Impact of Stain Normalization on Pathologist Assessment of Prostate Cancer: A Comparative Study
Abstract
Simple Summary
Abstract
1. Introduction
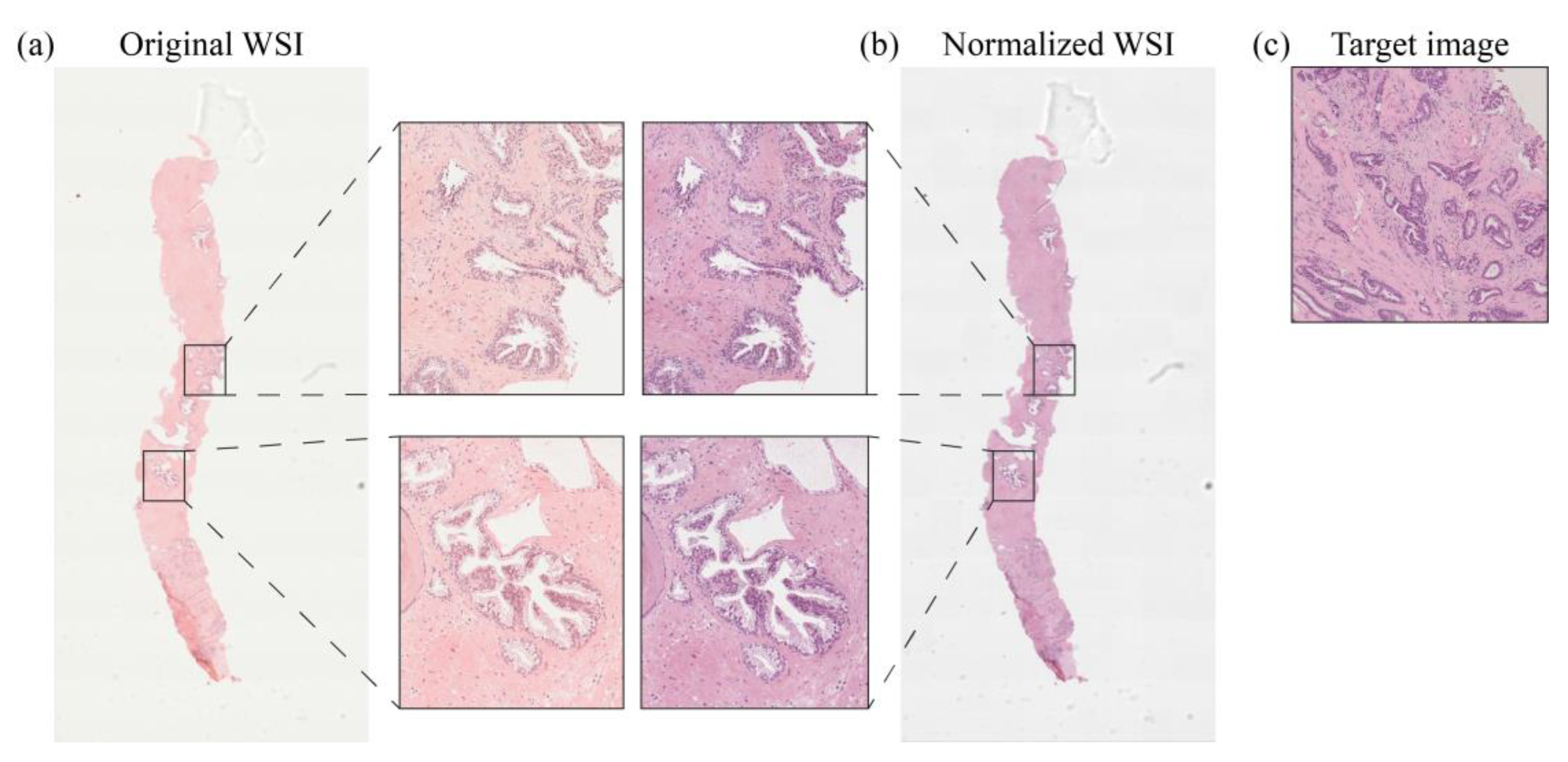
2. Materials and Methods
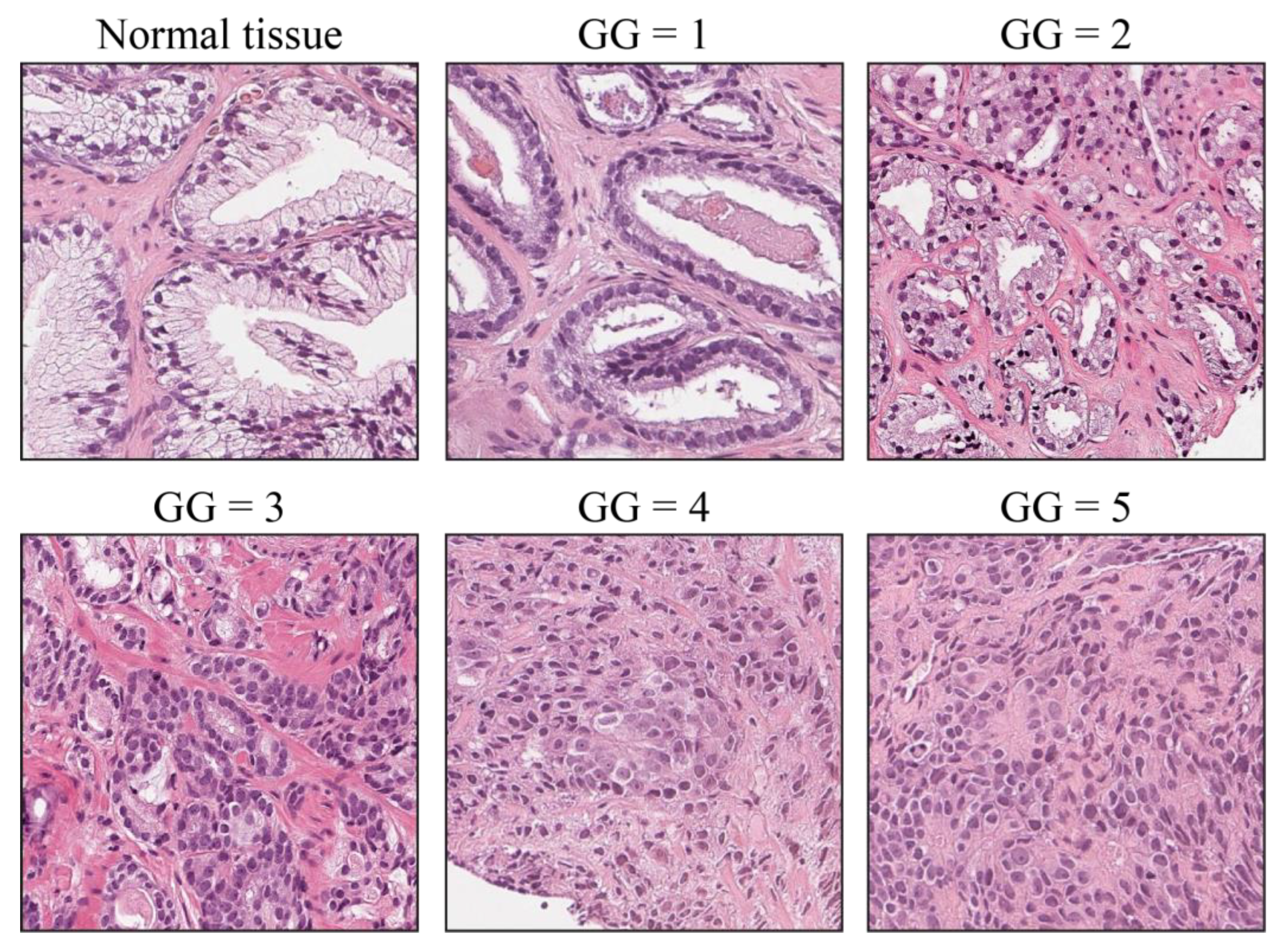
2.1. Dataset
2.2. Clinical Study Description
- Assessment of perceived stain color quality: it is quantified with a numerical scale from 1 to 10, where 1 indicates a low-quality image and 10 a high-quality image.
- Confidence in the given diagnosis: rated subjectively from 1 to 10, where 1 indicates a low degree of reliability in diagnosis and 10 denotes a high degree of confidence. Operatively, a high-confidence diagnosis occurs when the pathologist thinks that the given slide is sufficient to perform a diagnosis, whereas low confidence indicates that the pathologist is not fully convinced by the appearance of the examined slide and would resort to recuts or immunohistochemical analysis.
- Time required for diagnosis: it is expressed in seconds and indicates the time taken by the pathologist to examine the image in order to decide the diagnostic classification. It was measured from the time when the image is opened (i.e., when the pathologist starts examining the image) to the time when the diagnosis is formulated; after that, the pathologist stops examining the image and writes down the diagnosis. The time required for image loading, thus, was not considered.
3. Results
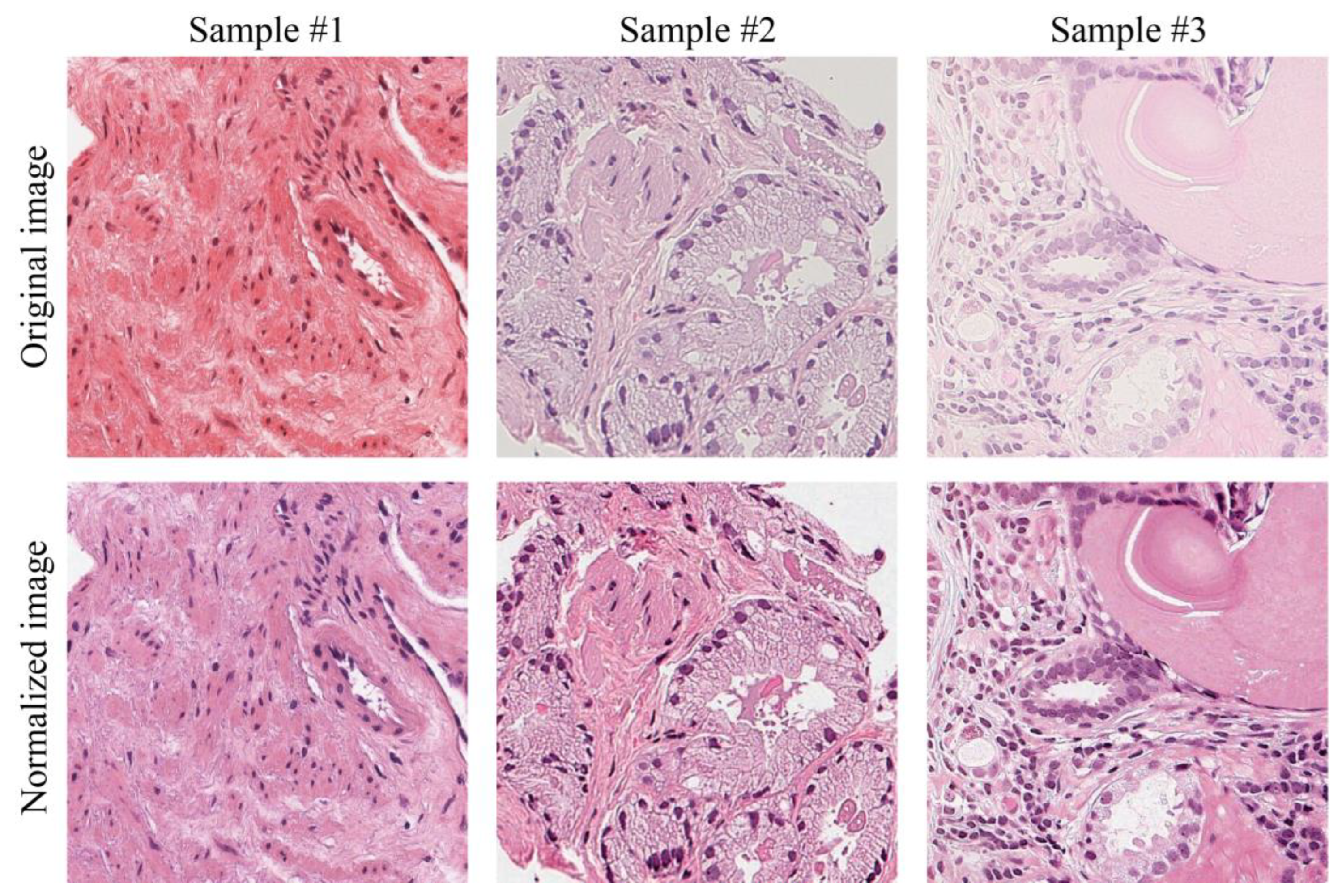
3.1. Evaluation of the Color Quality
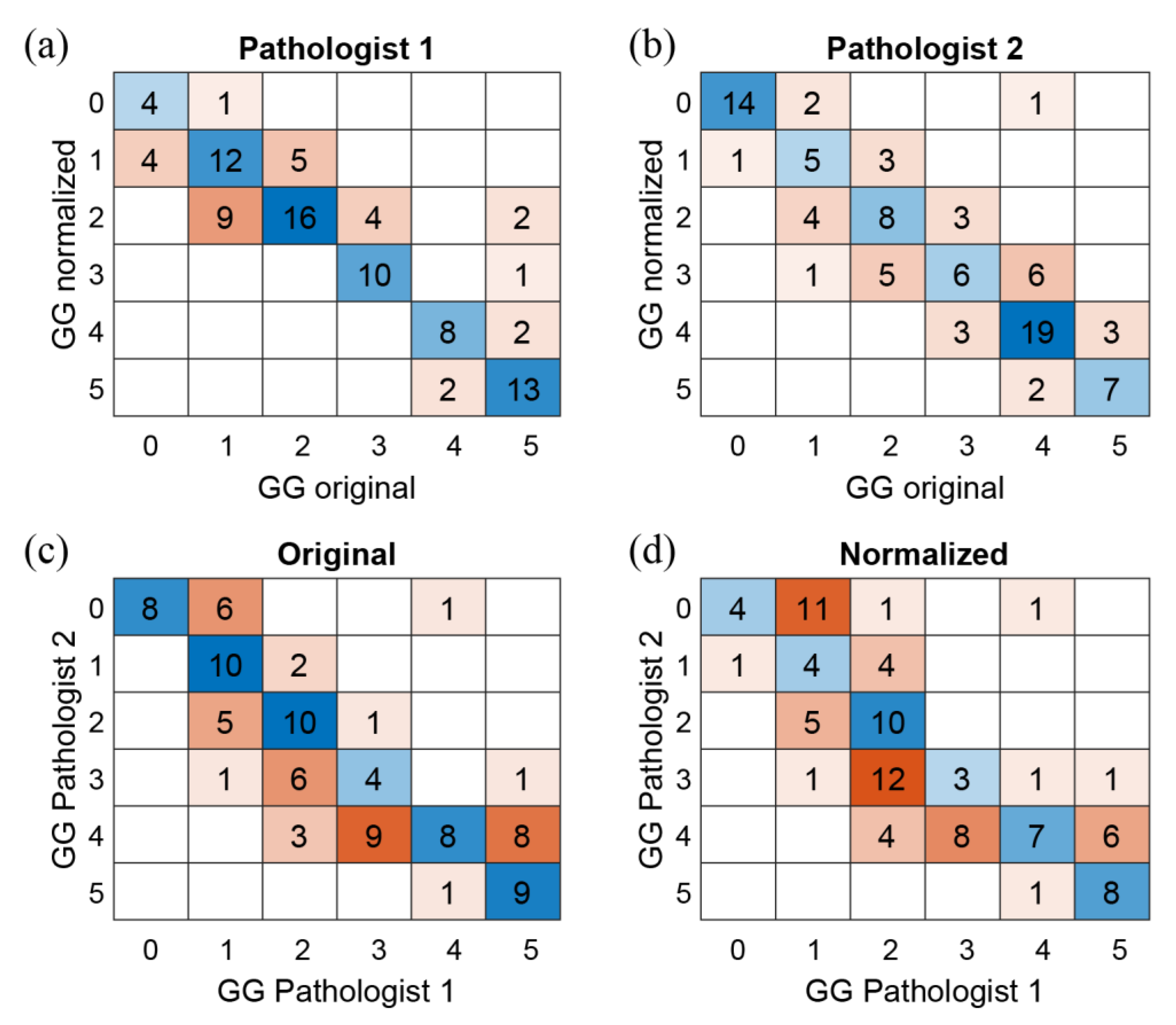
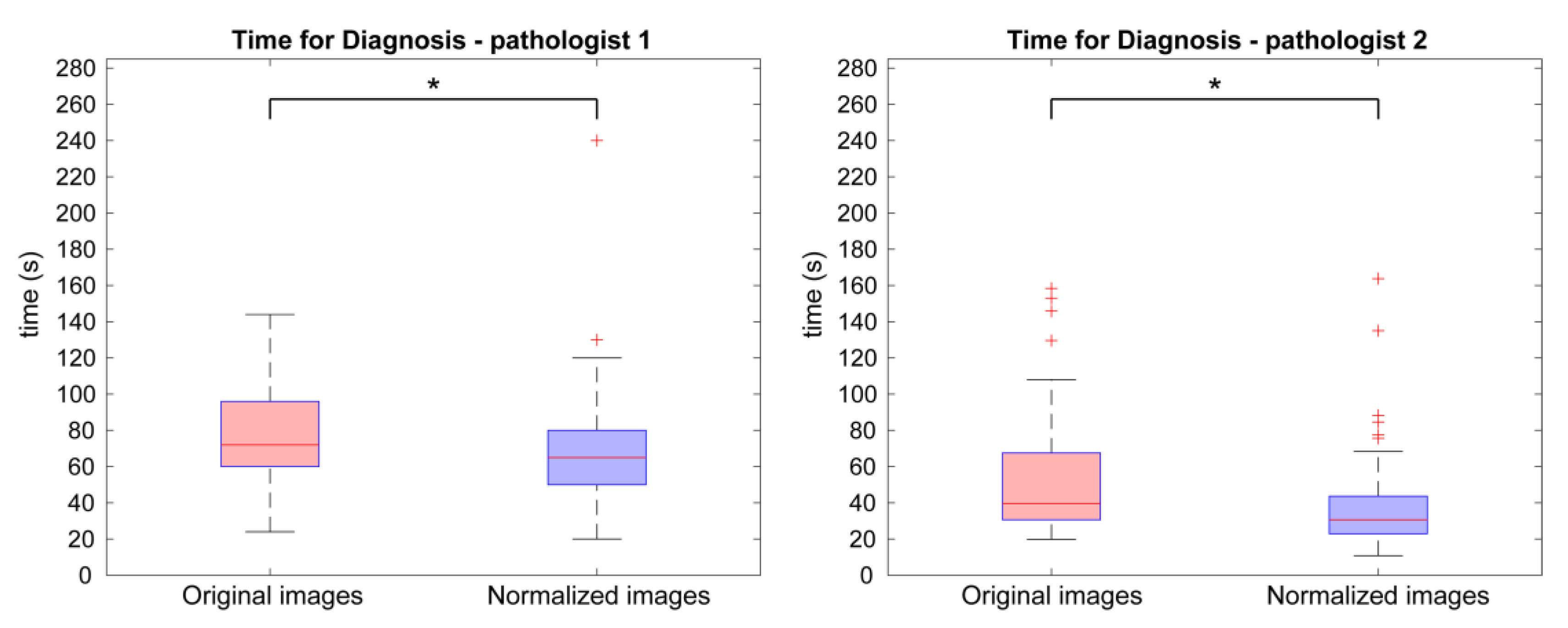
3.2. Assessment of Prostate Cancer: Diagnosis, Time and Confidence
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon. Fr. Int. Agency Res. Cancer 2018, 3, 2019. [Google Scholar]
- WHO. Classification of Tumours Editorial Board. In Urinary and Male Genital Tumours, 5th ed.; World Health Organization: Geneva, Switzerland, 2022; Volume 8, ISBN 978-92-832-4512-4. [Google Scholar]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Gleason, D.F. Classification of Prostatic Carcinomas. Cancer Chemother. Rep. 1966, 50, 125–128. [Google Scholar] [PubMed]
- Kirmiz, S.; Qi, J.; Babitz, S.K.; Linsell, S.; Denton, B.; Singh, K.; Auffenberg, G.; Montie, J.E.; Lane, B.R. Grade Groups Provide Improved Predictions of Pathological and Early Oncologic Outcomes Compared with Gleason Score Risk Groups. J. Urol. 2019, 201, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Knipper, S.; Karakiewicz, P.I.; Heinze, A.; Preisser, F.; Steuber, T.; Huland, H.; Graefen, M.; Tilki, D. Definition of High-Risk Prostate Cancer Impacts Oncological Outcomes after Radical Prostatectomy. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 184–190. [Google Scholar] [CrossRef]
- Leong, J.Y.; Herrera-Caceres, J.O.; Goldberg, H.; Tham, E.; Teplitsky, S.; Gomella, L.G.; Trabulsi, E.J.; Lallas, C.D.; Fleshner, N.E.; Tilki, D.; et al. Questioning the Status Quo: Should Gleason Grade Group 1 Prostate Cancer Be Considered a “Negative Core” in Pre-Radical Prostatectomy Risk Nomograms? An International Multicenter Analysis. Urology 2020, 137, 102–107. [Google Scholar] [CrossRef]
- Pukl, M.; Keyes, S.; Keyes, M.; Guillaud, M.; Volavšek, M. Multi-Scale Tissue Architecture Analysis of Favorable-Risk Prostate Cancer: Correlation with Biochemical Recurrence. Investig. Clin. Urol. 2020, 61, 482–490. [Google Scholar] [CrossRef]
- Wenzel, M.; Würnschimmel, C.; Chierigo, F.; Mori, K.; Tian, Z.; Terrone, C.; Shariat, S.F.; Saad, F.; Tilki, D.; Graefen, M.; et al. Pattern of Biopsy Gleason Grade Group 5 (4 + 5 vs. 5 + 4 vs. 5 + 5) Predicts Survival After Radical Prostatectomy or External Beam Radiation Therapy. Eur. Urol. Focus 2022, 8, 710–717. [Google Scholar] [CrossRef]
- Preisser, F.; Wang, N.; Abrams-Pompe, R.S.; Chun, F.K.-H.; Graefen, M.; Huland, H.; Tilki, D. Oncologic Outcomes of Organ-Confined Gleason Grade Group 4-5 Prostate Cancer after Radical Prostatectomy. Urol. Oncol. 2022, 40, 161.e9–161.e14. [Google Scholar] [CrossRef]
- Borowsky, A.D.; Glassy, E.F.; Wallace, W.D.; Kallichanda, N.S.; Behling, C.A.; Miller, D.V.; Oswal, H.N.; Feddersen, R.M.; Bakhtar, O.R.; Mendoza, A.E.; et al. Digital Whole Slide Imaging Compared With Light Microscopy for Primary Diagnosis in Surgical Pathology. Arch. Pathol. Lab. Med. 2020, 144, 1245–1253. [Google Scholar] [CrossRef]
- Camparo, P.; Egevad, L.; Algaba, F.; Berney, D.M.; Boccon-Gibod, L.; Compérat, E.; Evans, A.J.; Grobholz, R.; Kristiansen, G.; Langner, C.; et al. Utility of Whole Slide Imaging and Virtual Microscopy in Prostate Pathology. APMIS 2012, 120, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Feldman, M.D.; Abels, E.; Ashfaq, R.; Beltaifa, S.; Cacciabeve, N.G.; Cathro, H.P.; Cheng, L.; Cooper, K.; Dickey, G.E.; et al. Whole Slide Imaging Versus Microscopy for Primary Diagnosis in Surgical Pathology: A Multicenter Blinded Randomized Noninferiority Study of 1992 Cases (Pivotal Study). Am. J. Surg. Pathol. 2018, 42, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.; Allsbrook, W.C.J.; Kurokawa, K.; Matsuda, H.; Segawa, A.; Sano, T.; Suzuki, K.; Epstein, J.I. A Comparison of Interobserver Reproducibility of Gleason Grading of Prostatic Carcinoma in Japan and the United States. Arch. Pathol. Lab. Med. 2005, 129, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Allsbrook, W.C.; Mangold, K.A.; Johnson, M.H.; Lane, R.B.; Lane, C.G.; Amin, M.B.; Bostwick, D.G.; Humphrey, P.A.; Jones, E.C.; Reuter, V.E.; et al. Interobserver Reproducibility of Gleason Grading of Prostatic Carcinoma: Urologic Pathologists. Hum. Pathol. 2001, 32, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, T.A.; Eruyar, A.T.; Cebeci, O.O.; Memik, O.; Ozcan, L.; Kuskonmaz, I. Interobserver Variability in Gleason Histological Grading of Prostate Cancer. Scand. J. Urol. 2016, 50, 420–424. [Google Scholar] [CrossRef]
- Fraggetta, F.; L’Imperio, V.; Ameisen, D.; Carvalho, R.; Leh, S.; Kiehl, T.-R.; Serbanescu, M.; Racoceanu, D.; Della Mea, V.; Polonia, A.; et al. Best Practice Recommendations for the Implementation of a Digital Pathology Workflow in the Anatomic Pathology Laboratory by the European Society of Digital and Integrative Pathology (ESDIP). Diagnostics 2021, 11, 2167. [Google Scholar] [CrossRef]
- Rathore, S.; Iftikhar, M.A.; Chaddad, A.; Niazi, T.; Karasic, T.; Bilello, M. Segmentation and Grade Prediction of Colon Cancer Digital Pathology Images Across Multiple Institutions. Cancers 2019, 11, 1700. [Google Scholar] [CrossRef]
- Jang, H.-J.; Song, I.-H.; Lee, S.-H. Deep Learning for Automatic Subclassification of Gastric Carcinoma Using Whole-Slide Histopathology Images. Cancers 2021, 13, 1811. [Google Scholar] [CrossRef]
- Lyon, H.O.; De Leenheer, A.P.; Horobin, R.W.; Lambert, W.E.; Schulte, E.K.; Van Liedekerke, B.; Wittekind, D.H. Standardization of Reagents and Methods Used in Cytological and Histological Practice with Emphasis on Dyes, Stains and Chromogenic Reagents. Histochem. J. 1994, 26, 533–544. [Google Scholar] [CrossRef]
- Roy, S.; kumar Jain, A.; Lal, S.; Kini, J. A Study about Color Normalization Methods for Histopathology Images. Micron 2018, 114, 42–61. [Google Scholar] [CrossRef]
- Salehi, P.; Chalechale, A. Pix2pix-Based Stain-to-Stain Translation: A Solution for Robust Stain Normalization in Histopathology Images Analysis. In Proceedings of the 2020 International Conference on Machine Vision and Image Processing (MVIP), Qom, Iran, 18–20 February 2020; pp. 1–7. [Google Scholar]
- Gurcan, M.N.; Boucheron, L.E.; Can, A.; Madabhushi, A.; Rajpoot, N.M.; Yener, B. Histopathological Image Analysis: A Review. IEEE Rev. Biomed. Eng. 2009, 2, 147–171. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.R. Immunohistochemistry in Surgical Pathology: Principles and Practice. In Histopathology: Methods and Protocols; Day, C.E., Ed.; Springer: New York, NY, USA, 2014; pp. 81–109. ISBN 978-1-4939-1050-2. [Google Scholar]
- Michielli, N.; Caputo, A.; Scotto, M.; Mogetta, A.; Pennisi, O.A.M.; Molinari, F.; Balmativola, D.; Bosco, M.; Gambella, A.; Metovic, J.; et al. Stain Normalization in Digital Pathology: Clinical Multi-Center Evaluation of Image Quality. J. Pathol. Inform. 2022, 13, 100145. [Google Scholar] [CrossRef] [PubMed]
- Salvi, M.; Michielli, N.; Molinari, F. Stain Color Adaptive Normalization (SCAN) Algorithm: Separation and Standardization of Histological Stains in Digital Pathology. Comput. Methods Programs Biomed. 2020, 193, 105506. [Google Scholar] [CrossRef] [PubMed]
- Pontalba, J.T.; Gwynne-Timothy, T.; David, E.; Jakate, K.; Androutsos, D.; Khademi, A. Assessing the Impact of Color Normalization in Convolutional Neural Network-Based Nuclei Segmentation Frameworks. Front. Bioeng. Biotechnol. 2019, 7, 300. [Google Scholar] [CrossRef]
- Swiderska-Chadaj, Z.; de Bel, T.; Blanchet, L.; Baidoshvili, A.; Vossen, D.; van der Laak, J.; Litjens, G. Impact of Rescanning and Normalization on Convolutional Neural Network Performance in Multi-Center, Whole-Slide Classification of Prostate Cancer. Sci. Rep. 2020, 10, 14398. [Google Scholar] [CrossRef]
- Salvi, M.; Bosco, M.; Molinaro, L.; Gambella, A.; Papotti, M.; Acharya, U.R.; Molinari, F. A Hybrid Deep Learning Approach for Gland Segmentation in Prostate Histopathological Images. Artif. Intell. Med. 2021, 115, 102076. [Google Scholar] [CrossRef]
- Janowczyk, A.; Madabhushi, A. Deep Learning for Digital Pathology Image Analysis: A Comprehensive Tutorial with Selected Use Cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Salvi, M.; Molinari, F.; Acharya, U.R.; Molinaro, L.; Meiburger, K.M. Impact of Stain Normalization and Patch Selection on the Performance of Convolutional Neural Networks in Histological Breast and Prostate Cancer Classification. Comput. Methods Progr. Biomed. Updat. 2021, 1, 100004. [Google Scholar] [CrossRef]
- Liu, F.; Hernandez-Cabronero, M.; Sanchez, V.; Marcellin, M.W.; Bilgin, A. The Current Role of Image Compression Standards in Medical Imaging. Information 2017, 8, 131. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open Source Software for Digital Pathology Image Analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvi, M.; Caputo, A.; Balmativola, D.; Scotto, M.; Pennisi, O.; Michielli, N.; Mogetta, A.; Molinari, F.; Fraggetta, F. Impact of Stain Normalization on Pathologist Assessment of Prostate Cancer: A Comparative Study. Cancers 2023, 15, 1503. https://doi.org/10.3390/cancers15051503
Salvi M, Caputo A, Balmativola D, Scotto M, Pennisi O, Michielli N, Mogetta A, Molinari F, Fraggetta F. Impact of Stain Normalization on Pathologist Assessment of Prostate Cancer: A Comparative Study. Cancers. 2023; 15(5):1503. https://doi.org/10.3390/cancers15051503
Chicago/Turabian StyleSalvi, Massimo, Alessandro Caputo, Davide Balmativola, Manuela Scotto, Orazio Pennisi, Nicola Michielli, Alessandro Mogetta, Filippo Molinari, and Filippo Fraggetta. 2023. "Impact of Stain Normalization on Pathologist Assessment of Prostate Cancer: A Comparative Study" Cancers 15, no. 5: 1503. https://doi.org/10.3390/cancers15051503
APA StyleSalvi, M., Caputo, A., Balmativola, D., Scotto, M., Pennisi, O., Michielli, N., Mogetta, A., Molinari, F., & Fraggetta, F. (2023). Impact of Stain Normalization on Pathologist Assessment of Prostate Cancer: A Comparative Study. Cancers, 15(5), 1503. https://doi.org/10.3390/cancers15051503








