RBM15 Promates the Proliferation, Migration and Invasion of Pancreatic Cancer Cell Lines
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Dataset Sourcing and Pre-Processing of Healthy and Pancreatic Cancer Patients
2.2. Construction of m6A Gene Expression Differential Analysis
2.3. Construction of m6A Typing, Sample Gene Expression Differences, and Immune Cell Infiltration Enrichment Analysis
2.4. Construction of Correlation Analysis between m6A-Related Genes and Patients’ Clinical Data
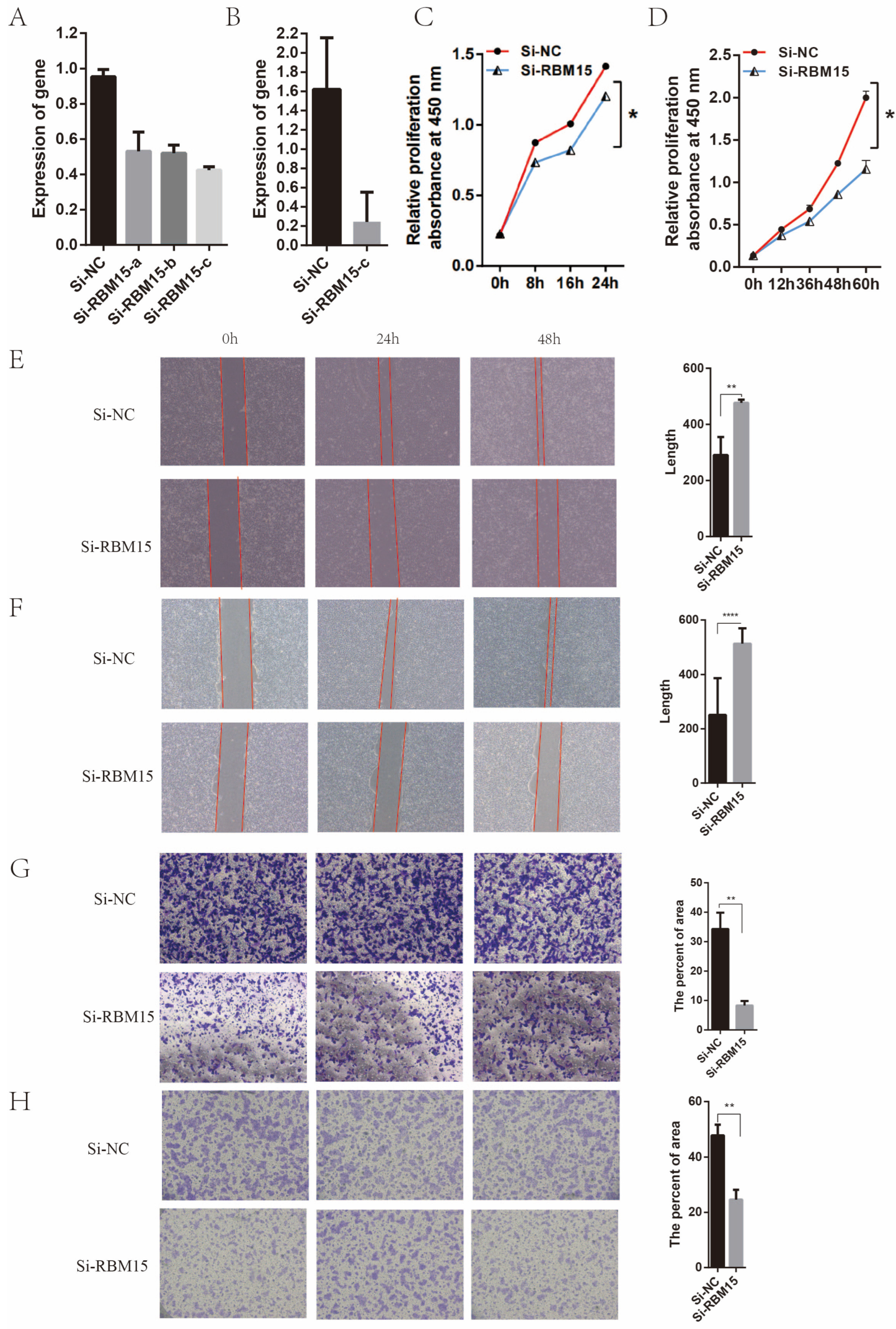
2.5. Experimental Testing
2.6. Statistical Analysis and Image Plotting
3. Results
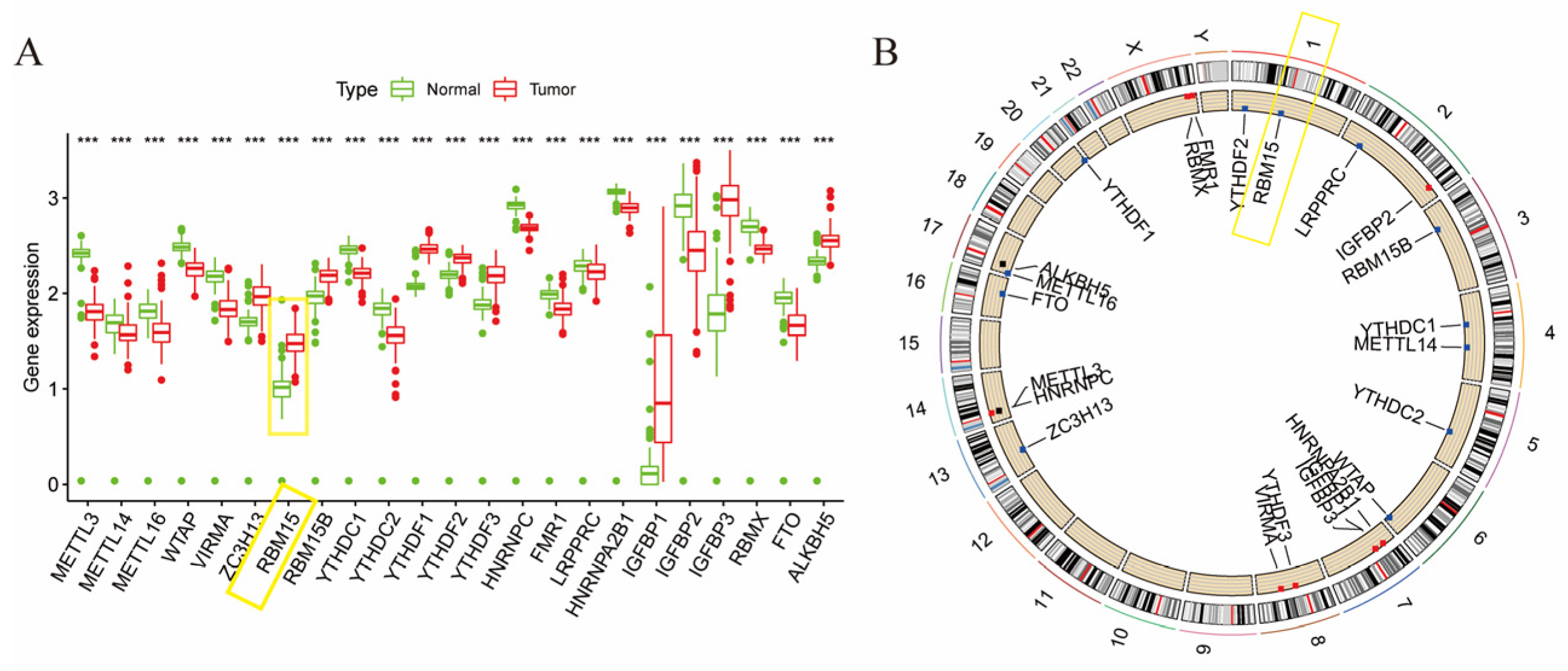
3.1. Expression of m6A Gene in Pancreatic Cancer
3.2. m6A Gene and Patient Prognosis in Pancreatic Cancer
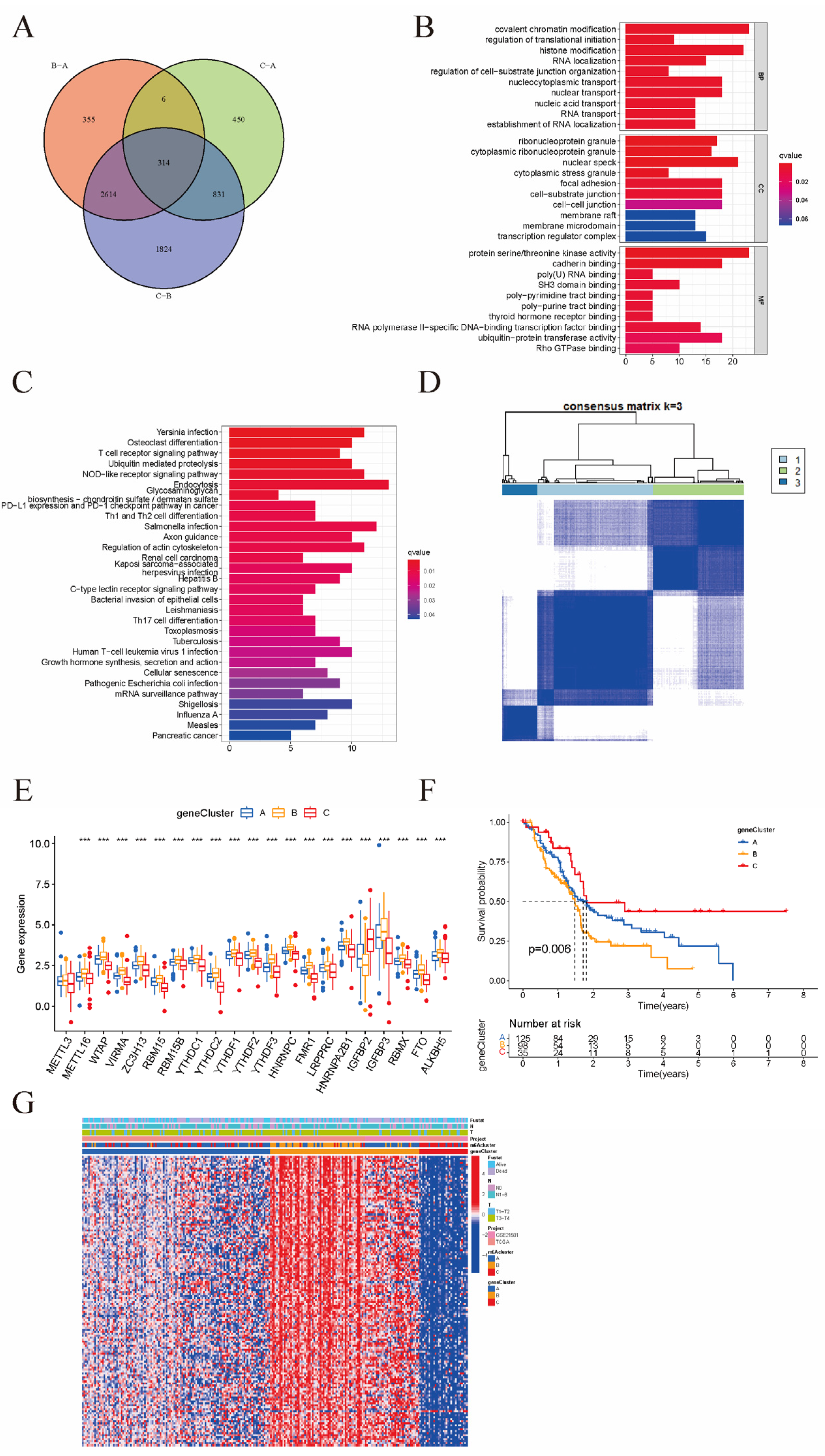
3.3. Patterns of m6A Methylation Modifications
3.4. Clinical and Transcriptomic Features of m6A-Related Phenotypes
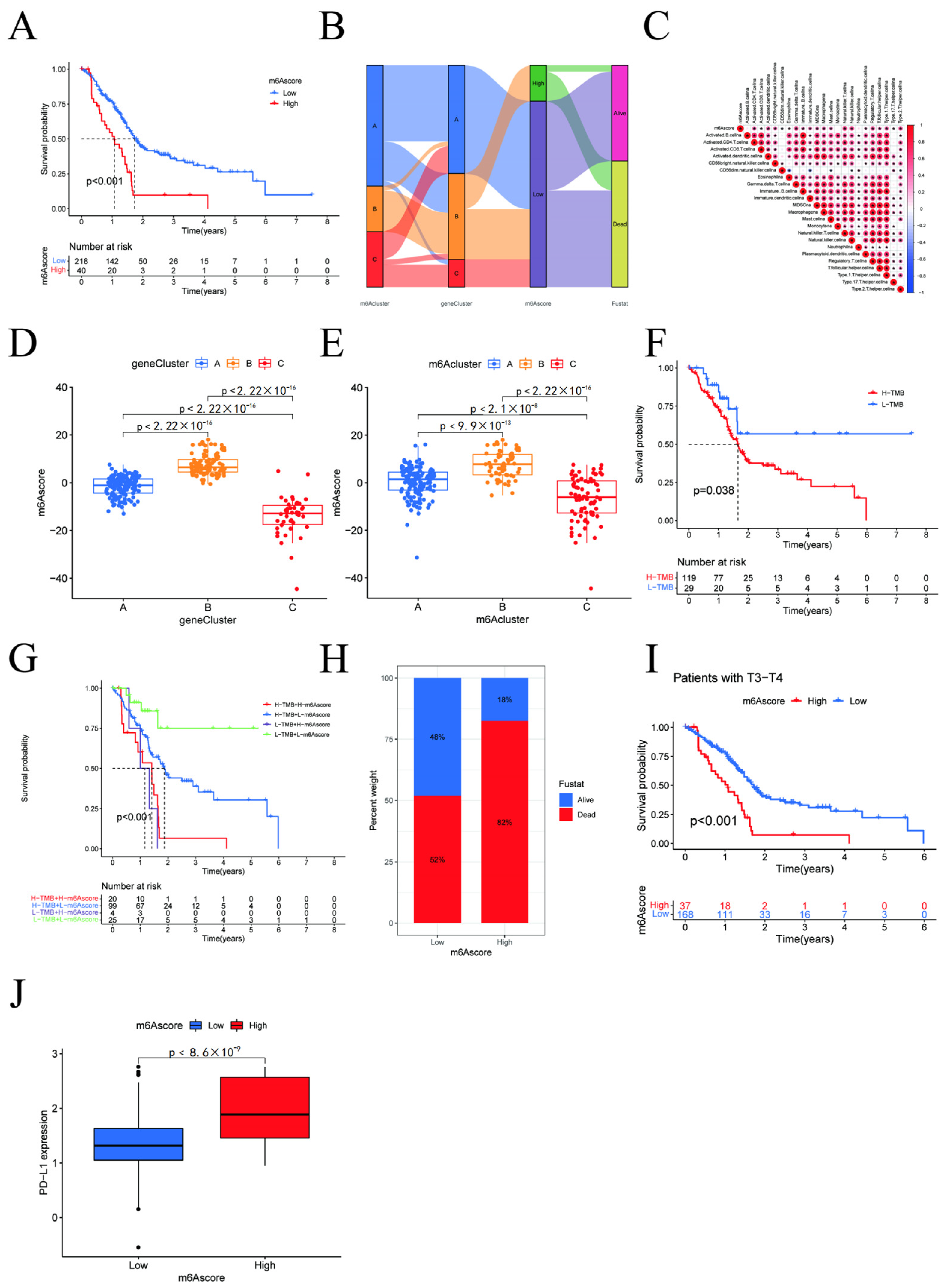
3.5. m6A Score and Clinicopathological Characteristics of Pancreatic Cancer Patients
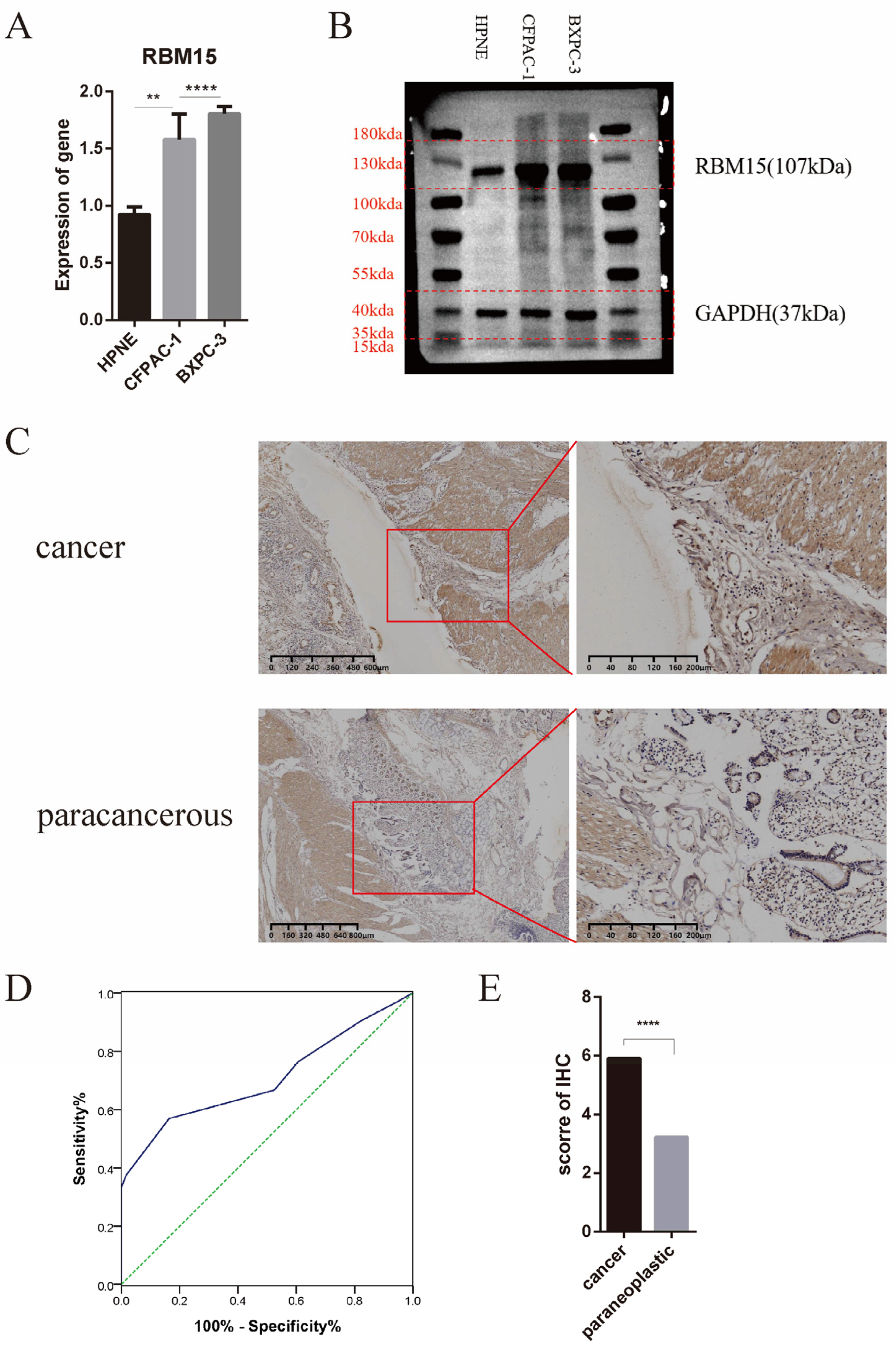
3.6. Clinical Significance of High Expression of RBM15 in Pancreatic Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Early Diagnosis and Treatment Group of the Chinese Medical Association Oncology Branch. Expert consensus on early diagnosis and treatment of pancreatic cancer of the Chinese Medical Association Oncology Branch. J. Clin. Hepatobiliary Dis. 2020, 36, 2675–2680. [Google Scholar]
- Siegel, R.; Miller, K.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wu, Q.; Li, B.; Wang, D.; Wang, L.; Zhou, Y.L. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 2020, 10, 1186. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell. Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.R.; Lee, H.; Goodarzi, H. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millán-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.G.; et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef]
- Chen, M.; Wei, L.; Law, C.T.; Tsang, F.H.; Shen, J.; Cheng, C.L.; Tsang, L.H.; Ho, D.W.; Chiu, D.K.; Lee, J.M.; et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018, 67, 2254–2270. [Google Scholar] [CrossRef]
- Cai, X.; Wang, X.; Cao, C.; Gao, Y.; Zhang, S.; Yang, Z.; Liu, Y.; Zhang, X.; Zhang, W.; Ye, L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018, 415, 11–19. [Google Scholar] [CrossRef]
- Su, H.R.; Liu, Y.Y.; Zhao, X.Y. Split end family RNA binding proteins: Novel Tumor Suppressors Coupling Transcriptional Regulation with RNA Processing. Cancer Transl. Med. 2015, 1, 21–25. [Google Scholar]
- Zhang, L.; Tran, N.-T.; Su, H.; Wang, R.; Lu, Y.; Tang, H.; Aoyagi, S.; Guo, A.; Khodadadi-Jamayran, A.; Zhou, D.; et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. eLife 2015, 4, e07938. [Google Scholar] [CrossRef]
- Horiuchi, K.; Kawamura, T.; Iwanari, H.; Ohashi, R.; Naito, M.; Kodama, T.; Hamakubo, T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013, 288, 33292–33302. [Google Scholar] [CrossRef]
- Niu, C.; Zhang, J.; Breslin, P.; Onciu, M.; Ma, Z.; Morris, S.W. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood 2009, 114, 2087–2096. [Google Scholar] [CrossRef]
- Hu, L.; Li, H.; Chi, Z.; He, J. Loss of the RNA-binding protein Rbm15 disrupts liver maturation in zebrafish. J. Biol. Chem. 2020, 295, 11466–11472. [Google Scholar] [CrossRef]
- Xu, F.; Guan, Y.; Ma, Y.; Xue, L.; Zhang, P.; Yang, X.; Chong, T. Bioinformatic analyses and experimental validation of the role of m6A RNA methylation regulators in progression and prognosis of adrenocortical carcinoma. Aging 2021, 13, 11919–11941. [Google Scholar] [CrossRef]
- Zhang, X.; Zhong, L.; Zou, Z.; Liang, G.; Tang, Z.; Li, K.; Tan, S.; Huang, Y.; Zhu, X. Clinical and Prognostic Pan-Cancer Analysis of N6-Methyladenosine Regulators in Two Types of Hematological Malignancies: A Retrospective Study Based on TCGA and GTEx Databases. Front. Oncol. 2021, 11, 623170. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, X.; Lu, Z. m6A RNA Methylation Regulators Act as Potential Prognostic Biomarkers in Lung Adenocarcinoma. Front. Genet. 2021, 12, 622233. [Google Scholar] [CrossRef]
- Wang, X.; Tian, L.; Li, Y.; Wang, J.; Yan, B.; Yang, L.; Li, Q.; Zhao, R.; Liu, M.; Wang, P.; et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J. Exp. Clin. Cancer Res. 2021, 40, 80. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, Z.; Liu, Y.; Yang, T.; Cao, M.; Jiang, Y.; Tang, Y.; Chen, H.; Zhang, W.; Gong, R.; et al. Novel Recurrent Altered Genes in Chinese Patients with Anaplastic Thyroid Cancer. J. Clin. Endocrinol. Metab. 2021, 106, 988–998. [Google Scholar] [CrossRef]
- Li, H.B.; Tong, J.; Zhu, S.; Batista, P.J.; Duffy, E.E.; Zhao, J.; Bailis, W.; Cao, G.; Kroehling, L.; Chen, Y.; et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017, 548, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yang, Y.; Guo, W.; Xu, L.; You, M.; Zhang, Y.C.; Sun, Z.; Cui, X.; Yu, G.; Qi, Z.; et al. METTL3-dependent m(6)A modification programs T follicular helper cell differentiation. Nat. Commun. 2021, 12, 1333. [Google Scholar] [CrossRef] [PubMed]
- McGrail, D.J.; Pilié, P.G.; Rashid, N.U.; Voorwerk, L.; Slagter, M.; Kok, M.; Jonasch, E.; Khasraw, M.; Heimberger, A.B.; Lim, B.; et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 2021, 32, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Addeo, A.; Banna, G.L.; Weiss, G.J. Tumor Mutation Burden—From Hopes to Doubts. JAMA Oncol. 2019, 5, 934–935. [Google Scholar] [CrossRef]
- Meiri, E.; Garrett-Mayer, E.; Halabi, S.; Mangat, P.K.; Shrestha, S.; Ahn, E.R.; Osayameh, O.; Perla, V.; Schilsky, R.L. Pembrolizumab (P) in patients (Pts) with colorectal cancer (CRC) with high tumor mutational burden (HTMB): Results from the Targeted Agent and Profiling Utilization Registry (TAPUR) Study. J. Clin. Oncol. 2020, 39, 2443–2451. [Google Scholar] [CrossRef]
- Strickler, J.H.; Hanks, B.A.; Khasraw, M. Tumor Mutational Burden as a Predictor of Immunotherapy Response: Is More Always Better? Clin. Cancer Res. 2021, 27, 1236–1241. [Google Scholar] [CrossRef]
- Lamkin, D.M.; Spitz, D.R.; Shahzad, M.M.; Zimmerman, B.; Lenihan, D.J.; DeGeest, K.; Lubaroff, D.M.; Shinn, E.H.; Sood, A.K.; Lutgendorf, S.K. Glucose as a prognostic factor in ovarian carcinoma. Cancer 2009, 115, 1021–1027. [Google Scholar] [CrossRef]



| No. of Patients | IHC Score | χ2 | p-Value | |||
|---|---|---|---|---|---|---|
| <5 | >5 | |||||
| Gender | Female | 30 | 17 (56.7) | 13 (43.3) | 3.886 | 0.049 |
| Male | 42 | 14 (33.3) | 28 (66.7) | |||
| Age | <70 years old | 45 | 20 (44.4) | 25 (55.6) | 0.094 | 0.759 |
| >70 years old | 27 | 11 (40.7) | 16 (59.3) | |||
| Smoking history | No | 58 | 23 (39.7) | 35 (60.3) | 1.407 | 0.236 |
| Yes | 14 | 8 (57.1) | 6 (42.9) | |||
| History of alcohol consumption | No | 62 | 26 (41.9) | 36 (58.1) | 0.228 | 0.633 |
| Yes | 10 | 5 (50.0) | 5 (50.0) | |||
| Family history | None | 72 | 31 (43.1) | 41 (56.9) | / | >0.05 |
| Venous blood glucose level | <7 mmol/L | 40 | 22 (55.0) | 18 (45.0) | 6.1 | 0.014 |
| >7 mmol/L | 31 | 8 (25.8) | 23 (74.2) | |||
| CA199 | <40 | 14 | 4 (28.6) | 10 (71.4) | 0.781 | 0.377 |
| >40 | 53 | 22 (41.5) | 31 (58.5) | |||
| Tumor with or without vascular invasion | No | 41 | 17 (41.5) | 24 (58.5) | 0.033 | 0.856 |
| Yes | 28 | 11 (39.3) | 17 (60.7) | |||
| Lymph node metastasis | No | 35 | 10 (28.6) | 25 (71.4) | 1.82 | 0.177 |
| Yes | 29 | 13 (44.8) | 16 (55.2) | |||
| Lymphocyte count | 1.1–3.2 | 58 | 21 (36.2) | 37 (63.8) | 7.157 | 0.007 |
| <1.1 | 13 | 10 (76.9) | 3 (23.1) | |||
| Tumor size | <3 cm | 20 | 8 (40.0) | 12 (60.0) | 0.038 | 0.846 |
| >3 cm | 47 | 20 (42.6) | 27 (57.4) | |||
| Neutrophil ratio | 40–75% | 62 | 27 (43.5) | 35 (56.5) | 0.12 | 0.73 |
| >75% | 8 | 4 (50.0) | 4 (50.0) | |||
| TNM | 0–II | 44 | 17 (29.3) | 27 (46.6) | 4.59 | 0.032 |
| III–IV | 14 | 10 (17.2) | 4 (6.9) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, H.; Zhang, H.; Mao, X.; Liu, S.; Xu, W.; Zhang, Y. RBM15 Promates the Proliferation, Migration and Invasion of Pancreatic Cancer Cell Lines. Cancers 2023, 15, 1084. https://doi.org/10.3390/cancers15041084
Dong H, Zhang H, Mao X, Liu S, Xu W, Zhang Y. RBM15 Promates the Proliferation, Migration and Invasion of Pancreatic Cancer Cell Lines. Cancers. 2023; 15(4):1084. https://doi.org/10.3390/cancers15041084
Chicago/Turabian StyleDong, Hui, Haidong Zhang, Xinyu Mao, Shiwei Liu, Wenjing Xu, and Yewei Zhang. 2023. "RBM15 Promates the Proliferation, Migration and Invasion of Pancreatic Cancer Cell Lines" Cancers 15, no. 4: 1084. https://doi.org/10.3390/cancers15041084
APA StyleDong, H., Zhang, H., Mao, X., Liu, S., Xu, W., & Zhang, Y. (2023). RBM15 Promates the Proliferation, Migration and Invasion of Pancreatic Cancer Cell Lines. Cancers, 15(4), 1084. https://doi.org/10.3390/cancers15041084






