Simple Summary
Endocrine tumors often cause hormonal imbalances and sometimes hormone syndrome. Generally, the prognosis of patients with metastatic endocrine malignancies is poor due to recurrence and treatment failure. Aberrant expression of m6A may disrupt the development and occurrence of malignancies. We provide a theoretical foundation for the development of new diagnostic markers and therapeutic techniques based on m6A alteration and regulators in endocrine cancers.
Abstract
With the development of RNA modification research, N6-methyladenosine (m6A) is regarded as one of the most important internal epigenetic modifications of eukaryotic mRNA. It is also regulated by methylase, demethylase, and protein preferentially recognizing the m6A modification. This dynamic and reversible post-transcriptional RNA alteration has steadily become the focus of cancer research. It can increase tumor stem cell self-renewal and cell proliferation. The m6A-modified genes may be the primary focus for cancer breakthroughs. Although some endocrine cancers are rare, they may have a high mortality rate. As a result, it is critical to recognize the significance of endocrine cancers and identify new therapeutic targets that will aid in improving disease treatment and prognosis. We summarized the latest experimental progress in the m6A modification in endocrine cancers and proposed the m6A alteration as a potential diagnostic marker for endocrine malignancies.
1. Introduction
Endocrine glands include the thyroid gland, adrenal gland, pancreas, parathyroid gland, ovary, and pituitary gland. Malignant tumors occurring in endocrine glands are called endocrine tumors [1,2]. Endocrine tumors often cause hormonal imbalances and sometimes hormone syndrome [3,4]. Although some endocrine tumors are rare in clinical practice, they can be fatal. Generally, the prognosis of patients with metastatic endocrine malignancies is poor due to recurrence and treatment failure [5].
In the 1970s, m6A was identified in eukaryotic messenger RNA (mRNA) [6,7] and viral nuclear RNA [8,9] for the first time. In eukaryotic mRNA, it is the most common dynamic internal genetic change, influencing RNA maturation, transcription, localization, translation, and metabolism [10]. It regulates many physiological and pathological processes [7]. The m6A modification has been proven to contain a preserved sequence “RRACH” ([G A] m6AC [U A C]) through m6A antibody-based affinity enrichment tests, high-throughput m6A sequencing, and methylated RNA m6A immunoprecipitation sequencing (MeRIP/m6A-seq) [11,12,13]. The m6A modification is not a random process. It is translated close to the 5’UTR or in the long exon, and it is enriched close to the termination codon and 3’UTRs [11,12]. Several enzymes catalyze the m6A modification, such as the m6A methyltransferase complex (MTC), also known as the m6A“writer”. The enzyme that removes m6A is called demethylase, also called the m6A“eraser” [8,9]. The enzyme that recognizes m6A and regulates the functions of activated m6A is called the m6A“reader” [10]. The expression of genes for writers, erasers, and readers influences m6A levels [11]. Aberrant m6A modification is associated with an increased risk of cancer.
Multiple studies have established a link between m6A variables and the development of endocrine cancers. Recent discoveries in relation to the m6A modification in endocrine cancers are discussed in this review.
2. M6A Methyltransferase
2.1. Writers
The m6A methyltransferase of RNA consists of “write in” proteins, including methyltransferase-like 3/5/14/16 (METTL3/5/14/16), the RNA binding motif protein 15/15B (RBM15/15B), Wilm’s tuner 1-associated protein (WTAP), Vir-like m6A methyltransferase-associated protein (VIRMA, also called KIAA1429), Zinc finger CCCH-type containing thirteen (ZC3H13) and Zinc finger CCHC-type containing four (ZCCHC4). The protein-based MTC is responsible for m6A installation [12] (Figure 1).
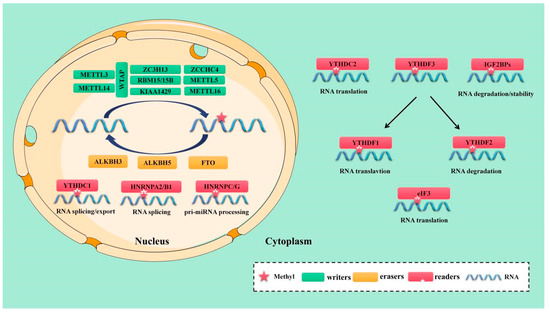
Figure 1.
The molecular mechanism of m6A mutation. m6A is installed by “writers” (METTL3/5/14/16, WTAP, RBM15/15B, KIAA1429, ZC3H13 and ZCCHC4,), removed by “erasers”(FTO, ALKBH5, and ALKBH3, termed “erasers”), and recognized by “readers”(YTHDC1/2, YTHDF1/2/3, IGF2BP1/2/3, HNRNPA2/B1, HNRNPC/G, and eIF3, termed “readers”.
Methyltransferase-like 3 (METTL3) is an S-adenosyl methyl (SAM) binding protein. In 1997, it was found that METTL3 is the main methyltransferase of m6A methylation and the most important component of the m6A MTC. Its aberrant expression can potentially alter the total methylation level of m6A [13]. METTL14 is another dynamic component of the m6A MTC. As the structural scaffold of METTL3, METTL14 can recognize specific RNA sequences (“RRACH”) and stabilize the MTC structure [13,14,15]. Both METTL3 and METTL14 are found in the nucleus as part of the central MTC and function to stabilize the complex [13]. They both cause m6A alteration in a one-to-one proportion. [13]. However, the only METTL3 serves as catalyst. To create S-adenosyl homocysteine, its internal SAM binding domain catalyzes the transfer of methyl from SAM to an adenine base in RNA (SAH). METTL16 was proposed in 2017 as an independent mRNA methyltransferase. Given that its binding site does not overlap with that of the METTL3/METTL14 methylation complex, METTL16 may function to control the stability and splicing of mRNA [16,17]. Overexpression of the METTL16 construct with a mutant catalytic domain initiated the splicing process [18]. In addition, it facilitated the attachment of m6A to the 3’UTR of mRNA and A43 to U6 MicroRNA (miRNA). A43 has been found in the ubiquitous U6”ACAGAGA”box and is known to regulate tumorigenesis by targeting forward mRNAs and non-coding RNA [19]. A previous study reported that METTL3/16 is a multifunctional enzyme with non-catalytic activity. In the absence of enzyme cofactors, the m6A writer acts as a reader and combines with the unmodified substrate constitutively to trigger the non-catalytic function [19]. A novel methyltransferase called METTL5 was recently discovered and is responsible for altering the 18S Ribosomal RNA (rRNA) m6A [20,21]. Like the METTL3/METTL14 complex, METTL5 is a co-activator of the TRNA methyltransferase activator subunit 11-2 (TRMT112). METTL5 can form heterodimers with TRMT112, which improves metabolic durability and the modified region of 18S rRNA precursors and mature forms [20].
The adaptor protein WTAP interacts with METTL3 and METTL14. Although it has no catalytic activity, it stabilizes the central complex. By attracting METTL3 and METTL14 to nuclear speckles, it mainly enhances the implantation of m6A [22,23]. METTL3 interacts with RNA-binding motif protein 15 (RBM15) and RBM15B in a WTAP-dependent way, directing these two proteins to particular RNA locations for methylation, but they do not have any enzymatic activity themselves [24,25]. In addition, immunoprecipitation studies have identified 26 core interaction factors between a large number of direct repeat erythroid-definitive (DREDs) of WTAP-binding proteins, and more than 100 proteins may bind to METTL3 or METTL14 [26]. MTC remains in the nuclear speckles after ZC3H13 interacts with WTAP via its low-complexity domain, which enhances its catalytic function [25,27]. After enlisting MTC, VIRMA discovered mRNA methylation close to the 3’ UTR and termination codon areas. Cleavage and polyadenylation-specific factor bundles 5 (CPSF5) and 6 (CPSF6) can also interact with it [28]. Recently, ZCCHC4 was discovered to function as a new methyltransferase that regulates the distribution and overall translation of rRNA ribosomal subunits by altering 28S rRNA [29].
2.2. Erasers
The enzyme known as m6A demethylase, which controls the reversible and dynamic modification of m6A, is referred to as an “eraser” [30,31]. The amount of m6A in cells is modulated by the m6A demethylase and m6A tags from mRNA can be removed selectively through complex intermediary processes, thus affecting several biological processes of tumor cells. m6A demethylase is mainly composed of fat mass and obesity-associated protein (FTO) [31] and Alk B homolog 5 (ALKBH5) [30]. These two proteins are predominantly found in the nucleus and belong to the α- ketoglutarate-dependent dioxygenase family, with Fe (II) and α-ketoglutarate catalyzing the demethylation of m6A in a dependent manner. Studies have shown that FTO was the first protein that can catalyze m6A demethylation [31]. Variations in the FTO gene have been linked to weight gain, short stature, and other health issues [32,33,34]. ALKBH5, the second RNA demethylase identified, can oxidize and reverse the m6A modification. ALKBH5 directly removes the methyl group from m6A-methylated adenosine, in contrast to the oxidative demethylation of FTO [35,36]. Moreover, recent research suggests that ALKBH3 is a novel m6A-modified demethylase. ALKBH3’s new substrate is m6A in mammalian transfer RNA (tRNA). ALKBH3 changes tRNA more frequently than mRNA and rRNA [37,38] (Figure 1).
2.3. Readers
The dynamic and reversible regulation of the m6A modification is driven by the functional interplay between the m6A writer and the eraser. However, different readers must recognize m6A for different downstream biological functions. The m6A reader can bind to RNA that contains m6A thanks to a conservative m6A binding domain, referred to as YTH domain family protein 1 (YTHDF1/2/3), YTH domain containing 1 (YTHDC1/2), the heterologous nuclear ribonucleoprotein (HNRNP) family (HNRNPA2B1, HNRNPC and HNRNPG), the m6A binding protein composed of insulin-like growth factor 2 mRNA binding proteins 1/2/3 (IGF2BP1/2/3) and eucaryotic initiation factor 3 (eIF3) [39,40] (Figure 1).
YTHDF2 was the first RNA discovered to recognize and bind m6A. Its transcript degrades m6A modification by directly recruiting the carbon catabolite repression 4-negative on TATA-less (CCR4-NOT) deadenylase complex [41,42]. The three YTHDF proteins participate in similar biological pathways [43]. Recent investigations have shown that YTHDF3 promotes the translation of methylated RNA by increasing the rate of protein synthesis in tandem with YTHDF1, and affects mRNA attenuation or translation mediated by YTHDF2 through an interaction with YTHDF2 [44,45]. Unlike YTHDF2, IGF2BPs identify m6A alteration under normal and stress conditions to alter gene regulation and tumor biology via their K homologous domains. To prevent m6A mRNAs from being degraded and to boost mRNA stability and translation, IGF2BPs interacts with the mRNA stabilizer HuR [46].
The YTHDC1 is a unique nuclear m6A reader protein which has recognition sites in the nucleus. YTHDC1 can interact with initiation factors and ribosomes, including eIF3/4E/4G, poly (A) binding protein (PABP), and 40S ribosomal subunits, to improve the translation efficiency of target RNA [47]. It can also regulate mRNA cleavage by coordinating and regulating pre-mRNA splicing factors, such as arginine-rich SR splicing factors (SRSFs) [48]. For example, by interacting with SRSF3, YTHDC1 facilitates the nuclear export of m6A methylated mRNA in concert with nuclear RNA export factor 1 (NXF1) and the three prime repair exonuclease (TREX) mRNA export complex [49,50]. In addition to interacting with YTHDC1, YTHDC2 can also bind to the 5’-3’ exoribonuclease 1 (XRN1) and meiosis-specific coiled-coil domain-containing protein (MEIOC) to increase target mRNA translation efficiency and decrease its expression [51,52]. Through its C-terminal region, certain m6A sites can also be located. To directly attract CCR4–NOT alkenylase complexes and transport RNA to the processor, the N-terminal portion of CCR4–NOT transfer complex subunit 1 (CNOT1) interacts with the Src homology domain. This promotes the rapid degradation of m6A-modified RNA. [41].
Members of the HNRNP family are important m6A readers. HNRNPA2/B1 can selectively detect changes in m6A on transcripts and trigger various splicing events. Studies have demonstrated that HNRNPA2/B1 can interact with drosha ribonuclease III (DROSHA) and the DiGeorge syndrome critical region 8 (DGCR8) to recognize m6A on a subset of primary miRNA transcripts, boosting primary miRNA processing [53]. Therefore, HNRNPA2/B1, which is a nuclear m6A binding protein, can affect the synthesis of miRNA [54]. In addition, m6A modification alters the RNA secondary structure. HNRNPC is a rich nuclear RNA binding protein that identifies the m6A-modified group, participates in pre-mRNA processing, and monitors the abundance of target transcripts and their alternative splicing [55]. By altering the m6A-modified RNA transcript, HNRNPG can induce similar effects to those of HNRNPC. In addition, it is known as the “m6A switch” because it controls the expression level of mRNA and its splicing mechanism [56].
3. m6A Modification and Cancer
3.1. Thyroid Carcinoma
Thyroid carcinoma (TC) is the most prevalent endocrine-related malignancy. [57]. It is also the sixth most prevalent type of malignant tumor in the world, according to the 2020 Global Cancer Statistics [58]. The incidence rate increases by about 4% annually, accounting for 1% of all new cancers. Papillary thyroid carcinoma (PTC) is the main pathological subtype of TC [59], accounting for about 80–90% of all TC cases [60,61,62]. Most patients with TC have a good prognosis. Although PTC is well-differentiated, its biological performance is quite different. The inert thyroid micropapillary carcinoma shows a slow progression and rarely invades other tissues, whereas PTC shows lymph node or distant metastasis and has high mortality rates [63]. Recurrence and distant metastasis occur in about 30% of individuals, which negatively impacts patient prognosis [64,65]. Similar to other malignant tumors, the occurrence of TC is affected by environmental and genetic factors [66,67]. However, the exact molecular mechanism underlying the prevalence of TC remains elusive. Thus, understanding the role of m6A in TC is important to determine the most effective therapeutic strategy for TC (Table 1, Figure 2).

Table 1.
The roles of m6A enzymes in endocrine cancer progression.
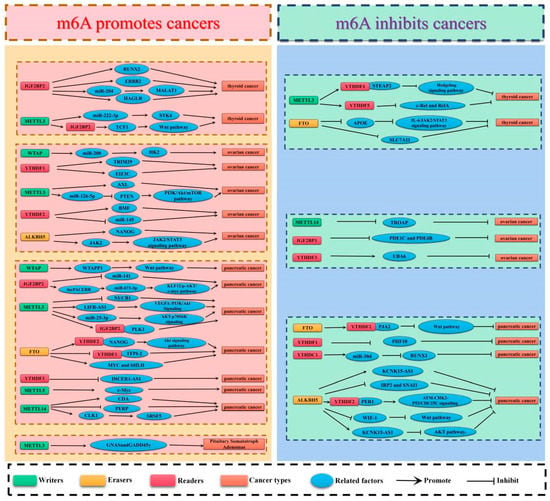
Figure 2.
The potential roles of m6A in endocrine cancer progression. The potential role of m6A in endocrine cancer progression is reflected in the regulation of tumor-associated gene expression. m6A modification promotes cancer progression by enhancing oncogene expression and inhibiting tumor suppressor gene expression. m6A modification hinders cancer progression by inhibiting oncogene expression and enhancing tumor suppressor gene expression.
The function of METTL3 in PTC is not well understood. Zhu et al. [68] reported that the oncogene METTL3 can inhibit tumor growth, metastasis, and the invasiveness of tumors. It enhanced the expression and translation of six-transmembrane epithelial antigen of the prostate 2 (STEAP2) mRNA mainly by recruiting the m6A “interpreter“ protein YTHDF1. Aberrant stimulation of the Hedgehog signaling pathway and epithelial-mesenchymal transition (EMT) were linked to decreased STEAP2 expression. EMT is a process through which cancer cells lose their epithelial properties and acquire stem cell-like invasive qualities which increases their capacity for local invasion and metastasis [114,115]. He et al. [69] found that c-RelA and Rel1 are m6A targets downstream of METTL3, and METTL3 can recognize c-Rel and RelA m6A modifications and cooperate with YTHDF2 to regulate the consistency of c-Rel1 and RelA mRNA, thereby triggering the nuclear factor kappa B (NF-κB) signaling pathway. In the chemotaxis test and mouse model experiments, it was found that an increased concentration of Interleukin-8 (IL-8) promoted the recruitment of tumor-associated neutrophils (TANs). Moreover, METTL3 deficiency increased the production of IL-8 in PTC cells, enhanced the recruitment of TAN, and promoted PTC progress. However, high METTL3 expression was associated with a worse prognosis in patients with TC in some studies. [70,71]. For example, Wang et al. [70] reported that increasing METTL3 expression promoted the migratory capacity and Wnt activity in TC cells through the regulation of m6A methylation on T cell factor 1 (TCF1). Moreover, the RIP assay showed that TCF1 interacted with METTL3 or IGF2BP2, and METTL3 positively regulated the abundance of TCF1, thereby decreasing IGFBP2 expression and accelerating the malignant behavior of the TC cell. Lin et al. [71] found that METTL3 mediated the m6A modification of pre-miR-222-3p to enhance the maturation of pre-miR-222-3p and increase miR-222-3p expression. PTC tumor suppressor serine/threonine Stress kinase 4 (STK4) is a target of miR-222-3p, which has an anti-regulatory effect on STK4. Thus, METTL3 knockdown can inhibit TC cell growth and malignant behavior.
Rescheduling energy metabolism through the activation of metabolic pathways such as glycolysis is a sign of cancer. Even in an aerobic environment, cancer cells revert to glycolysis for glucose metabolism rather than mitochondrial oxidative phosphorylation [116]. Huang et al. [72] postulated that PTC can be prevented by the activity of FTO, inhibiting the tumor development gene. Experimental studies showed that FTO inhibited the expression of Apolipoprotein E (APOE) through IGF2BP2-mediated m6A modification epigenetics. Furthermore, by modulating the IL-6/JAK2/STAT3 signal pathway, FTO and APOE may limit the glycolysis of PTC, which could ultimately affect the growth of the tumor. The toxic buildup of lipid-based reactive oxygen species (ROS) causes ferroptosis, an iron-dependent form of regulated cell death. Abnormal levels of iron can cause various diseases. Ji et al. [73] performed MeRIP-seq and RNA-seq analyses and found that FTO regulates PTC cell ferroptosis and tumor progression by regulating the epigenetic activation of the SLC7A11 gene.
Radioiodine (131-I) treatment can keep tumors in a “tumor dormancy” state for a long time, or even forever. It has few side effects and is a first-line treatment for a 131-I-avid disease with local recurrence and/or distant metastasis. However, if the dedifferentiation of PTC changes the mRNA or protein level of the iodine de-processing gene and loses the ability of iodine uptake and organization, then the tumor will not tolerate 131-I treatment. Radioactive iodine-resistant PTC is also referred to as radioactive iodine-refractory papillary thyroid carcinoma (RR-PTC) [117]. Clinically, the prognosis of RR-PTC is poor and researchers are attempting to develop effective interventions to improve patient outcomes. Some studies have proved that the m6A reader IGF2BP2 can be used as a post-transcriptional regulator to participate in the differentiation of cancer and cancer stem cells [118], such as hepatocellular carcinoma [119], liposarcoma [120], and glioblastoma [121]. IGF2BP2-dependent activation of the Erb-B2 receptor tyrosine kinase 2 (ERBB2) signal is the cause of acquired tyrosine kinase inhibitor (TKI) resistance. PTC cells can become resistant to TKIs because IGF2BP2 increases the expression of the ERBB2 protein by binding to the m6A methylation site of ERBB2 mRNA and thereby promoting the expression of ERBB2. This in turn increases the constitutive activation of signal pathways involved in the dedifferentiation process of PTC [74]. Another study by Sa et al. [75] found that metastatic PTC with high expression of IGF2BP2 was prone to 131-I incompatibility, and IGF2BP2 could integrate with 30-UTR of runt-related transcription factor 2 (RUNX2), thus damaging the stability of RUNX2 mRNA. RUNX2, as a transcription factor, can combine with the promoter of iodide binding gene NIS to regulate its activity. RUNX2 can inhibit the expression of NIS and then cause the differentiation of RR-PTC. Therefore, decreasing IGF2BP2 and increasing RUNX2 mRNA expression can restore the iodine uptake capacity of RR-PTC. In addition to overcoming the acquired drug resistance to RR-PTC, there is evidence from a few studies that IGF2BP2 is also strongly expressed in TC, which has been proven to be the target of miR-204. It can competitively bind miR-204 with metastasis-associated lung carcinoma transcript one, recognize and upregulate IGF2BP2 through m6A modification, and enhance myelocytomatosis oncogene (MYC) expression, thus stimulating the proliferation, migration, and invasion of TC cells [76]. Dong et al. [77] found that IGF2BP2 can also induce TC cell apoptosis by downregulating lncRNA-HAGLR and inhibiting TC cell proliferation, cell cycle progression, cell migration, and invasion.
3.2. Ovarian Cancer
For women, ovarian cancer (OC) has the highest fatality rate of any gynecological malignancy [122]. It is the fifth leading cause of cancer-related deaths among women worldwide [123,124], with a 5-year survival rate of less than 45% [122]. About half of OC patients are diagnosed at an advanced stage, which contributes to the poor overall survival rate [125,126]. OC is divided into three common types, including epithelial ovarian cancer (EOC), germ cell tumors, and sex cord-stromal tumors [127]. EOC accounts for more than 90% of all cases of OC [128]. Although surgery and targeted chemotherapy have made progress [129], the prognosis for most EOC patients is poor because of their inherent high recurrence rate and resistance to cisplatin [122]. Accumulating evidence shows that OC has a high degree of heterogeneity, which leads to the continuous growth and recurrence of chemotherapy-resistant cancer cells [130]. In addition, the primary cause of death in people with OC is tumor growth and spread, and there is currently no medication that can stop this from happening [131,132]. Here, we explore the role of m6A in regulating the development of OC through modulating protein translation. (Table 1, Figure 2).
Bi et al. [78] found that through m6A modification of pri-miR-126-5p, METTL3 stimulates miR-126-5P maturation. It was confirmed that miR-126-5p could activate the PI3K/Akt/mTOR pathway by directly targeting phosphatase and tensin homolog (PTEN). Inhibition of METTL3 via miR-126-5p knockdown slows the progression and carcinogenesis of OC by blocking the activation of the PI3K/Akt/mTOR pathway. Hua et al. [79] used a nude mouse model to show that steady overexpression of METTL3 significantly enhanced cell proliferation, lesion development, motility, and invasion in nude mice. METTL3 has also been found to promote EMT by up-regulating receiver tyrosine kinase AXL. Multiple studies have linked EMT to OC cell invasion and metastasis [133,134,135]. By promoting AXL translation and EMT, it is assumed that METTL3 plays a crucial carcinogenic role in the formation and/or invasion of OC. In addition to METTL3, Li et al. [80] found that in contrast to normal tissues, OC tissues have significantly reduced levels of METTL14 and m6A RNA methylation. The decreased expression of METTL14 was also associated with the change in the copy number variations (CNVs) and the low survival rate of OC patients. As a negative regulator, METTL14 can regulate the stability of the cell-proliferative gene troppin-associated protein (TROAP) mRNA and then prevent the growth of OC cells.
WTAP plays a crucial role as a “writer” in m6A modification. As a nuclear protein involved in cell function and cancer progression, it is frequently found close to splicing factors. Yet, WTAP’s role and mechanism in OC are largely mysterious. Lyu et al. [81] discovered that the hypoxic microenvironment influences the glycolysis pathway of cancer cells, hence driving the biological process of OC growth. They also, for the first time, reported the hypoxia-inducible factor-1α (HIF-1α). HIF-1α can actively regulate the increase in WTAP expression under hypoxia. A low survival rate is associated with high WTAP expression in OC patients. miR-200 expression in OC cells is regulated by WTAP via interactions with DGCR8, a component of the usual microprocessor complex of miRNA synthesis. miR-200 can positively regulate the key glycolytic enzyme hexokinase 2, significantly influence the intracellular Warburg effect, and promote tumor events and progress.
In addition, Nie et al. [82] discovered that upregulation of ALKBH5 in EOCs triggered resistance to cisplatin. Further analysis showed that homeobox A10 (HOXA10) is a transcription factor that promotes ALKBH5 transcription, and ALKBH5 can also control its activity. As a m6A-modified gene, Janus kinase 2 (JAK2) is one of ALKBH5’s primary targets. ALKBH5 maintains the stability of JAK2 mRNA in a YTHDF2-mediated manner. By persistently stimulating the signaling pathway of JAK2/STAT3, which is m6A-dependent, upregulation of the ALKBH5-HOXA10 loop enhances EOC tumor development and cisplatin resistance. To avoid cisplatin resistance in EOC, it may be possible to employ the technique of suppressing the expression of the ALKBH5-HOXA10 loop. Jiang et al. [83] reported higher levels of ALKBH5 expression in OC tissues compared with normal ovarian tissues, but the ALKBH5 expression levels were decreased in OC cell lines. Toll-like receptor 4 (TLR4), a molecule that participates in the tumor microenvironment (TME), showed a similar expression trend. Jiang et al. set up a macrophage and OC cell co-culture paradigm in vitro to examine the impact of aberrant TME on OC progression. An increased expression of ALKBH5 and TLR4 was observed in OC cells co-cultured with M2 macrophages, suggesting that TLR4 activates NF-κB. After mRNA demethylation, the expression of NANOG increased, which promoted the invasiveness of OC cells.
Huang et al. [84] found that tumorigenesis and cancer stem cell (CSC) self-renewal in ovarian cancer were both suppressed by FTO treatment, which is the first study to link FTO to the cell regulatory process mediated by the second messenger cyclic adenosine monophosphate (cAMP). The study showed that compared with non-CSCs, the cAMP level in ovarian CSCs was significantly decreased, and FTO overexpression induced an increase in the cAMP content, while its knockdown reduced the cAMP level. In addition, by reducing the m6A level at the 3′UTR and the mRNA stability of phosphodiesterase genes (PDE1C and PDE4B), FTO enhanced the cAMP signal and inhibited the stem cell characteristics of OC cells. In summary, this finding demonstrates that the cAMP pathway is a crucial FTO target in CSCs and reveals important insights for developing improved therapies for high-grade malignant ovarian cancer.
Due to the aggressive nature of EOC, chemotherapy resistance continues to be a major contributor to poor prognosis [85]. In cisplatin-resistant OC cells and clinical tissues, Hao et al. [86] found that tripartite motif-containing protein 29 (TRIM29) can enhance CSC-like characteristics of cisplatin-resistant OC cells as an oncogene. Moreover, YTHDF1 knockdown can inhibit the CSC-like characteristics of cisplatin-resistant OC cells and save this inhibition by increasing the expression of TRIM29. It is proved that YTHDF1, as an upstream molecule of TRIM29, can be recruited by TRIM29 to recognize its 3′UTR, participate in m6A modification, and promote its translation in cisplatin-resistant OC cells in an m6A-YTHDF-dependent manner.
A tumor suppressor factor named F-box and WD repeat domain-containing 7 (FBW7) recognizes substrates as part of the Skp1-cullin 1-F-box (SCF) E3 ubiquitin ligase complex, which catalyzes the degradation of many cancer-causing proteins. Xu et al. [87] found that compared with normal ovarian tissues, the expression of FBW7 was significantly downregulated in EOC tissues, whereas the expression of YTHDF2 was significantly elevated. YTHDF2 was identified as a new substrate of FBW7. By stimulating the proteasome degradation of YTHDF2 in OC, FBW7 mitigates YTHDF2’s tumor-promoting impact. In addition, YTHDF2 regulates the turnover of the m6A-modified mRNA apoptosis promoting gene Bcl2 modifying factor (BMF). It has been shown that by inhibiting YTHDF2-mediated BMF mRNA downregulation, FBW7 inhibits tumor growth and metastasis in OC. In addition to being identified as a new substrate of FBW7, YTHDF2 is also a target gene of miR-154. In OC, there is an inverse relationship between miR-145 and YTHDF2 expression levels. Overexpression of YTHDF2 has been shown to rescue miR-145-induced reductions in EOC proliferation and migration. Therefore, through m6A alteration, YTHDF2 and miR-145 can create a negative feedback mechanism that controls OC growth [88].
Wang et al. [89] demonstrated that lncRNA the ubiquitin-like modifier activating the enzyme 6 antisense RNA 1 (UBA6-AS1) was significantly associated with the prognosis of OC patients and was involved in the regulation of m6A. Positive correlations were also seen between UBA6-AS1 and UBA6 mRNA expression in OC tissues, and UBA6-AS1 was reported to prevent the degradation of UBA6 mRNA. By interacting with UBA6, UBA6-AS1 reduces the proliferation, migration, and invasion of OC cells. The reason for the positive correlation is that UBA6-AS1 increases the m6A methylation of UBA6 mRNA through RBM15. UBA6-AS1-RBM15-mediated m6A modification of UBA6 mRNA improved UBA6 mRNA stability, and IGF2BP1 was identified as the m6A reader protein.
3.3. Pancreatic Cancer
In terms of cancer-related mortality, pancreatic cancer (PC) is in the top seven of the worst diseases. [136,137]. The 5-year survival rate for pancreatic cancer is only 8%, and lowers to 3% at the distant metastatic stage [137]. Most patients have been diagnosed with unresectable local advanced or metastatic diseases because of the late onset of PC symptoms. Metastasis, recurrence, and chemotherapy resistance may lead to adverse outcomes. Over 90% of pancreatic illnesses can be attributed to pancreatic duct adenocarcinoma (PDAC), making it the most common form of PC [138,139]. The clinical prognosis of PDAC is poor [140]. Approximately 10% of people diagnosed will survive for 5 years after diagnosis [141,142]. Despite the fact that few patients were diagnosed with resectable local tumors, the 5-year survival rate following surgery was just 20% [143]. Therefore, m6a’s pathophysiological role and expression profile in PC must be determined (Table 1, Figure 2).
Through hypertranscriptional analysis, Tatekawa et al. [90] identified the polo-like kinase 1 (PLK1) gene in patients with PC which affects the prognosis. They found that METTL3 regulates the cell cycle of PC cells through the methylation of the 3′UTR of PLK1, which disrupts homeostastic balance and increases cell death by increasing replication stress. The study also found that IGF2BP2 binds to m6A in the 3’UTRs of PLK1, thereby increasing its expression. Demethylation of this region results in ataxia, telangiectasia, and increased radiosensitivity, which activates the Rad3-related protein pathway by elevating replication stress and formation of mitotic mutations. This demonstrates the critical role of PLK1 methylation in the PC cell cycle and shows its potential to be a new radiation and sensitization treatment target.
In addition, Hua et al. [91] noted that the METTL3 induced the m6A modification of Nucleobindin 1 (NUCB1) 5′UTR through the YTHDF2 reader. NUCB1 showed an additive effect with gemcitabine in vitro and in vivo. A decrease in PC cell proliferation was seen after NUCB1 overexpression. NUCB1 further inhibited the gemcitabine-induced unfolded protein response and autophagy by controlling activating transcription factor 6 activity and enhancing the effect of gemcitabine. Zhang et al. [92] reported the carcinogenic effect of miR-25-3p generated by cigarette smoke condensate via the m6A pathway in pancreatic cells. Cigarette smoke condensate can promote the excessive maturation of carcinogenic primary microRNA-25 (pri-miR-25) in pancreatic ductal epithelial cells through NF-kappaB-associated protein (NKAP)-mediated increased m6A modification. Because cigarette smoke condensate also causes hypomethylation of the METTL3 promoter, increased expression of this enzyme is required to catalyze this change. In terms of the splice site for pri-miR-25, the RNA-binding protein NKAP, as an m6A reader, preferentially binds to the common motif RGm6AC. By promoting the interaction between pri-miR-25 and the protein DGCR8 of the miRNA microprocessor complex, it enhances the maturation of miR-25-3P. Overexpression of miR-25-3p targets the pleckstrin homology domain and leucine-rich repeat protein phosphatase 2 (pHLPP2) mRNA and significantly inhibits its expression. This in turn stimulates AKT-p70S6K signal transduction. Therefore, the METTL3/miR –25–3p/pHLPP2/AKT axis plays a role in the initiation and progression of PDAC. Chen et al. [93] pointed out that METTL3 induced m6A methylation on the 3′UTR of leukemia inhibitory factor receptor antisense RNA 1 (LIFR-AS1) to enhance its mRNA stability, and also determined that LIFR-AS1 expression increased in PC cell lines and tumors. Through direct interaction with miR-150-5p, LIFR-AS1 increases expression of vascular endothelial growth factor A (VEGFA). It was found that LIFR-AS1 knockdown had adverse effects on the VEGFA/PI3K/Akt signal, which was reversed through the suppression of miR-150-5p expression. It is demonstrated that by regulating miR-150-5p/VEGFA, the novel METTL3/LIFR–AS1 axis stimulates PC development.
A pioneering study by Huang et al. [94] showed that the oncogene METTL5 promoted the proliferation, motility, invasion, and tumorigenesis of PC cells. Enhanced c-Myc translation may contribute to the carcinogenic role of METTL5. METTL5 is specifically regulated during translation thanks to the 5’untranslated region (5’UTR) of c-Myc mRNA and the m6A alteration of the coding DNA sequence region (near the 5’UTR). METTL5 and its cofactor TRMT112 synergistically promote the progression of PC.
Gemcitabine treatment dramatically upregulated m6A methyltransferase METTL14 expression in pancreatic cancer patients, as reported by Zhang et al. [95]. In addition, the reduction in METTL14 expression increased the sensitivity of drug-resistant cells to gemcitabine therapy. Mechanistically, they discovered that the transcription factor p65 can up-regulate the production of METTL14 by binding to its promoter region, which in turn increases the expression of the enzyme Cytidine deaminase (CDA), which inactivates gemcitabine. These findings suggest that reducing patient drug resistance by targeting METTL14 may be possible. Wang et al. [96] found that METTL14 overexpression directly targeted the downstream PERP mRNA (p53 effector related to PMP-22), which increased the turnover of PERP mRNA and reduced PERP expression in PC cells. There is evidence that PERP has a role in maintaining the integrity and homeostasis of epithelial cells, and that it regulates the adhesion subprogram (which influences cell death) [144]. In this way, it dramatically stimulates PC cells’ growth and migration. In addition, it participates in DNA damage-induced apoptosis through a mechanism dependent or independent of the p53 signaling pathway [145]. In a study on changes in the alternative splicing mode of m6A modification-related proteins in PC, Chen et al. [97] found that the alternative splicing mode of METTL14 regulates the process of m6A methylation. Another study showed that CLK1 altered the alternative splicing mode of METTL14 by activating SRSF5, promoted cyclin L2exon6.3 jumping, and inhibited METTL14 exon10 jumping, which accelerated cell proliferation, migration, and invasion.
Liu et al. [98] discovered that LncRNA-PACERR was increased in PC leading to enhanced tumor development, and LncRNA-PACERR promoted pancreatic cancer development by acting on IGF2BP2. PACERR collaborated with IGF2BP2 to increase KLF12 and c-myc expression. KLF12 and c-myc are both transcription factors; however, their precise regulatory locations are different. To stabilize KLF12 and c-myc, the PACERR interacts with a region on the secondary structure of the IGF2BP2 protein, which is located close to the KH1 and KH2 domains, in an m6A-dependent manner. By binding to miR-671-3p, the PACERR contributes to the occurrence of PDAC by activating the KLF12/p-AKT/c-myc signaling pathway. In addition, KLF12 specifically targets the promoter of LncRNA-PACERR. LncRNA-PACERR increases histone acetylation and promotes PC development through interactions with KLF12 in the nucleus when EP300 is recruited. Xu et al. [99] confirmed that the miR-141 genome amplification and silencing resulted in IGF2BP2 activation. Given that IGF2BP2 is a direct target of miR-141, the above finding shows a negative association between them. In addition, it was reported that an increased expression of IGF2BP2 stimulated the proliferation of PC cells via the PI3K/Akt signaling pathway.
Wang et al. [100] showed that FTO knockdown increased the methylation of TFPI-2 mRNA in PC cells, and the m6A reader YTHDF1 increased the stability of TFPI-2 mRNA, leading to the upregulation of TFPI-2 expression. This inhibited PC cell proliferation, colony formation, spheroid formation, migration, and invasion. Tang et al. [101] also observed reduced proliferation and DNA synthesis in PC cells following suppression of the FTO gene. Research has shown that FTO can decrease the quantity of the m6A protein, which stabilizes the MYC proto-oncogene bHLH transcription factor. This upregulates bHLH transcription factor expression, thereby reducing cell apoptosis and promoting tumor proliferation and growth. Tan et al. [102] found that FTO directly targeted the platelet-derived growth factor C (PDGFC), resulting in the formation of PDGFC 3ʹ in UTR, m6A modification was reduced and its mRNA expression was stabilized in an m6A-YTHDF2 dependent manner while the expression of PDGFC was increased. Consequently, the Akt signaling pathway was reactivated, which promoted cell growth, colony formation, and tumor progression. Zeng et al. [103] also found that FTO modified and demethylated m6A of the praja ring finger ubiquitin ligase 2 (PJA2) in an m6A-YTHDF2 dependent manner, enhancing the stability of PJA2 and stabilizing its mRNA and inhibiting the Wnt signaling pathway. In conclusion, these complex mechanisms regulate the proliferation, invasion, and metastasis of PC cells.
A study by Huang et al. [104] revealed a new DNA damage response mechanism involving the m6a modification of the chromatin remodeling complex subunit PHF10. They discovered that via increasing m6A alteration in PDAC cells, fluoxetine causes DSB and inhibits HR repair. The PBAF chromatin remodeling complex, which is thought to be the primary molecule impacted by non-sit-in therapy, contains the m6A writers ZC3H13 and PHF10. The ZC3H13-mediated m6A RNA modification inhibits PHF10 translation, allowing its participation in DNA damage response processes. The loss of PHF10 function increases the possibility of H2AX, RAD51, and 53BP1 recruitment to DSB sites, and the effectiveness of HR repair is reduced. Additionally, ZC3H13 regulates PHF10 translation and m6A methylation in a YTHDF1-dependent way. This discovery contributes to our knowledge of the mechanism of DNA repair and suggests potential treatments for PC.
Hu et al. [105] found that DICER1-AS1 induced transcription of its sense gene by promoting binding of the transcription factor YY1 to the DICER1 promoter. In addition, DICER1 enhanced the maturation of miR-5586-5p leading to the inhibition of the expression of glycolytic genes including LDHA, HK2, PGK1, and SLC2A1. These findings demonstrate that YTHDF3 is a key target of miR-5586-5p that regulates PC glycolysis by forming a negative feedback loop with DICER1-AS1. Glucose depletion enhances the interaction between YTHDF3 and DICER1-AS1 and glucose deprivation, leading to the degradation of DICER1-AS1.
Deng et al. [106] found that the mRNA of WTAPP1 was overexpressed in patients with PDAC, which correlated with poor survival. m6A modification of CCHC type zinc filament nuclear acid binding protein (CNBP) was reported to stabilize WTAPP1 RNA, thereby increasing the WTAPP1 RNA expression level in PDAC cells. The overexpressed WTAPP1 RNA can bind to its protein-coding counterpart WTAP mRNA, and several EIF3 translation initiation complexes are recruited to promote WTAP translation. The increase in WTAP protein levels in turn enhances the activation of the Wnt signaling pathway and induces the malignant phenotype of PDAC.
Hou et al. [107] found that YTHDC1 promotes the biosynthesis of mature miR-30d by regulating the m6A-mediated mRNA stability. Subsequently, miR-30d directly targets the transcription factor RUNX1, which binds to the promoters of the SLC2A1 gene and HK1 gene to inhibit aerobic glycolysis. The amount of m6A modified pri-miR-30d was dramatically decreased by the deletion of METTL3/14. To start miR-30d biosynthesis, YTHDC1 antagonizes the termination of MCPIP1’s miRNA biogenesis by preferentially recognizing m6A-modified pri-miR-30d and promoting its degradation. The axis of YTHDC1-miR-30-RUNX1-SLC2A1/HK1 is created.
Guo et al. [108] found that ALKBH5 activated PER1 by inducing m6A demethylation and transcription in an m6A-YTHDF2-dependent manner. The upregulation of PER1 triggered a reactivation of the ATM-CHK2-P53/CDC25C signaling pathway which inhibits tumor proliferation, migration, and invasion. PER1-induced P53 upregulation and P53-activated ALKBH5 transcription are important indicators of the feedback regulation of m6A modification in PC, creating an ALKBH5-PER1-P53-ALKBH5 feedback loop. Studies have demonstrated that ALKBH5 demethylates KCNK15-AS1 and participates in KCNK15-AS1-mediated cell migration and invasion, hence regulating tumor growth [109,110]. In a pioneering study by He et al. [109], it was confirmed that KCNK15-AS1 inhibits PC cell growth by regulating KCNK15 and PTEN. It was also found that ALKBH5-mediated m6A demethylation enhanced the stability of KCNK-AS1. The KCNK15-AS1 binds to KCNK15 5’UTR, leading to the inhibition of KCNK15 translation. In addition, KCNK15-AS1 recruits the MDM2 proto-oncogene (MDM2) and interacts with it to promote the ubiquitination of the RE1 silencing transfer factor (REST), transcriptionally upregulating phosphate and tensin homolog (PTEN) and thus inactivating the AKT pathway [110]. Huang et al. [111] revealed that ALKBH5 protects PDAC by regulating iron metabolic regulators. They also reported that the mRNA encoding ubiquitin ligase FBXL5 and mitochondrial iron introns SLC25A28 and SLC25A37 are potential substrates of ALKBH5 and that their stability is regulated by ALKBH5. The ALKBH5 overexpression that was triggered by FBXL5-mediated degradation led to a dramatic reduction in the levels of the iron-regulatory protein IRP2 and the EMT regulator SNAI1. Therefore, cellular iron levels and migratory and invasive capacities were significantly decreased. Tang et al. [112] showed that gemcitabine inhibited ALKBH5 expression in a patient-derived xenograft (PDX) model. The experiment proved that ALKBH5 deficiency promoted the proliferation, migration, and invasion of PDAC cells in vitro and in vivo, and its overexpression increased the sensitivity of PDAC cells to chemotherapy. Moreover, the m6A expression profile of some ALKBH5 target genes was altered, including that of Wnt aggression factor 1 (WIF-1).
4. Pituitary Adenoma
Near the base of the brain is a small gland called the pituitary, which is usually called the “main gland”. It is the most significant endocrine gland in the body, regulating the secretion of essential hormones. These hormones regulate vital bodily processes, including growth, blood pressure, reproduction, and metabolism [146,147]. 10–20% of all brain cancers are pituitary adenomas (PA), making them the second most prevalent type of brain tumor [148,149]. Previously, PA was classified according to its size. Those less than 10 mm were named microadenomas, and the rest were referred to as macroadenomas [150]. Currently, it is classified according to the type of hormone it excessively secretes into the blood, e.g., adrenocorticotropic hormone-secreting adenoma, growth hormone-secreting adenoma, prolactin-secreting adenoma, and thyroid hormone-secreting adenoma [151]. It is estimated that 35% of pituitary adenomas will show infiltration of the cavernous sinus and sphenoid sinus [152]. Transnasal transsphenoidal surgery is the main treatment for PAs [153]. In clinical practice, PAs that infiltrate the suprasellar or parasellar regions are hard to eradicate completely, and patients with a residual adenoma have a recurrence rate of 12–58% [149]. For instance, 10–20% of the adenoma will recur within 5–10 years, even when the adenoma is completely removed [154]. To achieve total tumor control or biochemical remission, drugs and stereotactic radiosurgery are suggested. [155,156]. Therefore, identification of drug targets through m6A modification research may generate strategies that will form the basis for clinical treatment (Table 1, Figure 2).
In a study of endocrine pituitary adenomas, it was found that the expression of METTL3 messenger RNA and protein in GH-PA samples was higher than that in normal pituitary tissue samples and non-secretory pituitary adenomas. The level of m6A modification in GH-PA was increased, and hypermethylated RNA was involved in hormone secretion and cell development. In GH3 cell lines, the absence of METTL3 decreased cell proliferation and GH secretion. They discovered that GNAS and GADD45 act as targets downstream of this mechanism. In addition, m6A methyltransferase METTL3 increased GNAS and GADD45 in an m6A-dependent manner, which promoted tumor growth and hormone secretion. These findings demonstrate that METTL3 and methylated RNA are potential targets for the clinical treatment of GH PAs [113].
5. The Future Perspectives on the Application of Targeted m6A Therapy in the Treatment of Endocrine Cancer
It should be noted that m6A modification is one kind of epigenetic modification to RNA. Studies into the mechanism of m6A alteration have revealed that it adds a layer of complexity to the regulation of RNA levels. The current hypothesis on the link between m6A and cancers is that m6A is neither good nor bad. It can either promote or inhibit cancer cell growth by affecting the mRNA level of related oncogenes or suppressor genes. The degree of m6A modification of RNA correlates closely with the level of expression of writing and clearing genes within cells. Many biological processes are initiated by attachment of reader proteins to the m6A modification site. In general, an aberrant expression of m6A may disrupt the normal RNA modification process and directly interfere with the processing, transport, translation, and destruction of mRNA, which may contribute to the development and occurrence of malignancies. In addition, besides m6a-modified mRNA, the role of m6a-modified non-coding RNA in endocrine cancer should be explored. A gene’s expression level can be modulated by ncRNA. The m6A alteration of ncRNA impacts its own expression level. The bidirectional regulation of m6A modification, particularly the regulation of ncRNA, may provide a new explanation for certain disorders. Mutations at the m6A site may disrupt the regulation of the m6A factor, especially “author” and “erasure”, leading to abnormal m6A modification levels in significant transcripts which is essential to the abnormal expression of related oncogenes. This calls for research focus on the mutation of the m6A locus, in various aspects such as single nucleotide polymorphism, which will improve the targeting of endocrine cancer. Based on their catalytic activity, numerous RNA-modifying enzymes have been shown to play key roles in the occurrence or maintenance of various forms of cancer [157]. Several m6a-related genes and proteins in tumors have the potential to be used as diagnostic markers and as therapeutic targets. Epigenetic mechanisms targeting has emerged as a promising new therapeutic approach. Targeting the m6A RNA modification pathway may prevent the onset and metastasis of cancer [158,159]. The development of RNA-modifying enzyme inhibitors will pave the way for a significant and novel approach to the treatment of tumors and other diseases. Small molecule inhibitors that target effector proteins involved in RNA methylation may hold tremendous potential as disease treatments. It is anticipated that RNA epigenetic medications may be applied in clinical practice to treat diseases. Given the numerous types of non-coding RNA, further research is advocated to identify novel therapeutic targets. Combining targeted mRNA and non-coding RNA therapy may offer an effective treatment for the seemingly intractable problem of endocrine cancer.
6. Conclusions
Abnormal changes in levels of RNA m6A modification or m6A regulatory factors have an important impact on the occurrence, development, metastasis, and prognosis of endocrine tumors. We provide a theoretical foundation for the development of new diagnostic markers and therapeutic techniques based on m6A alteration and regulators, and we discuss the knowledge about the role of m6A in endocrine cancers and its regulators and mechanisms. The data reviewed here provide a reference for further investigations into the carcinogenic or antitumor mechanism of the m6A regulators which may lead to the development of specific agonists or inhibitors of protein targets.
Author Contributions
The authors affirm that no part of the manuscript has been published or is under consideration elsewhere. The authors accept public accountabilities for all authors’ contributions. All authors have read and agreed to the published version of the manuscript.
Funding
National Natural Science Foundation of China (81902726), the Natural Science Foundation of Education Bureau of Liaoning Province (QNZR2020002), Natural Science Foundation of Education Bureau of Liaoning Province (QNZR2020009), the Natural Science Foundation of Liaoning Province (2020-MS-143), the Natural Science Foundation of Liaoning Province (2020-MS-186), the Natural Science Foundation of Liaoning Province (2021-MS-193) and Science and Technology Project of Shenyang City (21-173-9-31).
Conflicts of Interest
The authors have declared no potential conflict of interest.
References
- Schulte, K.M.; Gill, A.J.; Barczynski, M.; Karakas, E.; Miyauchi, A.; Knoefel, W.T.; Lombardi, C.P.; Talat, N.; Diaz-Cano, S.; Grant, C.S. Classification of parathyroid cancer. Ann. Surg. Oncol. 2012, 19, 2620–2628. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Fulp, W.J.; Strosberg, J.R.; Kvols, L.K.; Centeno, B.A.; Hodul, P.J. Predictors of lymph node metastases and impact on survival in resected pancreatic neuroendocrine tumors: A single-center experience. Am. J. Surg. 2014, 208, 775–780. [Google Scholar] [CrossRef]
- Sav, A.; Rotondo, F.; Syro, L.V.; Di Ieva, A.; Cusimano, M.D.; Kovacs, K. Invasive, atypical and aggressive pituitary adenomas and carcinomas. Endocrinol. Metab. Clin. N. Am. 2015, 44, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.Y.; Handy, B.; Michaelis, C.L.; Waguespack, S.G.; Hu, M.I.; Busaidy, N.; Jimenez, C.; Cabanillas, M.E.; Fritsche, H.J.; Cote, G.J.; et al. Detection and Prognostic Significance of Circulating Tumor Cells in Patients With Metastatic Thyroid Cancer. J. Clin. Endocrinol. Metab. 2016, 101, 4461–4467. [Google Scholar] [CrossRef] [PubMed]
- Scollo, C.; Russo, M.; Trovato, M.A.; Sambataro, D.; Giuffrida, D.; Manusia, M.; Sapuppo, G.; Malandrino, P.; Vigneri, R.; Pellegriti, G. Prognostic Factors for Adrenocortical Carcinoma Outcomes. Front. Endocrinol. 2016, 7, 99. [Google Scholar] [CrossRef]
- Roignant, J.Y.; Soller, M. m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trend Genet. 2017, 33, 380–390. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef]
- Oerum, S.; Meynier, V.; Catala, M.; Tisné, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021, 21, 7239–7255. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m(6)A(m) in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Wang, X.; He, C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014, 11, 669–672. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef]
- Bokar, J.A.; Rath-Shambaugh, M.E.; Ludwiczak, R.; Narayan, P.; Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994, 269, 17697–17704. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, J.; Zou, T.; Yin, P. Human m(6)A writers: Two subunits, 2 roles. RNA Biol. 2017, 14, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Warda, A.S.; Kretschmer, J.; Hackert, P.; Lenz, C.; Urlaub, H.; Höbartner, C.; Sloan, K.E.; Bohnsack, M.T. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017, 18, 2004–2014. [Google Scholar] [CrossRef] [PubMed]
- Shima, H.; Matsumoto, M.; Ishigami, Y.; Ebina, M.; Muto, A.; Sato, Y.; Kumagai, S.; Ochiai, K.; Suzuki, T.; Igarashi, K. S-Adenosylmethionine Synthesis Is Regulated by Selective N(6)-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 2017, 21, 3354–3363. [Google Scholar] [CrossRef]
- Thomas, J.M.; Batista, P.J.; Meier, J.L. Metabolic Regulation of the Epitranscriptome. ACS Chem. Biol. 2019, 14, 316–324. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835. [Google Scholar] [CrossRef]
- van Tran, N.; Ernst, F.; Hawley, B.R.; Zorbas, C.; Ulryck, N.; Hackert, P.; Bohnsack, K.E.; Bohnsack, M.T.; Jaffrey, S.R.; Graille, M.; et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019, 47, 7719–7733. [Google Scholar] [CrossRef]
- Richard, E.M.; Polla, D.L.; Assir, M.Z.; Contreras, M.; Shahzad, M.; Khan, A.A.; Razzaq, A.; Akram, J.; Tarar, M.N.; Blanpied, T.A.; et al. Bi-allelic Variants in METTL5 Cause Autosomal-Recessive Intellectual Disability and Microcephaly. Am. J. Hum. Genet. 2019, 105, 869–878. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Schöller, E.; Weichmann, F.; Treiber, T.; Ringle, S.; Treiber, N.; Flatley, A.; Feederle, R.; Bruckmann, A.; Meister, G. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 2018, 24, 499–512. [Google Scholar] [CrossRef]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3 ′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef]
- Ma, H.; Wang, X.; Cai, J.; Dai, Q.; Natchiar, S.K.; Lv, R.; Chen, K.; Lu, Z.; Chen, H.; Shi, Y.G.; et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019, 15, 88–94. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Dina, C.; Meyre, D.; Gallina, S.; Durand, E.; Körner, A.; Jacobson, P.; Carlsson, L.M.; Kiess, W.; Vatin, V.; Lecoeur, C.; et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007, 39, 724–726. [Google Scholar] [CrossRef]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Scuteri, A.; Sanna, S.; Chen, W.M.; Uda, M.; Albai, G.; Strait, J.; Najjar, S.; Nagaraja, R.; Orrú, M.; Usala, G.; et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007, 3, e115. [Google Scholar] [CrossRef]
- Aik, W.; Scotti, J.S.; Choi, H.; Gong, L.; Demetriades, M.; Schofield, C.J.; McDonough, M.A. Structure of human RNA N⁶-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014, 42, 4741–4754. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, L.; Zheng, G.; Fu, Y.; Ji, Q.; Liu, F.; Chen, H.; He, C. Crystal structure of the RNA demethylase ALKBH5 from zebrafish. FEBS Lett. 2014, 588, 892–898. [Google Scholar] [CrossRef]
- Sun, T.; Wu, R.; Ming, L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019, 112, 108613. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Ooshio, I.; Fusamae, Y.; Kitae, K.; Kawaguchi, M.; Jingushi, K.; Hase, H.; Harada, K.; Hirata, K.; Tsujikawa, K. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017, 7, 42271. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell. 2020, 25, 1582–1595. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m(6)A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Zhu, T.; Roundtree, I.A.; Wang, P.; Wang, X.; Wang, L.; Sun, C.; Tian, Y.; Li, J.; He, C.; Xu, Y. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014, 24, 1493–1496. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, Y.S.; Ping, X.L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Lesbirel, S.; Viphakone, N.; Parker, M.; Parker, J.; Heath, C.; Sudbery, I.; Wilson, S.A. The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci. Rep. 2018, 8, 13827. [Google Scholar] [CrossRef] [PubMed]
- Wojtas, M.N.; Pandey, R.R.; Mendel, M.; Homolka, D.; Sachidanandam, R.; Pillai, R.S. Regulation of m(6)A Transcripts by the 3′→5′ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol. Cell 2017, 68, 374–387. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Miranda-Filho, A.; Lortet-Tieulent, J.; Bray, F.; Cao, B.; Franceschi, S.; Vaccarella, S.; Dal Maso, L. Thyroid cancer incidence trends by histology in 25 countries: A population-based study. Lancet Diabetes Endocrinol. 2021, 9, 225–234. [Google Scholar] [CrossRef]
- LiVolsi, V.A. Papillary thyroid carcinoma: An update. Mod. Pathol. 2011, 24 (Suppl. S2), S1–S9. [Google Scholar] [CrossRef]
- Omur, O.; Baran, Y. An update on molecular biology of thyroid cancers. Crit. Rev. Oncol. Hematol. 2014, 90, 233–252. [Google Scholar] [CrossRef]
- Rosenbaum, M.A.; McHenry, C.R. Contemporary management of papillary carcinoma of the thyroid gland. Expert Rev. Anticancer Ther. 2009, 9, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Prasad, M.; Lemon, W.J.; Hampel, H.; Wright, F.A.; Kornacker, K.; LiVolsi, V.; Frankel, W.; Kloos, R.T.; Eng, C.; et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc. Natl. Acad. Sci. USA 2001, 98, 15044–15049. [Google Scholar] [CrossRef]
- Nixon, I.J.; Simo, R.; Newbold, K.; Rinaldo, A.; Suarez, C.; Kowalski, L.P.; Silver, C.; Shah, J.P.; Ferlito, A. Management of Invasive Differentiated Thyroid Cancer. Thyroid 2016, 26, 1156–1166. [Google Scholar] [CrossRef]
- Fröhlich, E.; Wahl, R. The current role of targeted therapies to induce radioiodine uptake in thyroid cancer. Cancer Treat. Rev. 2014, 40, 665–674. [Google Scholar] [CrossRef]
- Li, X.; Abdel-Mageed, A.B.; Kandil, E. BRAF mutation in papillary thyroid carcinoma. Int. J. Clin. Exp. Med. 2012, 5, 310–315. [Google Scholar]
- Li, X.; Abdel-Mageed, A.B.; Mondal, D.; Kandil, E. The nuclear factor kappa-B signaling pathway as a therapeutic target against thyroid cancers. Thyroid 2013, 23, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Peng, X.; Zhou, Q.; Tan, L.; Zhang, C.; Lin, S.; Long, M. METTL3-mediated m6A modification of STEAP2 mRNA inhibits papillary thyroid cancer progress by blocking the Hedgehog signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 2022, 13, 358. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhou, M.; Yin, J.; Wan, J.; Chu, J.; Jia, J.; Sheng, J.; Wang, C.; Yin, H.; He, F. METTL3 restrains papillary thyroid cancer progression via m(6)A/c-Rel/IL-8-mediated neutrophil infiltration. Mol. Ther. 2021, 29, 1821–1837. [Google Scholar] [CrossRef]
- Wang, K.; Jiang, L.; Zhang, Y.; Chen, C. Progression of Thyroid Carcinoma Is Promoted by the m6A Methyltransferase METTL3 Through Regulating m(6)A Methylation on TCF1. OncoTargets Ther. 2020, 13, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhu, Y.; Ji, C.; Yu, W.; Zhang, C.; Tan, L.; Long, M.; Luo, D.; Peng, X. METTL3-Induced miR-222-3p Upregulation Inhibits STK4 and Promotes the Malignant Behaviors of Thyroid Carcinoma Cells. J. Clin. Endocrinol. Metab. 2022, 107, 474–490. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sun, W.; Wang, Z.; Lv, C.; Zhang, T.; Zhang, D.; Dong, W.; Shao, L.; He, L.; Ji, X.; et al. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner. J. Exp. Clin. Cancer Res. 2022, 41, 42. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.H.; Fu, X.H.; Li, G.Q.; He, Q.; Qiu, X.G. FTO Prevents Thyroid Cancer Progression by SLC7A11 m6A Methylation in a Ferroptosis-Dependent Manner. Front. Endocrinol. 2022, 13, 857765. [Google Scholar] [CrossRef]
- Sa, R.; Liang, R.; Qiu, X.; He, Z.; Liu, Z.; Chen, L. IGF2BP2-dependent activation of ERBB2 signaling contributes to acquired resistance to tyrosine kinase inhibitor in differentiation therapy of radioiodine-refractory papillary thyroid cancer. Cancer Lett. 2022, 527, 10–23. [Google Scholar] [CrossRef]
- Sa, R.; Liang, R.; Qiu, X.; He, Z.; Liu, Z.; Chen, L. Targeting IGF2BP2 Promotes Differentiation of Radioiodine Refractory Papillary Thyroid Cancer via Destabilizing RUNX2 mRNA. Cancers 2022, 14, 1268. [Google Scholar] [CrossRef]
- Ye, M.; Dong, S.; Hou, H.; Zhang, T.; Shen, M. Oncogenic Role of Long Noncoding RNAMALAT1 in Thyroid Cancer Progression through Regulation of the miR-204/IGF2BP2/m6A-MYC Signaling. Mol. Ther. Nucleic Acids 2021, 23, 1–12. [Google Scholar] [CrossRef]
- Dong, L.; Geng, Z.; Liu, Z.; Tao, M.; Pan, M.; Lu, X. IGF2BP2 knockdown suppresses thyroid cancer progression by reducing the expression of long non-coding RNA HAGLR. Pathol. Res. Pract. 2021, 225, 153550. [Google Scholar] [CrossRef]
- Bi, X.; Lv, X.; Liu, D.; Guo, H.; Yao, G.; Wang, L.; Liang, X.; Yang, Y. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021, 28, 335–349. [Google Scholar] [CrossRef]
- Hua, W.; Zhao, Y.; Jin, X.; Yu, D.; He, J.; Xie, D.; Duan, P. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol. Oncol. 2018, 151, 356–365. [Google Scholar] [CrossRef]
- Li, Y.; Peng, H.; Jiang, P.; Zhang, J.; Zhao, Y.; Feng, X.; Pang, C.; Ren, J.; Zhang, H.; Bai, W.; et al. Downregulation of Methyltransferase-Like 14 Promotes Ovarian Cancer Cell Proliferation Through Stabilizing TROAP mRNA. Front. Oncol. 2022, 12, 824258. [Google Scholar] [CrossRef]
- Lyu, Y.; Zhang, Y.; Wang, Y.; Luo, Y.; Ding, H.; Li, P.; Ni, G. HIF-1α Regulated WTAP Overexpression Promoting the Warburg Effect of Ovarian Cancer by m6A-Dependent Manner. J. Immunol. Res. 2022, 2022, 6130806. [Google Scholar] [CrossRef]
- Nie, S.; Zhang, L.; Liu, J.; Wan, Y.; Jiang, Y.; Yang, J.; Sun, R.; Ma, X.; Sun, G.; Meng, H.; et al. ALKBH5-HOXA10 loop-mediated JAK2 m6A demethylation and cisplatin resistance in epithelial ovarian cancer. J. Exp. Clin. Cancer Res. 2021, 40, 284. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wan, Y.; Gong, M.; Zhou, S.; Qiu, J.; Cheng, W. RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-κB pathway. J. Cell. Mol. Med. 2020, 24, 6137–6148. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Y.; Kandpal, M.; Zhao, G.; Cardenas, H.; Ji, Y.; Chaparala, A.; Tanner, E.J.; Chen, J.; Davuluri, R.V.; et al. FTO-Dependent N (6)-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res. 2020, 80, 3200–3214. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wei, Q.; Jin, J.; Luo, Q.; Liu, Y.; Yang, Y.; Cheng, C.; Li, L.; Pi, J.; Si, Y.; et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020, 48, 3816–3831. [Google Scholar] [CrossRef]
- Hao, L.; Wang, J.M.; Liu, B.Q.; Yan, J.; Li, C.; Jiang, J.Y.; Zhao, F.Y.; Qiao, H.Y.; Wang, H.Q. m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118878. [Google Scholar] [CrossRef]
- Xu, F.; Li, J.; Ni, M.; Cheng, J.; Zhao, H.; Wang, S.; Zhou, X.; Wu, X. FBW7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein YTHDF2. Mol. Cancer 2021, 20, 45. [Google Scholar] [CrossRef]
- Li, J.; Wu, L.; Pei, M.; Zhang, Y. YTHDF2, a protein repressed by miR-145, regulates proliferation, apoptosis, and migration in ovarian cancer cells. J. Ovarian Res. 2020, 13, 111. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z. Long noncoding RNA UBA6-AS1 inhibits the malignancy of ovarian cancer cells via suppressing the decay of UBA6 mRNA. Bioengineered 2022, 13, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Tatekawa, S.; Tamari, K.; Chijimatsu, R.; Konno, M.; Motooka, D.; Mitsufuji, S.; Akita, H.; Kobayashi, S.; Murakumo, Y.; Doki, Y.; et al. N(6)-methyladenosine methylation-regulated polo-like kinase 1 cell cycle homeostasis as a potential target of radiotherapy in pancreatic adenocarcinoma. Sci. Rep. 2022, 12, 11074. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.Q.; Zhang, K.; Sheng, J.; Ning, Z.Y.; Li, Y.; Shi, W.D.; Liu, L.M. NUCB1 Suppresses Growth and Shows Additive Effects With Gemcitabine in Pancreatic Ductal Adenocarcinoma via the Unfolded Protein Response. Front. Cell Dev. Biol. 2021, 9, 641836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bai, R.; Li, M.; Ye, H.; Wu, C.; Wang, C.; Li, S.; Tan, L.; Mai, D.; Li, G.; et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 2019, 10, 1858. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Tao, Y.P.; Hong, Y.G.; Li, H.F.; Huang, Z.P.; Xu, X.F.; Zheng, H.; Hu, L.K. M(6)A-mediated up-regulation of LncRNA LIFR-AS1 enhances the progression of pancreatic cancer via miRNA-150-5p/ VEGFA/Akt signaling. Cell Cycle 2021, 20, 2507–2518. [Google Scholar] [CrossRef]
- Huang, H.; Li, H.; Pan, R.; Wang, S.; Khan, A.A.; Zhao, Y.; Zhu, H.; Liu, X. Ribosome 18S m(6)A methyltransferase METTL5 promotes pancreatic cancer progression by modulating c-Myc translation. Int. J. Oncol. 2022, 60, 9. [Google Scholar] [CrossRef]
- Zhang, C.; Ou, S.; Zhou, Y.; Liu, P.; Zhang, P.; Li, Z.; Xu, R.; Li, Y. m(6)A Methyltransferase METTL14-Mediated Upregulation of Cytidine Deaminase Promoting Gemcitabine Resistance in Pancreatic Cancer. Front. Oncol. 2021, 11, 696371. [Google Scholar] [CrossRef]
- Wang, M.; Liu, J.; Zhao, Y.; He, R.; Xu, X.; Guo, X.; Li, X.; Xu, S.; Miao, J.; Guo, J.; et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol. Cancer 2020, 19, 130. [Google Scholar] [CrossRef]
- Chen, S.; Yang, C.; Wang, Z.W.; Hu, J.F.; Pan, J.J.; Liao, C.Y.; Zhang, J.Q.; Chen, J.Z.; Huang, Y.; Huang, L.; et al. CLK1/SRSF5 pathway induces aberrant exon skipping of METTL14 and Cyclin L2 and promotes growth and metastasis of pancreatic cancer. J. Hematol. Oncol. 2021, 14, 60. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, M.; He, X.; Cao, Y.; Liu, P.; Li, F.; Zou, S.; Wen, C.; Zhan, Q.; Xu, Z.; et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2022, 15, 52. [Google Scholar] [CrossRef]
- Xu, X.; Yu, Y.; Zong, K.; Lv, P.; Gu, Y. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 497. [Google Scholar] [CrossRef]
- Wang, W.; He, Y.; Zhai, L.L.; Chen, L.J.; Yao, L.C.; Wu, L.; Tang, Z.G.; Ning, J.Z. m(6)A RNA demethylase FTO promotes the growth, migration and invasion of pancreatic cancer cells through inhibiting TFPI-2. Epigenetics 2022, 17, 1738–1752. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, S.; Chen, D.; Zhao, Z.; Zhou, J. The role of the fat mass and obesity-associated protein in the proliferation of pancreatic cancer cells. Oncol. Lett. 2019, 17, 2473–2478. [Google Scholar] [CrossRef]
- Tan, Z.; Shi, S.; Xu, J.; Liu, X.; Lei, Y.; Zhang, B.; Hua, J.; Meng, Q.; Wang, W.; Yu, X.; et al. RNA N6-methyladenosine demethylase FTO promotes pancreatic cancer progression by inducing the autocrine activity of PDGFC in an m(6)A-YTHDF2-dependent manner. Oncogene 2022, 41, 2860–2872. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Zhang, H.; Tan, Y.; Wang, Z.; Li, Y.; Yang, X. m6A demethylase FTO suppresses pancreatic cancer tumorigenesis by demethylating PJA2 and inhibiting Wnt signaling. Mol. Ther. Nucleic Acids 2021, 25, 277–292. [Google Scholar] [CrossRef]
- Huang, C.; Zhou, S.; Zhang, C.; Jin, Y.; Xu, G.; Zhou, L.; Ding, G.; Pang, T.; Jia, S.; Cao, L. ZC3H13-mediated N6-methyladenosine modification of PHF10 is impaired by fisetin which inhibits the DNA damage response in pancreatic cancer. Cancer Lett. 2022, 530, 16–28. [Google Scholar] [CrossRef]
- Hu, Y.; Tang, J.; Xu, F.; Chen, J.; Zeng, Z.; Han, S.; Wang, F.; Wang, D.; Huang, M.; Zhao, Y.; et al. A reciprocal feedback between N6-methyladenosine reader YTHDF3 and lncRNA DICER1-AS1 promotes glycolysis of pancreatic cancer through inhibiting maturation of miR-5586-5p. J. Exp. Clin. Cancer Res. 2022, 41, 69. [Google Scholar] [CrossRef]
- Deng, J.; Zhang, J.; Ye, Y.; Liu, K.; Zeng, L.; Huang, J.; Pan, L.; Li, M.; Bai, R.; Zhuang, L.; et al. N(6) -methyladenosine-Mediated Upregulation of WTAPP1 Promotes WTAP Translation and Wnt Signaling to Facilitate Pancreatic Cancer Progression. Cancer Res. 2021, 81, 5268–5283. [Google Scholar] [CrossRef]
- Hou, Y.; Zhang, Q.; Pang, W.; Hou, L.; Liang, Y.; Han, X.; Luo, X.; Wang, P.; Zhang, X.; Li, L.; et al. YTHDC1-mediated augmentation of miR-30d in repressing pancreatic tumorigenesis via attenuation of RUNX1-induced transcriptional activation of Warburg effect. Cell Death Differ. 2021, 28, 3105–3124. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, K.; Jiang, W.; Hu, Y.; Xiao, W.; Huang, Y.; Feng, Y.; Pan, Q.; Wan, R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol. Cancer 2020, 19, 91. [Google Scholar] [CrossRef]
- He, Y.; Hu, H.; Wang, Y.; Yuan, H.; Lu, Z.; Wu, P.; Liu, D.; Tian, L.; Yin, J.; Jiang, K.; et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell. Physiol. Biochem. 2018, 48, 838–846. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yue, H.; Cheng, Y.; Ding, Z.; Xu, Z.; Lv, C.; Wang, Z.; Wang, J.; Yin, C.; Hao, H.; et al. ALKBH5-mediated m(6)A demethylation of KCNK15-AS1 inhibits pancreatic cancer progression via regulating KCNK15 and PTEN/AKT signaling. Cell Death Dis. 2021, 12, 1121. [Google Scholar] [CrossRef]
- Huang, R.; Yang, L.; Zhang, Z.; Liu, X.; Fei, Y.; Tong, W.M.; Niu, Y.; Liang, Z. RNA m(6)A Demethylase ALKBH5 Protects Against Pancreatic Ductal Adenocarcinoma via Targeting Regulators of Iron Metabolism. Front. Cell Dev. Biol. 2021, 9, 724282. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Yang, Y.; Kang, M.; Wang, Y.; Wang, Y.; Bi, Y.; He, S.; Shimamoto, F. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol. Cancer 2020, 19, 3. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Wang, Z.; Gao, J.; Yang, C.; Feng, M.; Niu, Y.; Tong, W.M.; Bao, X.; Wang, R. METTL3-mediated RNA m6A Hypermethylation Promotes Tumorigenesis and GH Secretion of Pituitary Somatotroph Adenomas. J. Clin. Endocrinol. Metab. 2022, 107, 136–149. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Schlumberger, M.; Brose, M.; Elisei, R.; Leboulleux, S.; Luster, M.; Pitoia, F.; Pacini, F. Definition and management of radioactive iodine-refractory differentiated thyroid cancer. Lancet Diabetes Endocrinol. 2014, 2, 356–358. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Mu, Q.; Huang, H. The Roles of Insulin-Like Growth Factor 2 mRNA-Binding Protein 2 in Cancer and Cancer Stem Cells. Stem Cells Int. 2018, 2018, 4217259. [Google Scholar] [CrossRef]
- Su, T.S.; Liu, W.Y.; Han, S.H.; Jansen, M.; Yang-Fen, T.L.; P’Eng, F.K.; Chou, C.K. Transcripts of the insulin-like growth factors I and II in human hepatoma. Cancer Res. 1989, 49, 1773–1777. [Google Scholar]
- Cleynen, I.; Brants, J.R.; Peeters, K.; Deckers, R.; Debiec-Rychter, M.; Sciot, R.; Van de Ven, W.J.; Petit, M.M. HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-kappaB. Mol. Cancer Res. 2007, 5, 363–372. [Google Scholar] [CrossRef]
- Janiszewska, M.; Suvà, M.L.; Riggi, N.; Houtkooper, R.H.; Auwerx, J.; Clément-Schatlo, V.; Radovanovic, I.; Rheinbay, E.; Provero, P.; Stamenkovic, I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012, 26, 1926–1944. [Google Scholar] [CrossRef]
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef]
- Luvero, D.; Plotti, F.; Aloisia, A.; Montera, R.; Terranova, C.; Carlo, D.C.N.; Scaletta, G.; Lopez, S.; Miranda, A.; Capriglione, S.; et al. Ovarian cancer relapse: From the latest scientific evidence to the best practice. Crit. Rev. Oncol. Hematol. 2019, 140, 28–38. [Google Scholar] [CrossRef]
- Kleinschmidt, E.G.; Miller, N.; Ozmadenci, D.; Tancioni, I.; Osterman, C.D.; Barrie, A.M.; Taylor, K.N.; Ye, A.; Jiang, S.; Connolly, D.C.; et al. Rgnef promotes ovarian tumor progression and confers protection from oxidative stress. Oncogene 2019, 38, 6323–6337. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.J.; Hennessy, B.; Mills, G.B. The biology of ovarian cancer: New opportunities for translation. Nat. Rev. Cancer 2009, 9, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.T.; Webber, E.M.; Sawaya, G.F. Screening for Ovarian Cancer: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2018, 319, 595–606. [Google Scholar] [CrossRef]
- Furuya, M. Ovarian cancer stroma: Pathophysiology and the roles in cancer development. Cancers 2012, 4, 701–724. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Bitler, B.G.; Watson, Z.L.; Wheeler, L.J.; Behbakht, K. PARP inhibitors: Clinical utility and possibilities of overcoming resistance. Gynecol. Oncol. 2017, 147, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, E.A.; Sargent, D.J.; Grothey, A.; Kerbel, R.S. Drug rechallenge and treatment beyond progression--implications for drug resistance. Nat. Rev. Clin. Oncol. 2013, 10, 571–587. [Google Scholar] [CrossRef] [PubMed]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, W.; Wang, X.; Xu, C.; Zhang, N.; Di, W. Cisplatin-stimulated macrophages promote ovarian cancer migration via the CCL20-CCR6 axis. Cancer Lett. 2020, 472, 59–69. [Google Scholar] [CrossRef]
- Cao, L.; Shao, M.; Schilder, J.; Guise, T.; Mohammad, K.S.; Matei, D. Tissue transglutaminase links TGF-β, epithelial to mesenchymal transition and a stem cell phenotype in ovarian cancer. Oncogene 2012, 31, 2521–2534. [Google Scholar] [CrossRef]
- Takai, M.; Terai, Y.; Kawaguchi, H.; Ashihara, K.; Fujiwara, S.; Tanaka, T.; Tsunetoh, S.; Tanaka, Y.; Sasaki, H.; Kanemura, M.; et al. The EMT (epithelial-mesenchymal-transition)-related protein expression indicates the metastatic status and prognosis in patients with ovarian cancer. J. Ovarian Res. 2014, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Mitra, R.; Chen, X.; Greenawalt, E.J.; Maulik, U.; Jiang, W.; Zhao, Z.; Eischen, C.M. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat. Commun. 2017, 8, 1604. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Chandana, S.; Babiker, H.M.; Mahadevan, D. Therapeutic trends in pancreatic ductal adenocarcinoma (PDAC). Expert Opin. Investig. Drugs 2019, 28, 161–177. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Orth, M.; Metzger, P.; Gerum, S.; Mayerle, J.; Schneider, G.; Belka, C.; Schnurr, M.; Lauber, K. Pancreatic ductal adenocarcinoma: Biological hallmarks, current status, and future perspectives of combined modality treatment approaches. Radiat. Ther. 2019, 14, 141. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Connor, A.A.; Gallinger, S. Pancreatic cancer evolution and heterogeneity: Integrating omics and clinical data. Nat. Rev. Cancer 2022, 22, 131–142. [Google Scholar] [CrossRef]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Ihrie, R.A.; Marques, M.R.; Nguyen, B.T.; Horner, J.S.; Papazoglu, C.; Bronson, R.T.; Mills, A.A.; Attardi, L.D. Perp is a p63-regulated gene essential for epithelial integrity. Cell 2005, 120, 843–856. [Google Scholar] [CrossRef]
- Awais, R.; Spiller, D.G.; White, M.R.; Paraoan, L. p63 is required beside p53 for PERP-mediated apoptosis in uveal melanoma. Br. J. Cancer 2016, 115, 983–992. [Google Scholar] [CrossRef]
- Yeh, P.J.; Chen, J.W. Pituitary tumors: Surgical and medical management. J. Surg. Oncol. 1997, 6, 67–92. [Google Scholar] [CrossRef] [PubMed]
- Dorton, A.M. The pituitary gland: Embryology, physiology, and pathophysiology. Neonatal Netw. 2000, 19, 9–17. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Liao, P.; Rouse, C.; Chen, Y.; Dowling, J.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro-Oncol. 2014, 16 (Suppl. S4), iv1–iv63. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, C.D.; Su, Z.P.; Chen, Y.X.; Cai, L.; Zhuge, Q.C.; Wu, Z.B. Natural history of postoperative nonfunctioning pituitary adenomas: A systematic review and meta-analysis. Neuroendocrinology 2012, 96, 333–342. [Google Scholar] [CrossRef]
- Ezzat, S.; Asa, S.L.; Couldwell, W.T.; Barr, C.E.; Dodge, W.E.; Vance, M.L.; McCutcheon, I.E. The prevalence of pituitary adenomas: A systematic review. Am. Cancer Soc. 2004, 101, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Melmed, S. Pituitary-Tumor Endocrinopathies. N. Engl. J. Med. 2020, 382, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.M.; Batra, S.; Lim, M.; Gallia, G.L.; Burger, P.C.; Salvatori, R.; Wand, G.; Quinones-Hinojosa, A.; Kleinberg, L.; Redmond, K.J. Invasive adenoma and pituitary carcinoma: A SEER database analysis. Neurosurg. Rev. 2014, 37, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.W.; Bodach, M.E.; Tumialan, L.M.; Oyesiku, N.M.; Patil, C.G.; Litvack, Z.; Aghi, M.K.; Zada, G. Congress of Neurological Surgeons Systematic Review and Evidence-Based Guideline on Primary Management of Patients With Nonfunctioning Pituitary Adenomas. Neurosurgery 2016, 79, E533–E535. [Google Scholar] [CrossRef]
- Karki, M.; Sun, J.; Yadav, C.P.; Zhao, B. Large and giant pituitary adenoma resection by microscopic trans-sphenoidal surgery: Surgical outcomes and complications in 123 consecutive patients. J. Clin. Neurosci. 2017, 44, 310–314. [Google Scholar] [CrossRef]
- Salomon, M.P.; Wang, X.; Marzese, D.M.; Hsu, S.C.; Nelson, N.; Zhang, X.; Matsuba, C.; Takasumi, Y.; Ballesteros-Merino, C.; Fox, B.A.; et al. The Epigenomic Landscape of Pituitary Adenomas Reveals Specific Alterations and Differentiates Among Acromegaly, Cushing’s Disease and Endocrine-Inactive Subtypes. Clin. Cancer Res. 2018, 24, 4126–4136. [Google Scholar] [CrossRef]
- Reddy, R.; Cudlip, S.; Byrne, J.V.; Karavitaki, N.; Wass, J.A. Can we ever stop imaging in surgically treated and radiotherapy-naive patients with non-functioning pituitary adenoma? Eur. J. Endocrinol. 2011, 165, 739–744. [Google Scholar] [CrossRef]
- Tzelepis, K.; Rausch, O.; Kouzarides, T. RNA-modifying enzymes and their function in a chromatin context. Nat. Struct. Mol. Biol. 2019, 26, 858–862. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
- Burgess, H.M.; Depledge, D.P.; Thompson, L.; Srinivas, K.P.; Grande, R.C.; Vink, E.I.; Abebe, J.S.; Blackaby, W.P.; Hendrick, A.; Albertella, M.R.; et al. Targeting the m(6)A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev. 2021, 35, 1005–1019. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).