Utility of Comprehensive Genomic Profiling Tests for Patients with Incurable Pancreatic Cancer in Clinical Practice
Abstract
Simple Summary
Abstract
1. Background
2. Material and Methods
2.1. Study Design, Patients and Collecting Data
- 1.
- Patients’ characteristics
- 2.
- Results of CGP tests
- 3.
- Survival outcomes according to HRD related gene mutations
2.2. Statistical Analysis
3. Results
3.1. Characteristics of CGP Tests
3.2. Results of CGP Tests
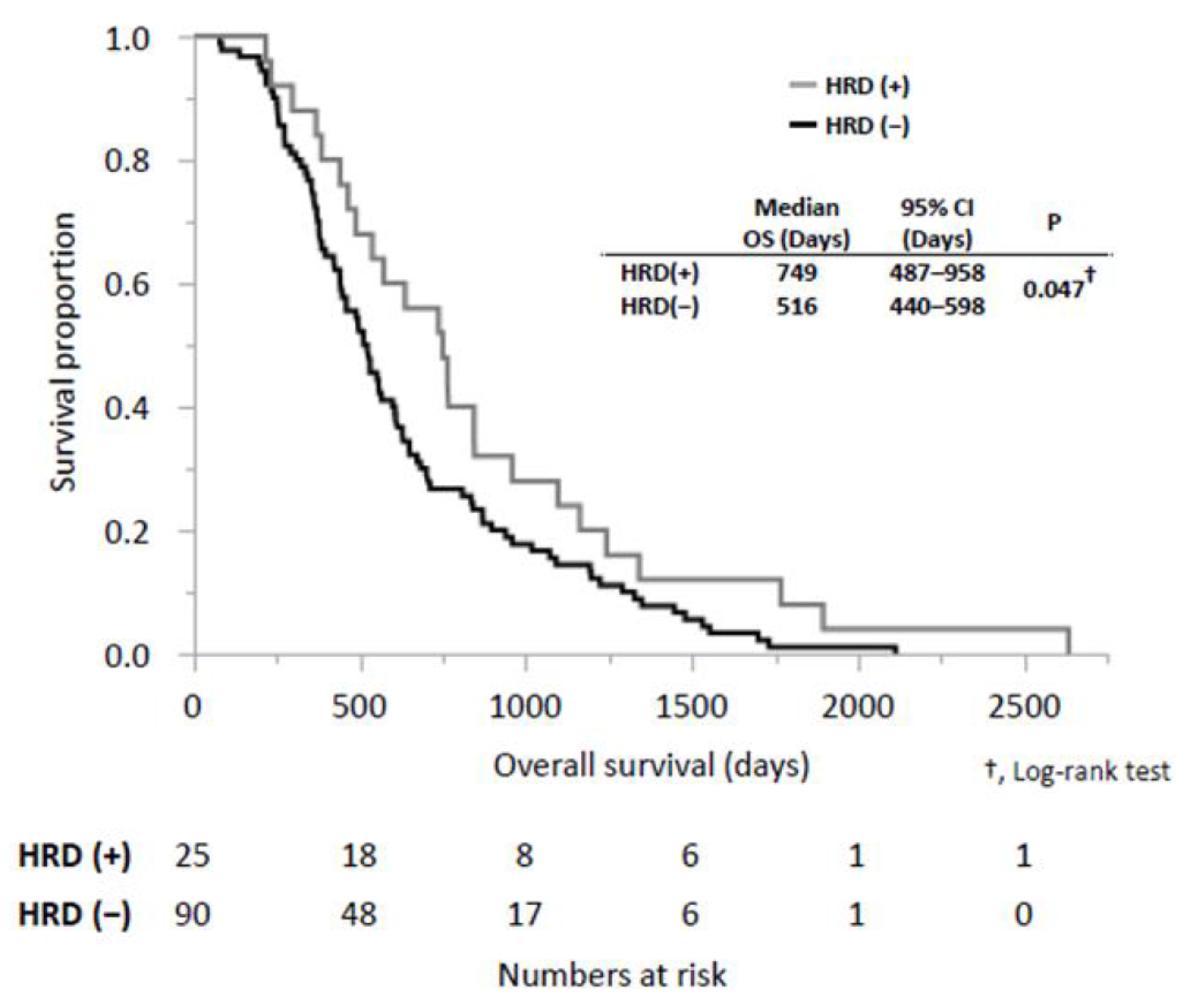
3.3. Survival Outcomes According to HRD-Related Gene Mutations
3.4. Treatment Outcomes According to HRD-Related Genetic Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Gheorghe, G.; Bungau, S.; Ilie, M.; Behl, T.; Vesa, C.M.; Brisc, C.; Bacalbasa, N.; Turi, V.; Costache, R.S.; Diaconu, C.C. Early Diagnosis of Pancreatic Cancer: The Key for Survival. Diagnostics 2020, 10, 869. [Google Scholar] [CrossRef]
- Fukuda, J.; Ikezawa, K.; Nakao, M.; Okagaki, S.; Ashida, R.; Ioka, T.; Takada, R.; Yamai, T.; Fukutake, N.; Uehara, H.; et al. Predictive Factors for Pancreatic Cancer and Its Early Detection Using Special Pancreatic Ultrasonography in High-Risk Individuals. Cancers 2021, 13, 502. [Google Scholar] [CrossRef]
- Ilic, M.; Ilic, I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 2016, 22, 9694–9705. [Google Scholar] [CrossRef]
- Takada, R.; Ikezawa, K.; Kiyota, R.; Imai, T.; Abe, Y.; Kai, Y.; Yamai, T.; Fukutake, N.; Uehara, H.; Ashida, R.; et al. Microsatellite instability status of pancreatic cancer and experience with pembrolizumab treatment. Suizo 2021, 36, 120–127. [Google Scholar] [CrossRef]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Moon, J.J.; Lu, A.; Moon, C. Role of genomic instability in human carcinogenesis. Exp. Biol. Med. 2019, 244, 227–240. [Google Scholar] [CrossRef]
- Luchini, C.; Brosens, L.A.A.; Wood, L.D.; Chatterjee, D.; Shin, J.I.; Sciammarella, C.; Fiadone, G.; Malleo, G.; Salvia, R.; Kryklyva, V.; et al. Comprehensive characterisation of pancreatic ductal adenocarcinoma with microsatellite instability: Histology, molecular pathology and clinical implications. Gut 2021, 70, 148–156. [Google Scholar] [CrossRef]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef]
- Golan, T.; O’Kane, G.M.; Denroche, R.E.; Raitses-Gurevich, M.; Grant, R.C.; Holter, S.; Wang, Y.; Zhang, A.; Jang, G.H.; Stossel, C.; et al. Genomic Features and Classification of Homologous Recombination Deficient Pancreatic Ductal Adenocarcinoma. Gastroenterology 2021, 160, 2119–2132. [Google Scholar] [CrossRef]
- Casolino, R.; Paiella, S.; Azzolina, D.; Beer, P.A.; Corbo, V.; Lorenzoni, G.; Gregori, D.; Golan, T.; Braconi, C.; Froeling, F.E.M.; et al. Homologous Recombination Deficiency in Pancreatic Cancer: A Systematic Review and Prevalence Meta-Analysis. J. Clin. Oncol. 2021, 39, 2617–2631. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Ikezawa, K.; Kiyota, R.; Takada, R.; Daiku, K.; Maeda, S.; Imai, T.; Abe, Y.; Kai, Y.; Yamai, T.; Fukutake, N.; et al. Efficacy and safety of modified fluorouracil/leucovorin plus irinotecan and oxaliplatin (mFOLFIRINOX) compared with S-1 as second-line chemotherapy in metastatic pancreatic cancer. JGH Open 2021, 5, 679–685. [Google Scholar] [CrossRef]
- Yamai, T.; Ikezawa, K.; Kawamoto, Y.; Hirao, T.; Higashi, S.; Daiku, K.; Maeda, S.; Abe, Y.; Urabe, M.; Kai, Y.; et al. 5-Fluorouracil/L-Leucovorin Plus Oxaliplatin (FOLFOX) Regimen as Salvage Chemotherapy for Patients with Unresectable Pancreatic Cancer Receiving Gemcitabine and Nab-Paclitaxel and 5-Fluorouracil/L-Leucovorin Plus Nanoliposomal Irinotecan: Preliminary Results from Clinical Practice. Curr. Oncol. 2022, 29, 2644–2649. [Google Scholar] [CrossRef]
- Mukai, Y.; Ueno, H. Establishment and implementation of Cancer Genomic Medicine in Japan. Cancer Sci. 2021, 112, 970–977. [Google Scholar] [CrossRef]
- Inagaki, C.; Maeda, D.; Hatake, K.; Sato, Y.; Hashimoto, K.; Sakai, D.; Yachida, S.; Nonomura, N.; Satoh, T. Clinical Utility of Next-Generation Sequencing-Based Panel Testing under the Universal Health-Care System in Japan: A Retrospective Analysis at a Single University Hospital. Cancers 2021, 13, 1121. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology: Pancreatic Adenocarcinoma Version 1.2022. Available online: https://www.nccn.org/guidelines/guidelines-with-evidence-blocks (accessed on 23 December 2022).
- Kunimasa, K.; Sugimoto, N.; Kawamura, T.; Yamasaki, T.; Honma, K.; Nagata, S.; Kukita, Y.; Fujisawa, F.; Inoue, T.; Yamaguchi, Y.; et al. Clinical application of comprehensive genomic profiling panel to thoracic malignancies: A single-center retrospective study. Thorac. Cancer 2022, 13, 2970–2977. [Google Scholar] [CrossRef]
- Park, W.; Chen, J.; Chou, J.F.; Varghese, A.M.; Yu, K.H.; Wong, W.; Capanu, M.; Balachandran, V.; McIntyre, C.A.; El Dika, I.; et al. Genomic Methods Identify Homologous Recombination Deficiency in Pancreas Adenocarcinoma and Optimize Treatment Selection. Clin. Cancer Res. 2020, 26, 3239–3247. [Google Scholar] [CrossRef]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.; Kaestner, V.; Mailankody, S. Cancer Drugs Approved Based on Biomarkers and Not Tumor Type-FDA Approval of Pembrolizumab for Mismatch Repair-Deficient Solid Cancers. JAMA Oncol. 2018, 4, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Eso, Y.; Shimizu, T.; Takeda, H.; Takai, A.; Marusawa, H. Microsatellite instability and immune checkpoint inhibitors: Toward precision medicine against gastrointestinal and hepatobiliary cancers. J. Gastroenterol. 2020, 55, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Kai, Y.; Ikezawa, K.; Takada, R.; Daiku, K.; Maeda, S.; Abe, Y.; Yamai, T.; Fukutake, N.; Nakabori, T.; Uehara, H.; et al. Success rate of microsatellite instability examination and complete response with pembrolizumab in biliary tract cancer. JGH Open 2021, 5, 712–716. [Google Scholar] [CrossRef]
- Eso, Y.; Seno, H. Current status of treatment with immune checkpoint inhibitors for gastrointestinal, hepatobiliary, and pancreatic cancers. Therap. Adv. Gastroenterol. 2020, 13, 1756284820948773. [Google Scholar] [CrossRef]
- Naito, Y.; Aburatani, H.; Amano, T.; Baba, E.; Furukawa, T.; Hayashida, T.; Hiyama, E.; Ikeda, S.; Kanai, M.; Kato, M.; et al. Clinical practice guidance for next-generation sequencing in cancer diagnosis and treatment (edition 2.1). Int. J. Clin. Oncol. 2021, 26, 233–283. [Google Scholar] [CrossRef]
- Wang-Gillam, A.; Li, C.P.; Bodoky, G.; Dean, A.; Shan, Y.S.; Jameson, G.; Macarulla, T.; Lee, K.H.; Cunningham, D.; Blanc, J.F.; et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): A global, randomised, open-label, phase 3 trial. Lancet 2016, 387, 545–557. [Google Scholar] [CrossRef]
- Oettle, H.; Riess, H.; Stieler, J.M.; Heil, G.; Schwaner, I.; Seraphin, J.; Gorner, M.; Molle, M.; Greten, T.F.; Lakner, V.; et al. Second-line oxaliplatin, folinic acid, and fluorouracil versus folinic acid and fluorouracil alone for gemcitabine-refractory pancreatic cancer: Outcomes from the CONKO-003 trial. J. Clin. Oncol. 2014, 32, 2423–2429. [Google Scholar] [CrossRef]
- Gill, S.; Ko, Y.J.; Cripps, C.; Beaudoin, A.; Dhesy-Thind, S.; Zulfiqar, M.; Zalewski, P.; Do, T.; Cano, P.; Lam, W.Y.H.; et al. PANCREOX: A Randomized Phase III Study of Fluorouracil/Leucovorin With or Without Oxaliplatin for Second-Line Advanced Pancreatic Cancer in Patients Who Have Received Gemcitabine-Based Chemotherapy. J. Clin. Oncol. 2016, 34, 3914–3920. [Google Scholar] [CrossRef]
- Park, H.S.; Kang, B.; Chon, H.J.; Im, H.S.; Lee, C.K.; Kim, I.; Kang, M.J.; Hwang, J.E.; Bae, W.K.; Cheon, J.; et al. Liposomal irinotecan plus fluorouracil/leucovorin versus FOLFIRINOX as the second-line chemotherapy for patients with metastatic pancreatic cancer: A multicenter retrospective study of the Korean Cancer Study Group (KCSG). ESMO Open 2021, 6, 100049. [Google Scholar] [CrossRef] [PubMed]
- Sehdev, A.; Gbolahan, O.; Hancock, B.A.; Stanley, M.; Shahda, S.; Wan, J.; Wu, H.H.; Radovich, M.; O’Neil, B.H. Germline and Somatic DNA Damage Repair Gene Mutations and Overall Survival in Metastatic Pancreatic Adenocarcinoma Patients Treated with FOLFIRINOX. Clin. Cancer Res. 2018, 24, 6204–6211. [Google Scholar] [CrossRef] [PubMed]
| Number of patients, n | 115 |
| Median age (range), y.o | 63 (37–80) |
| Sex | |
| Male, n (%) | 63 (54.8) |
| Female, n (%) | 52 (45.2) |
| Diagnosis at the start of treatment | |
| R or BR, n (%) | 26 (22.6) |
| UR-LA, n (%) | 24 (20.9) |
| UR-M, n (%) | 65 (56.5) |
| Timing of submitting cancer gene panel tests | |
| During 1st-line treatment, n (%) | 30 (26.1) |
| After disease progression of 1st-line treatment, n (%) | 59 (51.3) |
| After disease progression of 2nd-line treatment, n (%) | 26 (22.6) |
| Samples for CGP tests | |
| Pancreas, n (%) | 51 (44.4) |
| Liver, n (%) | 45 (39.1) |
| Lymph node, n (%) | 7 (6.1) |
| Gastrointestinal tract, n (%) | 5 (4.3) |
| Lung, n (%) | 4 (3.5) |
| Peritoneum, n (%) | 3 (2.6) |
| Sampling methods | |
| EUS-FNA, n (%) | 44 (38.3) |
| Percutaneous needle biopsy, n (%) | 42 (36.5) |
| Surgery, n (%) | 24 (20.9) |
| Forceps biopsy, n (%) | 5 (4.3) |
| Kinds of cancer gene panel tests | |
| FoundationOne CDx, n (%) | 93 (80.9) |
| NCC Oncopanel, n (%) | 22 (19.1) |
| Quality control | |
| Pass/met the criteria, n (%) | 98 (85.2) |
| Qualified, n (%) | 17 (14.8) |
| TMB-H or MSI-H, n (%) | 8 (6.9) |
| Pathological gene mutations (range), n | 4 (1–10) |
| KRAS mutations, n (%) | 107 (93.0) |
| TP53 mutations, n (%) | 96 (83.0) |
| CDKN2A mutations, n (%) | 61 (53.0) |
| SMAD4 mutations, n (%) | 29 (45.2) |
| HRD-related genes mutations, n (%) | 25 (21.7) |
| BRCA1/2 mutations, n (%) | 11 (9.6) |
| ATM mutations, n (%) | 4 (3.5) |
| RAD51C mutations, n (%) | 3 (2.6) |
| FANCA mutations, n (%) | 2 (1.7) |
| Others mutations, n (%) | 5 (4.3) |
| Administration of pembrolizumab, n (%) | 4 (3.5) |
| Clinical Trial Participation, n (%) | 2 (1.7) |
| HRD (+) | HRD (−) | p-Value | |
|---|---|---|---|
| Number of patients, n | 25 | 90 | |
| Median age (range), y.o. | 61 (38–78) | 64 (37–80) | 0.585 † |
| Sex | 0.822 § | ||
| Male, n (%) | 13 (52.0) | 50 (55.6) | |
| Female, n (%) | 12 (48.0) | 40 (44.4) | |
| Operability at the time of diagnosis | |||
| R or BR, n (%) | 5 (20.0) | 22 (24.4) | 0.658 § |
| UR-LA, n (%) | 6 (42.0) | 18 (20.0) | |
| UR-M, n (%) | 14 (56.0) | 50 (55.6) | |
| TMB-H or MSI-H, n (%) | 1 (4.0) | 7 (7.8) | 1.000 § |
| Performance status | 0.691 § | ||
| 0 | 16 (64.0) | 60 (66.7) | |
| 1- | 5 (20.0) | 21 (23.3) | |
| NA | 4 (16.0) | 9 (10.0) | |
| Platinum-based regimen, n (%) | 21 (84.0) | 60 (66.7) | 0.136 § |
| Curative resection, n (%) | 4 (16.0) | 24 (26.7) | 0.429 § |
| Median WBC (range), /μL | 5380 (2970–10,490) | 5420 (2530–19,910) | 0.929 † |
| Median Hb (range), g/dL | 12.8 (9.4–15.0) | 12.5 (8.2–15.1) | 0.265 † |
| Median Platelet (range), 104/μL | 20.5 (15.1–44.9) | 22.8 (9.7–49.7) | 0.610 † |
| Median Albumin (range), mg/dL | 3.8 (2.8–4.5) | 3.8 (2.3–5.5) | 0.187 † |
| Median CA19-9 (range), mg/dL | 503 (2–100,000) | 368 (2–100,000) | 0.741 † |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Age (>70 vs. ≤70, y.o.) | 0.65 | 0.43–1.00 | 0.051 | |||
| Male vs. Female | 0.73 | 0.51–1.08 | 0.114 | |||
| PS (1- and NA vs. 0) | 1.43 | 0.96–2.11 | 0.078 | |||
| Operability at the time of diagnosis. (UR-M vs. R, BR, and UR-LA) | 1.82 | 1.25–2.66 | 0.002 | 1.97 | 1.34–2.92 | <0.001 |
| HRD related gene mutation (HRD (+) vs. HRD (−)) | 0.63 | 0.40–0.99 | 0.049 | 0.60 | 0.34–0.96 | 0.035 |
| Platinum-based regimen (Y vs. N) | 0.97 | 0.64–1.50 | 0.876 | |||
| Baseline WBC (>5420 vs. ≤5420, /μL) | 0.92 | 0.63–1.34 | 0.673 | |||
| Baseline Hb (<12.7 vs. ≥12.7, g/dL) | 1.58 | 1.08–2.30 | 0.018 | 1.54 | 1.06–2.23 | 0.025 |
| Baseline Platelet (<22.6 vs. ≥22.6, ×104/μL) | 0.98 | 0.67–1.43 | 0.915 | |||
| Baseline albumin (<3.8 vs. ≥3.8, U/mL) | 1.45 | 0.99–2.12 | 0.056 | |||
| Baseline CA19-9 (>400 vs. <400, U/mL) | 1.41 | 0.96–2.06 | 0.078 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamai, T.; Ikezawa, K.; Sugimoto, N.; Urabe, M.; Kai, Y.; Takada, R.; Nakabori, T.; Uehara, H.; Kawamura, T.; Kunimasa, K.; et al. Utility of Comprehensive Genomic Profiling Tests for Patients with Incurable Pancreatic Cancer in Clinical Practice. Cancers 2023, 15, 970. https://doi.org/10.3390/cancers15030970
Yamai T, Ikezawa K, Sugimoto N, Urabe M, Kai Y, Takada R, Nakabori T, Uehara H, Kawamura T, Kunimasa K, et al. Utility of Comprehensive Genomic Profiling Tests for Patients with Incurable Pancreatic Cancer in Clinical Practice. Cancers. 2023; 15(3):970. https://doi.org/10.3390/cancers15030970
Chicago/Turabian StyleYamai, Takuo, Kenji Ikezawa, Naotoshi Sugimoto, Makiko Urabe, Yugo Kai, Ryoji Takada, Tasuku Nakabori, Hiroyuki Uehara, Takahisa Kawamura, Kei Kunimasa, and et al. 2023. "Utility of Comprehensive Genomic Profiling Tests for Patients with Incurable Pancreatic Cancer in Clinical Practice" Cancers 15, no. 3: 970. https://doi.org/10.3390/cancers15030970
APA StyleYamai, T., Ikezawa, K., Sugimoto, N., Urabe, M., Kai, Y., Takada, R., Nakabori, T., Uehara, H., Kawamura, T., Kunimasa, K., Yamamoto, S., Wakamatsu, T., Hayashi, T., Kukita, Y., Fujisawa, F., Inoue, T., Yamaguchi, Y., Yamasaki, T., Honma, K., & Ohkawa, K. (2023). Utility of Comprehensive Genomic Profiling Tests for Patients with Incurable Pancreatic Cancer in Clinical Practice. Cancers, 15(3), 970. https://doi.org/10.3390/cancers15030970







