zheng, F.; Han, W. Identification of a Novel Nine-SnoRNA Signature with Potential Prognostic and Therapeutic Value in Ovarian Cancer. Cancer Med. 2022, 11, 2159–2170. [Google Scholar] [CrossRef]
- Zhang, P.-F.; Wu, J.; Luo, J.-H.; Li, K.-S.; Wang, F.; Huang, W.; Wu, Y.; Gao, S.-P.; Zhang, X.-M.; Zhang, P.-N. SNHG22 Overexpression Indicates Poor Prognosis and Induces Chemotherapy Resistance via the MiR-2467/Gal-1 Signaling Pathway in Epithelial Ovarian Carcinoma. Aging 2019, 11, 8204–8216. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef]
- Rutenberg-Schoenberg, M.; Sexton, A.N.; Simon, M.D. The Properties of Long Noncoding RNAs That Regulate Chromatin. Annu. Rev. Genom. Hum. Genet. 2016, 17, 69–94. [Google Scholar] [CrossRef]
- Salamini-Montemurri, M.; Lamas-Maceiras, M.; Barreiro-Alonso, A.; Vizoso-Vázquez, Á.; Rodríguez-Belmonte, E.; Quindós-Varela, M.; Cerdán, M.E. The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use. Cancers 2020, 12, 1020. [Google Scholar] [CrossRef]
- Hosseini, E.S.; Zarei, M.A.; Kashani, H.H.; Milajerdi, A.; Dehghanani, Z.Z.; Bafrani, H.H.; Nikzad, H. The Role of Altered Long Noncoding RNAs in Overall Survival of Ovarian Cancer: A Systematic Review and Meta-Analysis. Pathol. Res. Pract. 2021, 219, 153363. [Google Scholar] [CrossRef]
- Park, S.-A.; Kim, L.K.; Kim, Y.T.; Heo, T.-H.; Kim, H.J. Long Noncoding RNA E2F4as Promotes Progression and Predicts Patient Prognosis in Human Ovarian Cancer. Cancers 2020, 12, 3626. [Google Scholar] [CrossRef]
- Chu, Z.-P.; Dai, J.; Jia, L.-G.; Li, J.; Zhang, Y.; Zhang, Z.-Y.; Yan, P. Increased Expression of Long Noncoding RNA HMMR-AS1 in Epithelial Ovarian Cancer: An Independent Prognostic Factor. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8145–8150. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.; Xu, Y.; Zhu, Y.; Hu, Y.; Yan, Y.; Yan, H. LncRNA FAM83H-AS1 Contributes to the Radioresistance, Proliferation, and Metastasis in Ovarian Cancer through Stabilizing HuR Protein. Eur. J. Pharmacol. 2019, 852, 134–141. [Google Scholar] [CrossRef]
- Pan, L.; Meng, Q.; Li, H.; Liang, K.; Li, B. LINC00339 Promotes Cell Proliferation, Migration, and Invasion of Ovarian Cancer Cells via MiR-148a-3p/ROCK1 Axes. Biomed. Pharmacother. 2019, 120, 109423. [Google Scholar] [CrossRef]
- Yu, H.; Xu, Y.; Zhang, D.; Liu, G. Long Noncoding RNA LUCAT1 Promotes Malignancy of Ovarian Cancer through Regulation of MiR-612/HOXA13 Pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2095–2100. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-N.; Zhang, H.-Y. Serum LncRNA LOXL1-AS1 Is a Diagnostic and Prognostic Marker for Epithelial Ovarian Cancer. J. Gene. Med. 2020, 22, e3233. [Google Scholar] [CrossRef]
- Barwal, T.S.; Sharma, U.; Rana, M.K.; Bazala, S.; Singh, I.; Murmu, M.; Kapoor, H.S.; Thakur, S.; Jain, M.; Jain, A. A Diagnostic and Prognostic Value of Blood-Based Circulating Long Non-Coding RNAs in Thyroid, Pancreatic and Ovarian Cancer. Crit. Rev. Oncol. Hematol. 2022, 171, 103598. [Google Scholar] [CrossRef]
- Chen, Q.; Su, Y.; He, X.; Zhao, W.; Wu, C.; Zhang, W.; Si, X.; Dong, B.; Zhao, L.; Gao, Y.; et al. Plasma Long Non-Coding RNA MALAT1 Is Associated with Distant Metastasis in Patients with Epithelial Ovarian Cancer. Oncol. Lett. 2016, 12, 1361–1366. [Google Scholar] [CrossRef]
- Gong, J.; Xu, X.; Zhang, X.; Zhou, Y. LncRNA MIR4435-2HG Is a Potential Early Diagnostic Marker for Ovarian Carcinoma. Acta Biochim. Biophys. Sin. 2019, 51, 953–959. [Google Scholar] [CrossRef]
- Xie, W.; Sun, H.; Li, X.; Lin, F.; Wang, Z.; Wang, X. Ovarian Cancer: Epigenetics, Drug Resistance, and Progression. Cancer Cell Int. 2021, 21, 434. [Google Scholar] [CrossRef]
- Soda, N.; Umer, M.; Kashaninejad, N.; Kasetsirikul, S.; Kline, R.; Salomon, C.; Nguyen, N.-T.; Shiddiky, M.J.A. PCR-Free Detection of Long Non-Coding HOTAIR RNA in Ovarian Cancer Cell Lines and Plasma Samples. Cancers 2020, 12, 2233. [Google Scholar] [CrossRef]
- Clark, K.A.; Paquette, A.; Tao, K.; Bell, R.; Boyle, J.L.; Rosenthal, J.; Snow, A.K.; Stark, A.W.; Thompson, B.A.; Unger, J.; et al. Comprehensive Evaluation and Efficient Classification of BRCA1 RING Domain Missense Substitutions. Am. J. Hum. Genet. 2022, 109, 1153–1174. [Google Scholar] [CrossRef]
- Godoy, P.M.; Barczak, A.J.; DeHoff, P.; Srinivasan, S.; Etheridge, A.; Galas, D.; Das, S.; Erle, D.J.; Laurent, L.C. Comparison of Reproducibility, Accuracy, Sensitivity, and Specificity of MiRNA Quantification Platforms. Cell Rep. 2019, 29, 4212–4222.e5. [Google Scholar] [CrossRef]
- El-Mogy, M.; Lam, B.; Haj-Ahmad, T.A.; McGowan, S.; Yu, D.; Nosal, L.; Rghei, N.; Roberts, P.; Haj-Ahmad, Y. Diversity and Signature of Small RNA in Different Bodily Fluids Using next Generation Sequencing. BMC Genom. 2018, 19, 408. [Google Scholar] [CrossRef]
- Li, L. Non-Coding RNA in the Exosome of the Epithelia Ovarian Cancer; Peking Union Medical College Hospital: Beijing, China, 2018. [Google Scholar]
- ZIWIG. Evaluation of Salivary MiRNAs in the Presence of an Adnexal Mass of Ovarian Origin—OVAmiARN Study. 2022. Available online: https://ichgcp.net/clinical-trials-registry/NCT05514028 (accessed on 14 January 2023).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabi, Y.; Favier, A.; Razakamanantsoa, L.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Touboul, C.; Bendifallah, S.; Daraï, E. Insight on Non-Coding RNAs from Biofluids in Ovarian Tumors. Cancers 2023, 15, 1539. https://doi.org/10.3390/cancers15051539
Dabi Y, Favier A, Razakamanantsoa L, Delbos L, Poilblanc M, Descamps P, Golfier F, Touboul C, Bendifallah S, Daraï E. Insight on Non-Coding RNAs from Biofluids in Ovarian Tumors. Cancers. 2023; 15(5):1539. https://doi.org/10.3390/cancers15051539
Chicago/Turabian StyleDabi, Yohann, Amélia Favier, Léo Razakamanantsoa, Léa Delbos, Mathieu Poilblanc, Philippe Descamps, Francois Golfier, Cyril Touboul, Sofiane Bendifallah, and Emile Daraï. 2023. "Insight on Non-Coding RNAs from Biofluids in Ovarian Tumors" Cancers 15, no. 5: 1539. https://doi.org/10.3390/cancers15051539
APA StyleDabi, Y., Favier, A., Razakamanantsoa, L., Delbos, L., Poilblanc, M., Descamps, P., Golfier, F., Touboul, C., Bendifallah, S., & Daraï, E. (2023). Insight on Non-Coding RNAs from Biofluids in Ovarian Tumors. Cancers, 15(5), 1539. https://doi.org/10.3390/cancers15051539






