Innate Immune Program in Formation of Tumor-Initiating Cells from Cells-of-Origin of Breast, Prostate, and Ovarian Cancers
Abstract
Simple Summary
Abstract
1. Brief Summary of TICs/CSCs
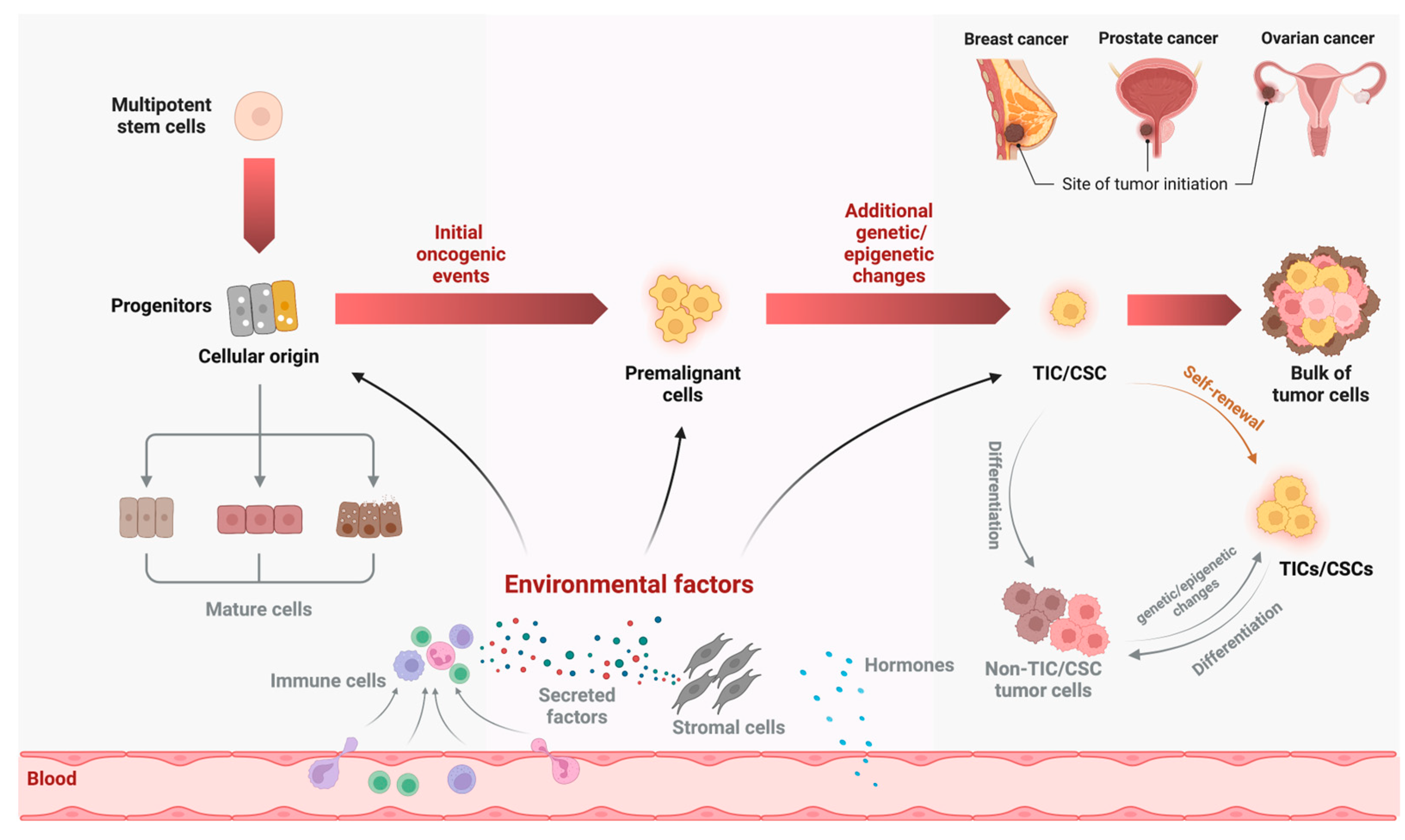
2. Formation of TIC/CSC from the Cellular Origin of Cancer
3. Luminal Progenitors as Cellular Origins of Breast, Prostate, and Ovarian Cancers
4. Common Innate Immune Program in LPs
5. Presence of Microbes in Normal and Cancerous Breast, Prostate, and FT Tissues
5.1. Microbes in Breast Tissue and Cancer
5.2. Microbes in Prostate Tissue and Cancer
5.3. Microbes in FT Tissue and Ovarian Cancer
6. Immune Programs in Formation of TICs/CSCs
7. Perspective of Targeting TICs/CSCs by Enhancing Immunotherapy
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 2014, 15, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Herschkowitz, J.I.; Zhao, W.; Zhang, M.; Usary, J.; Murrow, G.; Edwards, D.; Knezevic, J.; Greene, S.B.; Darr, D.; Troester, M.A.; et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc. Natl. Acad. Sci. USA 2012, 109, 2778–2783. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef]
- Dalerba, P.; Dylla, S.J.; Park, I.K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef]
- Chen, X.; Li, Q.; Liu, X.; Liu, C.; Liu, R.; Rycaj, K.; Zhang, D.; Liu, B.; Jeter, C.; Calhoun-Davis, T.; et al. Defining a Population of Stem-like Human Prostate Cancer Cells That Can Generate and Propagate Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2016, 22, 4505–4516. [Google Scholar] [CrossRef]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef]
- Jiao, J.; Hindoyan, A.; Wang, S.; Tran, L.M.; Goldstein, A.S.; Lawson, D.; Chen, D.; Li, Y.; Guo, C.; Zhang, B.; et al. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS ONE 2012, 7, e42564. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Liu, X.; Laffin, B.; Chen, X.; Choy, G.; Jeter, C.R.; Calhoun-Davis, T.; Li, H.; Palapattu, G.S.; Pang, S.; et al. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell 2012, 10, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Bapat, S.A.; Mali, A.M.; Koppikar, C.B.; Kurrey, N.K. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005, 65, 3025–3029. [Google Scholar] [CrossRef] [PubMed]
- Foster, R.; Buckanovich, R.J.; Rueda, B.R. Ovarian cancer stem cells: Working towards the root of stemness. Cancer Lett. 2013, 338, 147–157. [Google Scholar] [CrossRef]
- Lupia, M.; Cavallaro, U. Ovarian cancer stem cells: Still an elusive entity? Mol. Cancer 2017, 16, 64. [Google Scholar] [CrossRef]
- Motohara, T.; Yoshida, G.J.; Katabuchi, H. The hallmarks of ovarian cancer stem cells and niches: Exploring their harmonious interplay in therapy resistance. Semin. Cancer Biol. 2021, 77, 182–193. [Google Scholar] [CrossRef]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M.; Zhan, Q.; Jordan, S.; Duncan, L.M.; Weishaupt, C.; et al. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef]
- Quintana, E.; Shackleton, M.; Sabel, M.S.; Fullen, D.R.; Johnson, T.M.; Morrison, S.J. Efficient tumour formation by single human melanoma cells. Nature 2008, 456, 593–598. [Google Scholar] [CrossRef]
- Driessens, G.; Beck, B.; Caauwe, A.; Simons, B.D.; Blanpain, C. Defining the mode of tumour growth by clonal analysis. Nature 2012, 488, 527–530. [Google Scholar] [CrossRef]
- Schepers, A.G.; Snippert, H.J.; Stange, D.E.; van den Born, M.; van Es, J.H.; van de Wetering, M.; Clevers, H. Lineage Tracing Reveals Lgr5+ Stem Cell Activity in Mouse Intestinal Adenomas. Science 2012, 337, 730–735. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Lodestijn, S.C.; Miedema, D.M.; Lenos, K.J.; Nijman, L.E.; Belt, S.C.; El Makrini, K.; Lecca, M.C.; Waasdorp, C.; van den Bosch, T.; Bijlsma, M.F.; et al. Marker-free lineage tracing reveals an environment-instructed clonogenic hierarchy in pancreatic cancer. Cell Rep. 2021, 37, 109852. [Google Scholar] [CrossRef] [PubMed]
- Nik-Zainal, S.; Van Loo, P.; Wedge, D.C.; Alexandrov, L.B.; Greenman, C.D.; Lau, K.W.; Raine, K.; Jones, D.; Marshall, J.; Ramakrishna, M.; et al. The life history of 21 breast cancers. Cell 2012, 149, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Arya, R.K.; Maheshwari, S.; Singh, A.; Meena, S.; Pandey, P.; Dormond, O.; Datta, D. Tumor heterogeneity and cancer stem cell paradigm: Updates in concept, controversies and clinical relevance. Int. J. Cancer 2015, 136, 1991–2000. [Google Scholar] [CrossRef]
- Rycaj, K.; Tang, D.G. Cell-of-Origin of Cancer versus Cancer Stem Cells: Assays and Interpretations. Cancer Res. 2015, 75, 4003–4011. [Google Scholar] [CrossRef]
- Shackleton, M.; Vaillant, F.; Simpson, K.J.; Stingl, J.; Smyth, G.K.; Asselin-Labat, M.L.; Wu, L.; Lindeman, G.J.; Visvader, J.E. Generation of a functional mammary gland from a single stem cell. Nature 2006, 439, 84–88. [Google Scholar] [CrossRef]
- Stingl, J.; Eirew, P.; Ricketson, I.; Shackleton, M.; Vaillant, F.; Choi, D.; Li, H.I.; Eaves, C.J. Purification and unique properties of mammary epithelial stem cells. Nature 2006, 439, 993–997. [Google Scholar] [CrossRef]
- Foulkes, W.D. BRCA1 functions as a breast stem cell regulator. J. Med. Genet. 2004, 41, 1–5. [Google Scholar] [CrossRef]
- Liu, S.; Ginestier, C.; Charafe-Jauffret, E.; Foco, H.; Kleer, C.G.; Merajver, S.D.; Dontu, G.; Wicha, M.S. BRCA1 regulates human mammary stem/progenitor cell fate. Proc. Natl. Acad. Sci. USA 2008, 105, 1680–1685. [Google Scholar] [CrossRef]
- Visvader, J.E.; Stingl, J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes Dev. 2014, 28, 1143–1158. [Google Scholar] [CrossRef]
- Skibinski, A.; Kuperwasser, C. The origin of breast tumor heterogeneity. Oncogene 2015, 34, 5309–5316. [Google Scholar] [CrossRef] [PubMed]
- Asselin-Labat, M.L.; Sutherland, K.D.; Barker, H.; Thomas, R.; Shackleton, M.; Forrest, N.C.; Hartley, L.; Robb, L.; Grosveld, F.G.; van der Wees, J.; et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 2007, 9, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.; Teschendorff, A.; Sharp, G.; Novcic, N.; Russell, A.; Avril, S.; Prater, M.; Eirew, P.; Caldas, C.; Watson, C.J.; et al. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012, 14, R134. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.H.; Kunasegaran, K.; Tarulli, G.A.; De Silva, D.; Voorhoeve, P.M.; Pietersen, A.M. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Res. 2014, 16, R1. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; van Bragt, M.P.; Li, Z. A Long-Lived Luminal Subpopulation Enriched with Alveolar Progenitors Serves as Cellular Origin of Heterogeneous Mammary Tumors. Stem Cell Rep. 2015, 5, 60–74. [Google Scholar] [CrossRef]
- Lim, E.; Vaillant, F.; Wu, D.; Forrest, N.C.; Pal, B.; Hart, A.H.; Asselin-Labat, M.L.; Gyorki, D.E.; Ward, T.; Partanen, A.; et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009, 15, 907–913. [Google Scholar] [CrossRef]
- Molyneux, G.; Geyer, F.C.; Magnay, F.A.; McCarthy, A.; Kendrick, H.; Natrajan, R.; Mackay, A.; Grigoriadis, A.; Tutt, A.; Ashworth, A.; et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 2010, 7, 403–417. [Google Scholar] [CrossRef]
- Molyneux, G.; Smalley, M.J. The cell of origin of BRCA1 mutation-associated breast cancer: A cautionary tale of gene expression profiling. J. Mammary Gland Biol. Neoplasia. 2011, 16, 51–55. [Google Scholar] [CrossRef]
- Proia, T.A.; Keller, P.J.; Gupta, P.B.; Klebba, I.; Jones, A.D.; Sedic, M.; Gilmore, H.; Tung, N.; Naber, S.P.; Schnitt, S.; et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 2011, 8, 149–163. [Google Scholar] [CrossRef]
- Wang, H.; Xiang, D.; Liu, B.; He, A.; Randle, H.J.; Zhang, K.X.; Dongre, A.; Sachs, N.; Clark, A.P.; Tao, L.; et al. Inadequate DNA Damage Repair Promotes Mammary Transdifferentiation, Leading to BRCA1 Breast Cancer. Cell 2019, 178, 135–151.e119. [Google Scholar] [CrossRef]
- Pfefferle, A.D.; Herschkowitz, J.I.; Usary, J.; Harrell, J.C.; Spike, B.T.; Adams, J.R.; Torres-Arzayus, M.I.; Brown, M.; Egan, S.E.; Wahl, G.M.; et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 2013, 14, R125. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.M.; Abate-Shen, C. Molecular genetics of prostate cancer: New prospects for old challenges. Genes Dev. 2010, 24, 1967–2000. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.S.; Huang, J.; Guo, C.; Garraway, I.P.; Witte, O.N. Identification of a cell of origin for human prostate cancer. Science 2010, 329, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.A.; Zong, Y.; Memarzadeh, S.; Xin, L.; Huang, J.; Witte, O.N. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc. Natl. Acad. Sci. USA 2010, 107, 2610–2615. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.A.; Shen, M.M. Revisiting the concept of cancer stem cells in prostate cancer. Oncogene 2011, 30, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Choi, N.; Zhang, B.; Zhang, L.; Ittmann, M.; Xin, L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 2012, 21, 253–265. [Google Scholar] [CrossRef]
- Wang, Z.A.; Mitrofanova, A.; Bergren, S.K.; Abate-Shen, C.; Cardiff, R.D.; Califano, A.; Shen, M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013, 15, 274–283. [Google Scholar] [CrossRef]
- Goldstein, A.S.; Lawson, D.A.; Cheng, D.; Sun, W.; Garraway, I.P.; Witte, O.N. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc. Natl. Acad. Sci. USA 2008, 105, 20882–20887. [Google Scholar] [CrossRef]
- Liu, J.; Pascal, L.E.; Isharwal, S.; Metzger, D.; Ramos Garcia, R.; Pilch, J.; Kasper, S.; Williams, K.; Basse, P.H.; Nelson, J.B.; et al. Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol. Endocrinol. 2011, 25, 1849–1857. [Google Scholar] [CrossRef]
- Ousset, M.; Van Keymeulen, A.; Bouvencourt, G.; Sharma, N.; Achouri, Y.; Simons, B.D.; Blanpain, C. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol. 2012, 14, 1131–1138. [Google Scholar] [CrossRef]
- Wang, X.; Kruithof-de Julio, M.; Economides, K.D.; Walker, D.; Yu, H.; Halili, M.V.; Hu, Y.P.; Price, S.M.; Abate-Shen, C.; Shen, M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 2009, 461, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Hynes, P.G.; Tillman, H.S.; Lake, R.; Abou-Kheir, W.G.; Fang, L.; Casey, O.M.; Ameri, A.H.; Martin, P.L.; Yin, J.J.; et al. Identification of Different Classes of Luminal Progenitor Cells within Prostate Tumors. Cell Rep. 2015, 13, 2147–2158. [Google Scholar] [CrossRef] [PubMed]
- Chua, C.W.; Shibata, M.; Lei, M.; Toivanen, R.; Barlow, L.J.; Bergren, S.K.; Badani, K.K.; McKiernan, J.M.; Benson, M.C.; Hibshoosh, H.; et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 2014, 16, 951–961. [Google Scholar] [CrossRef]
- Karthaus, W.R.; Iaquinta, P.J.; Drost, J.; Gracanin, A.; van Boxtel, R.; Wongvipat, J.; Dowling, C.M.; Gao, D.; Begthel, H.; Sachs, N.; et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 2014, 159, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Barros-Silva, J.D.; Linn, D.E.; Steiner, I.; Guo, G.; Ali, A.; Pakula, H.; Ashton, G.; Peset, I.; Brown, M.; Clarke, N.W.; et al. Single-Cell Analysis Identifies LY6D as a Marker Linking Castration-Resistant Prostate Luminal Cells to Prostate Progenitors and Cancer. Cell Rep. 2018, 25, 3504–3518.e3506. [Google Scholar] [CrossRef]
- Kwon, O.J.; Zhang, L.; Xin, L. Stem Cell Antigen-1 Identifies a Distinct Androgen-Independent Murine Prostatic Luminal Cell Lineage with Bipotent Potential. Stem Cells 2016, 34, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.; Cambuli, F.; Aparicio, L.; Shibata, M.; Robinson, B.D.; Xuan, S.; Li, W.; Hibshoosh, H.; Loda, M.; Rabadan, R.; et al. A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors. eLife 2020, 9, e59465. [Google Scholar] [CrossRef]
- Guo, W.; Li, L.; He, J.; Liu, Z.; Han, M.; Li, F.; Xia, X.; Zhang, X.; Zhu, Y.; Wei, Y.; et al. Single-cell transcriptomics identifies a distinct luminal progenitor cell type in distal prostate invagination tips. Nat. Genet. 2020, 52, 908–918. [Google Scholar] [CrossRef]
- Karthaus, W.R.; Hofree, M.; Choi, D.; Linton, E.L.; Turkekul, M.; Bejnood, A.; Carver, B.; Gopalan, A.; Abida, W.; Laudone, V.; et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science 2020, 368, 497–505. [Google Scholar] [CrossRef]
- Wang, Z.A.; Toivanen, R.; Bergren, S.K.; Chambon, P.; Shen, M.M. Luminal Cells Are Favored as the Cell of Origin for Prostate Cancer. Cell Rep. 2014, 8, 1339–1346. [Google Scholar] [CrossRef]
- Chene, G.; Dauplat, J.; Radosevic-Robin, N.; Cayre, A.; Penault-Llorca, F. Tu-be or not tu-be: That is the question…about serous ovarian carcinogenesis. Crit. Rev. Oncol. Hematol. 2013, 88, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Flesken-Nikitin, A.; Hwang, C.I.; Cheng, C.Y.; Michurina, T.V.; Enikolopov, G.; Nikitin, A.Y. Ovarian surface epithelium at the junction area contains a cancer-prone stem cell niche. Nature 2013, 495, 241–245. [Google Scholar] [CrossRef]
- Bowtell, D.D.; Bohm, S.; Ahmed, A.A.; Aspuria, P.J.; Bast, R.C., Jr.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Ducie, J.; Dao, F.; Considine, M.; Olvera, N.; Shaw, P.A.; Kurman, R.J.; Shih, I.M.; Soslow, R.A.; Cope, L.; Levine, D.A. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nat. Commun. 2017, 8, 990. [Google Scholar] [CrossRef] [PubMed]
- Labidi-Galy, S.I.; Papp, E.; Hallberg, D.; Niknafs, N.; Adleff, V.; Noe, M.; Bhattacharya, R.; Novak, M.; Jones, S.; Phallen, J.; et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 2017, 8, 1093. [Google Scholar] [CrossRef] [PubMed]
- Perets, R.; Wyant, G.A.; Muto, K.W.; Bijron, J.G.; Poole, B.B.; Chin, K.T.; Chen, J.Y.; Ohman, A.W.; Stepule, C.D.; Kwak, S.; et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell 2013, 24, 751–765. [Google Scholar] [CrossRef]
- Sherman-Baust, C.A.; Kuhn, E.; Valle, B.L.; Shih Ie, M.; Kurman, R.J.; Wang, T.L.; Amano, T.; Ko, M.S.; Miyoshi, I.; Araki, Y.; et al. A genetically engineered ovarian cancer mouse model based on fallopian tube transformation mimics human high-grade serous carcinoma development. J. Pathol. 2014, 233, 228–237. [Google Scholar] [CrossRef]
- Wu, R.; Zhai, Y.; Kuick, R.; Karnezis, A.N.; Garcia, P.; Naseem, A.; Hu, T.C.; Fearon, E.R.; Cho, K.R. Impact of oviductal versus ovarian epithelial cell of origin on ovarian endometrioid carcinoma phenotype in the mouse. J. Pathol. 2016, 240, 341–351. [Google Scholar] [CrossRef]
- Ghosh, A.; Syed, S.M.; Tanwar, P.S. In vivo genetic cell lineage tracing reveals that oviductal secretory cells self-renew and give rise to ciliated cells. Development 2017, 144, 3031–3041. [Google Scholar] [CrossRef]
- Park, E.S.; Xiang, D.; Xie, Y.; Bronson, R.T.; Li, Z. Oncogenic Events Dictate the Types and Locations of Gynecological Malignancies Originating from Krt8(+) Mesothelial and Mullerian-Derived Epithelial Cells. Cancers 2022, 14, 841. [Google Scholar] [CrossRef]
- Kessler, M.; Fotopoulou, C.; Meyer, T. The molecular fingerprint of high grade serous ovarian cancer reflects its fallopian tube origin. Int. J. Mol. Sci. 2013, 14, 6571–6596. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, E.; Kurman, R.J.; Vang, R.; Sehdev, A.S.; Han, G.; Soslow, R.; Wang, T.L.; Shih Ie, M. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma—Evidence supporting the clonal relationship of the two lesions. J. Pathol. 2012, 226, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Lohmussaar, K.; Kopper, O.; Korving, J.; Begthel, H.; Vreuls, C.P.H.; van Es, J.H.; Clevers, H. Assessing the origin of high-grade serous ovarian cancer using CRISPR-modification of mouse organoids. Nat. Commun. 2020, 11, 2660. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Dolgalev, I.; Zhang, T.; Ran, H.; Levine, D.A.; Neel, B.G. Both fallopian tube and ovarian surface epithelium are cells-of-origin for high-grade serous ovarian carcinoma. Nat. Commun. 2019, 10, 5367. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Park, E.S.; Chen, X.; Han, S.; Xiang, D.; Ren, F.; Liu, G.; Chen, H.; Yuan, G.C.; Li, Z. Distinct niche structures and intrinsic programs of fallopian tube and ovarian surface epithelial cells. iScience 2023, 26, 105861. [Google Scholar] [CrossRef]
- Lim, E.; Wu, D.; Pal, B.; Bouras, T.; Asselin-Labat, M.L.; Vaillant, F.; Yagita, H.; Lindeman, G.J.; Smyth, G.K.; Visvader, J.E. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010, 12, R21. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Y.; Zhang, Q.; Wang, F.; Zhang, D. Toll-like receptors and prostate cancer. Front. Immunol. 2014, 5, 352. [Google Scholar] [CrossRef]
- Akashi, S.; Saitoh, S.; Wakabayashi, Y.; Kikuchi, T.; Takamura, N.; Nagai, Y.; Kusumoto, Y.; Fukase, K.; Kusumoto, S.; Adachi, Y.; et al. Lipopolysaccharide interaction with cell surface Toll-like receptor 4-MD-2: Higher affinity than that with MD-2 or CD14. J. Exp. Med. 2003, 198, 1035–1042. [Google Scholar] [CrossRef]
- Ou, T.; Lilly, M.; Jiang, W. The Pathologic Role of Toll-Like Receptor 4 in Prostate Cancer. Front. Immunol. 2018, 9, 1188. [Google Scholar] [CrossRef]
- Lee, S.H.; Hong, B.; Sharabi, A.; Huang, X.F.; Chen, S.Y. Embryonic stem cells and mammary luminal progenitors directly sense and respond to microbial products. Stem Cells 2009, 27, 1604–1615. [Google Scholar] [CrossRef]
- Mielcarska, M.B.; Bossowska-Nowicka, M.; Toka, F.N. Cell Surface Expression of Endosomal Toll-Like Receptors-A Necessity or a Superfluous Duplication? Front. Immunol. 2020, 11, 620972. [Google Scholar] [CrossRef] [PubMed]
- Komal, A.; Noreen, M.; El-Kott, A.F. TLR3 agonists: RGC100, ARNAX, and poly-IC: A comparative review. Immunol. Res. 2021, 69, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Artibani, M.; Alsaadi, A.; Wietek, N.; Morotti, M.; Shi, T.; Zhong, Z.; Santana Gonzalez, L.; El-Sahhar, S.; KaramiNejadRanjbar, M.; et al. The Repertoire of Serous Ovarian Cancer Non-genetic Heterogeneity Revealed by Single-Cell Sequencing of Normal Fallopian Tube Epithelial Cells. Cancer Cell 2020, 37, 226–242.e227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, S.; Li, X.; Kirk, J.S.; Tang, D.G. Prostate Luminal Progenitor Cells in Development and Cancer. Trends Cancer 2018, 4, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Zhou, P.; Chen, A.X.; Liu, G.Y.; Yu, K.D.; Shao, Z.M. Toll-like receptor 3 -926T>A increased the risk of breast cancer through decreased transcriptional activity. Oncoimmunology 2019, 8, e1673126. [Google Scholar] [CrossRef]
- Shuang, C.; Weiguang, Y.; Zhenkun, F.; Yike, H.; Jiankun, Y.; Jing, X.; Xinghan, L.; Yue, L.; Dalin, L. Toll-like receptor 5 gene polymorphism is associated with breast cancer susceptibility. Oncotarget 2017, 8, 88622–88629. [Google Scholar] [CrossRef]
- Wan, G.X.; Cao, Y.W.; Li, W.Q.; Li, Y.C.; Zhang, W.J.; Li, F. Associations between TLR9 polymorphisms and cancer risk: Evidence from an updated meta-analysis of 25,685 subjects. Asian Pac. J. Cancer Prev. 2014, 15, 8279–8285. [Google Scholar] [CrossRef]
- Resler, A.J.; Malone, K.E.; Johnson, L.G.; Malkki, M.; Petersdorf, E.W.; McKnight, B.; Madeleine, M.M. Genetic variation in TLR or NFkappaB pathways and the risk of breast cancer: A case-control study. BMC Cancer 2013, 13, 219. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, F.; Zhang, H.; Zhu, Y.; Wu, K.; Tan, G. Significance of TLR4/MyD88 expression in breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 7034–7039. [Google Scholar]
- Ma, F.J.; Liu, Z.B.; Hu, X.; Ling, H.; Li, S.; Wu, J.; Shao, Z.M. Prognostic value of myeloid differentiation primary response 88 and Toll-like receptor 4 in breast cancer patients. PLoS ONE 2014, 9, e111639. [Google Scholar] [CrossRef]
- Chen, Y.C.; Giovannucci, E.; Lazarus, R.; Kraft, P.; Ketkar, S.; Hunter, D.J. Sequence variants of Toll-like receptor 4 and susceptibility to prostate cancer. Cancer Res. 2005, 65, 11771–11778. [Google Scholar] [CrossRef] [PubMed]
- Cheng, I.; Plummer, S.J.; Casey, G.; Witte, J.S. Toll-like receptor 4 genetic variation and advanced prostate cancer risk. Cancer Epidemiol. Biomark. Prev. 2007, 16, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Bae, J.S.; Chang, I.H.; Kim, K.D.; Lee, J.; Shin, H.D.; Lee, J.Y.; Kim, W.J.; Kim, W.; Myung, S.C. Sequence variants of Toll-like receptor 4 (TLR4) and the risk of prostate cancer in Korean men. World J. Urol. 2012, 30, 225–232. [Google Scholar] [CrossRef]
- Zheng, S.L.; Augustsson-Balter, K.; Chang, B.; Hedelin, M.; Li, L.; Adami, H.O.; Bensen, J.; Li, G.; Johnasson, J.E.; Turner, A.R.; et al. Sequence variants of toll-like receptor 4 are associated with prostate cancer risk: Results from the CAncer Prostate in Sweden Study. Cancer Res. 2004, 64, 2918–2922. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Z.; Miao, C.H. Pooled analysis of association between a genetic variant in the 3’-untranslated region of Toll-like receptor 4 and cancer risk. Genet. Mol. Res. 2015, 14, 17847–17855. [Google Scholar] [CrossRef]
- Lindstrom, S.; Hunter, D.J.; Gronberg, H.; Stattin, P.; Wiklund, F.; Xu, J.; Chanock, S.J.; Hayes, R.; Kraft, P. Sequence variants in the TLR4 and TLR6-1-10 genes and prostate cancer risk. Results based on pooled analysis from three independent studies. Cancer Epidemiol. Biomark. Prev. 2010, 19, 873–876. [Google Scholar] [CrossRef]
- Shui, I.M.; Stark, J.R.; Penney, K.L.; Schumacher, F.R.; Epstein, M.M.; Pitt, M.J.; Stampfer, M.J.; Tamimi, R.M.; Lindstrom, S.; Sesso, H.D.; et al. Genetic variation in the toll-like receptor 4 and prostate cancer incidence and mortality. Prostate 2012, 72, 209–216. [Google Scholar] [CrossRef]
- Li, Z.; Block, M.S.; Vierkant, R.A.; Fogarty, Z.C.; Winham, S.J.; Visscher, D.W.; Kalli, K.R.; Wang, C.; Goode, E.L. The inflammatory microenvironment in epithelial ovarian cancer: A role for TLR4 and MyD88 and related proteins. Tumour Biol. 2016, 37, 13279–13286. [Google Scholar] [CrossRef]
- Kania, K.D.; Hareza, D.; Wilczynski, J.R.; Wilczynski, M.; Jarych, D.; Malinowski, A.; Paradowska, E. The Toll-like Receptor 4 Polymorphism Asp299Gly Is Associated with an Increased Risk of Ovarian Cancer. Cells 2022, 11, 3137. [Google Scholar] [CrossRef]
- Pradere, J.P.; Dapito, D.H.; Schwabe, R.F. The Yin and Yang of Toll-like receptors in cancer. Oncogene 2014, 33, 3485–3495. [Google Scholar] [CrossRef]
- Morales-Sanchez, A.; Fuentes-Panana, E.M. Human viruses and cancer. Viruses 2014, 6, 4047–4079. [Google Scholar] [CrossRef] [PubMed]
- Senchukova, M.A. Helicobacter Pylori and Gastric Cancer Progression. Curr. Microbiol. 2022, 79, 383. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Douglass, J.; Prasath, V.; Neace, M.; Atrchian, S.; Manjili, M.H.; Shokouhi, S.; Habibi, M. The microbiome and breast cancer: A review. Breast Cancer Res. Treat. 2019, 178, 493–496. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chan, C.H.; Lim, Y.B.; Yang, S.F.; Yeh, L.T.; Wang, Y.H.; Chou, M.C.; Yeh, C.B. Risk of Breast Cancer in Women with Mastitis: A Retrospective Population-Based Cohort Study. Medicina 2020, 56, 372. [Google Scholar] [CrossRef] [PubMed]
- Lambe, M.; Johansson, A.L.; Altman, D.; Eloranta, S. Mastitis and the risk of breast cancer. Epidemiology 2009, 20, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Papakonstantinou, A.; Nuciforo, P.; Borrell, M.; Zamora, E.; Pimentel, I.; Saura, C.; Oliveira, M. The conundrum of breast cancer and microbiome—A comprehensive review of the current evidence. Cancer Treat. Rev. 2022, 111, 102470. [Google Scholar] [CrossRef]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e1326. [Google Scholar] [CrossRef]
- Yang, Z.; Tang, X.; McMullen, T.P.W.; Brindley, D.N.; Hemmings, D.G. PDGFRalpha Enhanced Infection of Breast Cancer Cells with Human Cytomegalovirus but Infection of Fibroblasts Increased Prometastatic Inflammation Involving Lysophosphatidate Signaling. Int. J. Mol. Sci. 2021, 22, 9817. [Google Scholar] [CrossRef]
- Amarante, M.K.; Watanabe, M.A. The possible involvement of virus in breast cancer. J. Cancer Res. Clin. Oncol. 2009, 135, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chang, P.; Wang, L.; Yao, Q.; Guo, W.; Chen, J.; Yan, T.; Cao, C. The role of human papillomavirus infection in breast cancer. Med. Oncol. 2012, 29, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. Urinary tract infection syndromes: Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. N. Am. 2014, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Harding, G.K.; Ronald, A.R. The management of urinary infections: What have we learned in the past decade? Int. J. Antimicrob. Agents 1994, 4, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Krieger, J.N.; Lee, S.W.; Jeon, J.; Cheah, P.Y.; Liong, M.L.; Riley, D.E. Epidemiology of prostatitis. Int. J. Antimicrob. Agents 2008, 31 (Suppl. S1), S85–S90. [Google Scholar] [CrossRef]
- Etienne, M.; Chavanet, P.; Sibert, L.; Michel, F.; Levesque, H.; Lorcerie, B.; Doucet, J.; Pfitzenmeyer, P.; Caron, F. Acute bacterial prostatitis: Heterogeneity in diagnostic criteria and management. Retrospective multicentric analysis of 371 patients diagnosed with acute prostatitis. BMC Infect. Dis. 2008, 8, 12. [Google Scholar] [CrossRef]
- Kim, S.H.; Ha, U.S.; Yoon, B.I.; Kim, S.W.; Sohn, D.W.; Kim, H.W.; Cho, S.Y.; Cho, Y.H. Microbiological and clinical characteristics in acute bacterial prostatitis according to lower urinary tract manipulation procedure. J. Infect. Chemother. 2014, 20, 38–42. [Google Scholar] [CrossRef]
- Nagy, V.; Kubej, D. Acute bacterial prostatitis in humans: Current microbiological spectrum, sensitivity to antibiotics and clinical findings. Urol. Int. 2012, 89, 445–450. [Google Scholar] [CrossRef]
- Yoon, B.I.; Kim, S.; Han, D.S.; Ha, U.S.; Lee, S.J.; Kim, H.W.; Han, C.H.; Cho, Y.H. Acute bacterial prostatitis: How to prevent and manage chronic infection? J. Infect. Chemother. 2012, 18, 444–450. [Google Scholar] [CrossRef]
- Krieger, J.N.; Nyberg, L., Jr.; Nickel, J.C. NIH consensus definition and classification of prostatitis. JAMA 1999, 282, 236–237. [Google Scholar] [CrossRef]
- Trinchieri, A.; Abdelrahman, K.M.; Bhatti, K.H.; Bello, J.O.; Das, K.; Gatsev, O.; Gergova, I.; Magri, V.; Mourmouras, N.; Mourmouris, P.; et al. Spectrum of Causative Pathogens and Resistance Rates to Antibacterial Agents in Bacterial Prostatitis. Diagnostics 2021, 11, 1333. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, E.; White, J.R.; Yu, S.H.; Kulac, I.; Ertunc, O.; De Marzo, A.M.; Yegnasubramanian, S.; Mangold, L.A.; Partin, A.W.; Sfanos, K.S. Profiling the Urinary Microbiome in Men with Positive versus Negative Biopsies for Prostate Cancer. J. Urol. 2018, 199, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Meng, H.; Zhou, F.; Ni, X.; Shen, S.; Das, U.N. Urinary microbiota in patients with prostate cancer and benign prostatic hyperplasia. Arch. Med. Sci. 2015, 11, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Jaratlerdsiri, W.; Patrick, S.M.; Lyons, R.J.; Haynes, A.M.; Collins, C.C.; Stricker, P.D.; Bornman, M.S.R.; Hayes, V.M. Metagenomic analysis reveals a rich bacterial content in high-risk prostate tumors from African men. Prostate 2019, 79, 1731–1738. [Google Scholar] [CrossRef]
- Lawson, J.S.; Glenn, W.K. Evidence for a causal role by human papillomaviruses in prostate cancer—A systematic review. Infect. Agent Cancer 2020, 15, 41. [Google Scholar] [CrossRef]
- Lawson, J.S.; Glenn, W.K. Multiple pathogens and prostate cancer. Infect. Agent Cancer 2022, 17, 23. [Google Scholar] [CrossRef]
- Moghoofei, M.; Keshavarz, M.; Ghorbani, S.; Babaei, F.; Nahand, J.S.; Tavakoli, A.; Mortazavi, H.S.; Marjani, A.; Mostafaei, S.; Monavari, S.H. Association between human papillomavirus infection and prostate cancer: A global systematic review and meta-analysis. Asia Pac. J. Clin. Oncol. 2019, 15, e59–e67. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, L.; Han, L.; Fu, G.; Tuo, X.; Ma, S.; Li, Q.; Wang, Y.; Liang, D.; Tang, M.; et al. The differential distribution of bacteria between cancerous and noncancerous ovarian tissues in situ. J. Ovarian Res. 2020, 13, 8. [Google Scholar] [CrossRef]
- Brewster, W.R.; Burkett, W.C.; Ko, E.M.; Bae-Jump, V.; Nicole McCoy, A.; Keku, T.O. An evaluation of the microbiota of the upper reproductive tract of women with and without epithelial ovarian cancer. Gynecol. Oncol. Rep. 2022, 42, 101017. [Google Scholar] [CrossRef]
- Zhou, B.; Sun, C.; Huang, J.; Xia, M.; Guo, E.; Li, N.; Lu, H.; Shan, W.; Wu, Y.; Li, Y.; et al. The biodiversity Composition of Microbiome in Ovarian Carcinoma Patients. Sci. Rep. 2019, 9, 1691. [Google Scholar] [CrossRef]
- Sipos, A.; Ujlaki, G.; Miko, E.; Maka, E.; Szabo, J.; Uray, K.; Krasznai, Z.; Bai, P. The role of the microbiome in ovarian cancer: Mechanistic insights into oncobiosis and to bacterial metabolite signaling. Mol. Med. 2021, 27, 33. [Google Scholar] [CrossRef] [PubMed]
- Paradowska, E.; Jablonska, A.; Studzinska, M.; Wilczynski, M.; Wilczynski, J.R. Detection and genotyping of CMV and HPV in tumors and fallopian tubes from epithelial ovarian cancer patients. Sci. Rep. 2019, 9, 19935. [Google Scholar] [CrossRef] [PubMed]
- Shanmughapriya, S.; Senthilkumar, G.; Vinodhini, K.; Das, B.C.; Vasanthi, N.; Natarajaseenivasan, K. Viral and bacterial aetiologies of epithelial ovarian cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2311–2317. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Z.K.; Hafez, M.M.; Kamel, M.M.; Zekri, A.R. Human Papillomavirus Genotypes and Methylation of CADM1, PAX1, MAL and ADCYAP1 Genes in Epithelial Ovarian Cancer Patients. Asian Pac. J. Cancer Prev. 2017, 18, 169–176. [Google Scholar] [CrossRef]
- Wu, Q.J.; Guo, M.; Lu, Z.M.; Li, T.; Qiao, H.Z.; Ke, Y. Detection of human papillomavirus-16 in ovarian malignancy. Br. J. Cancer 2003, 89, 672–675. [Google Scholar] [CrossRef]
- Idahl, A.; Lundin, E.; Elgh, F.; Jurstrand, M.; Moller, J.K.; Marklund, I.; Lindgren, P.; Ottander, U. Chlamydia trachomatis, Mycoplasma genitalium, Neisseria gonorrhoeae, human papillomavirus, and polyomavirus are not detectable in human tissue with epithelial ovarian cancer, borderline tumor, or benign conditions. Am. J. Obstet. Gynecol. 2010, 202, 71.e1–71.e6. [Google Scholar] [CrossRef]
- Tamboli, C.P.; Neut, C.; Desreumaux, P.; Colombel, J.F. Dysbiosis in inflammatory bowel disease. Gut 2004, 53, 1–4. [Google Scholar] [CrossRef]
- Khademalhosseini, M.; Arababadi, M.K. Toll-like receptor 4 and breast cancer: An updated systematic review. Breast Cancer 2019, 26, 265–271. [Google Scholar] [CrossRef]
- Kelly, M.G.; Alvero, A.B.; Chen, R.; Silasi, D.A.; Abrahams, V.M.; Chan, S.; Visintin, I.; Rutherford, T.; Mor, G. TLR-4 signaling promotes tumor growth and paclitaxel chemoresistance in ovarian cancer. Cancer Res. 2006, 66, 3859–3868. [Google Scholar] [CrossRef]
- Bayik, D.; Lathia, J.D. Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer 2021, 21, 526–536. [Google Scholar] [CrossRef]
- Chen, P.; Hsu, W.H.; Han, J.; Xia, Y.; DePinho, R.A. Cancer Stemness Meets Immunity: From Mechanism to Therapy. Cell Rep. 2021, 34, 108597. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Chao, M.P.; Majeti, R.; Weissman, I.L. Macrophages as mediators of tumor immunosurveillance. Trends Immunol. 2010, 31, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Barkal, A.A.; Brewer, R.E.; Markovic, M.; Kowarsky, M.; Barkal, S.A.; Zaro, B.W.; Krishnan, V.; Hatakeyama, J.; Dorigo, O.; Barkal, L.J.; et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 2019, 572, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Chua, A.C.; Hodson, L.J.; Moldenhauer, L.M.; Robertson, S.A.; Ingman, W.V. Dual roles for macrophages in ovarian cycle-associated development and remodelling of the mammary gland epithelium. Development 2010, 137, 4229–4238. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Williams, C.B.; Yeh, E.S.; Soloff, A.C. Tumor-associated macrophages: Unwitting accomplices in breast cancer malignancy. NPJ breast cancer 2016, 2, 15025. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef]
- Guery, L.; Hugues, S. Th17 Cell Plasticity and Functions in Cancer Immunity. Biomed. Res. Int. 2015, 2015, 314620. [Google Scholar] [CrossRef]
- Xiang, T.; Long, H.; He, L.; Han, X.; Lin, K.; Liang, Z.; Zhuo, W.; Xie, R.; Zhu, B. Interleukin-17 produced by tumor microenvironment promotes self-renewal of CD133+ cancer stem-like cells in ovarian cancer. Oncogene 2015, 34, 165–176. [Google Scholar] [CrossRef]
- Xu, Y.; Dong, X.; Qi, P.; Ye, Y.; Shen, W.; Leng, L.; Wang, L.; Li, X.; Luo, X.; Chen, Y.; et al. Sox2 Communicates with Tregs Through CCL1 to Promote the Stemness Property of Breast Cancer Cells. Stem Cells 2017, 35, 2351–2365. [Google Scholar] [CrossRef]
- Weng, Y.S.; Tseng, H.Y.; Chen, Y.A.; Shen, P.C.; Al Haq, A.T.; Chen, L.M.; Tung, Y.C.; Hsu, H.L. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer 2019, 18, 42. [Google Scholar] [CrossRef]
- Raghavan, S.; Mehta, P.; Xie, Y.; Lei, Y.L.; Mehta, G. Ovarian cancer stem cells and macrophages reciprocally interact through the WNT pathway to promote pro-tumoral and malignant phenotypes in 3D engineered microenvironments. J. Immunother. Cancer 2019, 7, 190. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Cheng, X.; Chen, H.; Chen, C.; Xie, S.; Zhao, M.; Liu, D.; Deng, Q.; Liu, Y.; Wang, X.; et al. Induction of breast cancer stem cells by M1 macrophages through Lin-28B-let-7-HMGA2 axis. Cancer Lett. 2019, 452, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Wang, S.; Wang, N.; Zheng, Y.; Zhou, J.; Yang, B.; Wang, X.; Zhang, J.; Guo, L.; Wang, S.; et al. CCL5 derived from tumor-associated macrophages promotes prostate cancer stem cells and metastasis via activating beta-catenin/STAT3 signaling. Cell Death Dis. 2020, 11, 234. [Google Scholar] [CrossRef] [PubMed]
- Valeta-Magara, A.; Gadi, A.; Volta, V.; Walters, B.; Arju, R.; Giashuddin, S.; Zhong, H.; Schneider, R.J. Inflammatory Breast Cancer Promotes Development of M2 Tumor-Associated Macrophages and Cancer Mesenchymal Cells through a Complex Chemokine Network. Cancer Res. 2019, 79, 3360–3371. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Medzhitov, R. Toll-like receptors and cancer. Nat. Rev. Cancer 2009, 9, 57–63. [Google Scholar] [CrossRef]
- Abreu, M.T. Toll-like receptor signalling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010, 10, 131–144. [Google Scholar] [CrossRef]
- Fukata, M.; Chen, A.; Klepper, A.; Krishnareddy, S.; Vamadevan, A.S.; Thomas, L.S.; Xu, R.; Inoue, H.; Arditi, M.; Dannenberg, A.J.; et al. Cox-2 is regulated by Toll-like receptor-4 (TLR4) signaling: Role in proliferation and apoptosis in the intestine. Gastroenterology 2006, 131, 862–877. [Google Scholar] [CrossRef]
- Macedo, L.; Pinhal-Enfield, G.; Alshits, V.; Elson, G.; Cronstein, B.N.; Leibovich, S.J. Wound healing is impaired in MyD88-deficient mice: A role for MyD88 in the regulation of wound healing by adenosine A2A receptors. Am. J. Pathol. 2007, 171, 1774–1788. [Google Scholar] [CrossRef]
- Scheeren, F.A.; Kuo, A.H.; van Weele, L.J.; Cai, S.; Glykofridis, I.; Sikandar, S.S.; Zabala, M.; Qian, D.; Lam, J.S.; Johnston, D.; et al. A cell-intrinsic role for TLR2-MYD88 in intestinal and breast epithelia and oncogenesis. Nat. Cell Biol. 2014, 16, 1238–1248. [Google Scholar] [CrossRef]
- Miranda, A.; Hamilton, P.T.; Zhang, A.W.; Pattnaik, S.; Becht, E.; Mezheyeuski, A.; Bruun, J.; Micke, P.; de Reynies, A.; Nelson, B.H. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc. Natl. Acad. Sci. USA 2019, 116, 9020–9029. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.M.; Messer, K.S.; Ralainirina, N.; Li, H.; Leem, C.J.; Gorjestani, S.; Woo, G.; Nguyen, A.V.; Figueiredo, C.C.; Foubert, P.; et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature 2016, 539, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, J. Functions of Immune Checkpoint Molecules Beyond Immune Evasion. Adv. Exp. Med. Biol. 2020, 1248, 201–226. [Google Scholar] [CrossRef]
- Morad, G.; Helmink, B.A.; Sharma, P.; Wargo, J.A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 2021, 184, 5309–5337. [Google Scholar] [CrossRef]
- McDonald, K.A.; Kawaguchi, T.; Qi, Q.; Peng, X.; Asaoka, M.; Young, J.; Opyrchal, M.; Yan, L.; Patnaik, S.; Otsuji, E.; et al. Tumor Heterogeneity Correlates with Less Immune Response and Worse Survival in Breast Cancer Patients. Ann. Surg. Oncol. 2019, 26, 2191–2199. [Google Scholar] [CrossRef]
- Rouzbahani, E.; Majidpoor, J.; Najafi, S.; Mortezaee, K. Cancer stem cells in immunoregulation and bypassing anti-checkpoint therapy. Biomed. Pharmacother. 2022, 156, 113906. [Google Scholar] [CrossRef]
- Abdou, Y.; Goudarzi, A.; Yu, J.X.; Upadhaya, S.; Vincent, B.; Carey, L.A. Immunotherapy in triple negative breast cancer: Beyond checkpoint inhibitors. NPJ Breast Cancer 2022, 8, 121. [Google Scholar] [CrossRef]
- Zhu, H.; Du, C.; Yuan, M.; Fu, P.; He, Q.; Yang, B.; Cao, J. PD-1/PD-L1 counterattack alliance: Multiple strategies for treating triple-negative breast cancer. Drug Discov. Today 2020, 25, 1762–1771. [Google Scholar] [CrossRef]
- Kandalaft, L.E.; Dangaj Laniti, D.; Coukos, G. Immunobiology of high-grade serous ovarian cancer: Lessons for clinical translation. Nat. Rev. Cancer 2022, 22, 640–656. [Google Scholar] [CrossRef]
- Sharma, P.; Pachynski, R.K.; Narayan, V.; Flechon, A.; Gravis, G.; Galsky, M.D.; Mahammedi, H.; Patnaik, A.; Subudhi, S.K.; Ciprotti, M.; et al. Nivolumab Plus Ipilimumab for Metastatic Castration-Resistant Prostate Cancer: Preliminary Analysis of Patients in the CheckMate 650 Trial. Cancer Cell 2020, 38, 489–499.e483. [Google Scholar] [CrossRef]
- Taghizadeh, H.; Marhold, M.; Tomasich, E.; Udovica, S.; Merchant, A.; Krainer, M. Immune checkpoint inhibitors in mCRPC—Rationales, challenges and perspectives. Oncoimmunology 2019, 8, e1644109. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Kees, T.; Almeida, A.S.; Liu, B.; He, X.Y.; Ng, D.; Han, X.; Spector, D.L.; McNeish, I.A.; Gimotty, P.; et al. Activating a collaborative innate-adaptive immune response to control metastasis. Cancer Cell 2021, 39, 1361–1374.e1369. [Google Scholar] [CrossRef] [PubMed]
- Mata-Haro, V.; Cekic, C.; Martin, M.; Chilton, P.M.; Casella, C.R.; Mitchell, T.C. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science 2007, 316, 1628–1632. [Google Scholar] [CrossRef]
- Schiffman, M.; Wacholder, S. Success of HPV vaccination is now a matter of coverage. Lancet Oncol. 2012, 13, 10–12. [Google Scholar] [CrossRef]
- Kendrick, H.; Regan, J.L.; Magnay, F.A.; Grigoriadis, A.; Mitsopoulos, C.; Zvelebil, M.; Smalley, M.J. Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genom. 2008, 9, 591. [Google Scholar] [CrossRef]
- Toroghian, Y.; Khayyami, R.; Hassanian, S.M.; Nassiri, M.; Ferns, G.A.; Khazaei, M.; Avan, A. The Therapeutic Potential of Targeting the Toll-like Receptor Pathway in Breast Cancer. Curr. Pharm. Des. 2022, 28, 2203–2210. [Google Scholar] [CrossRef]
- Farooq, M.; Batool, M.; Kim, M.S.; Choi, S. Toll-Like Receptors as a Therapeutic Target in the Era of Immunotherapies. Front. Cell Dev. Biol. 2021, 9, 756315. [Google Scholar] [CrossRef]
- Vindevogel, E.; Baert, T.; Van Hoylandt, A.; Verbist, G.; Vande Velde, G.; Garg, A.D.; Agostinis, P.; Vergote, I.; Coosemans, A. The Use of Toll-like Receptor 4 Agonist to Reshape the Immune Signature in Ovarian Cancer. Anticancer Res. 2016, 36, 5781–5792. [Google Scholar] [CrossRef] [PubMed]
- Roby, K.F.; Taylor, C.C.; Sweetwood, J.P.; Cheng, Y.; Pace, J.L.; Tawfik, O.; Persons, D.L.; Smith, P.G.; Terranova, P.F. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis 2000, 21, 585–591. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.; Chen, X.; Li, Z. Innate Immune Program in Formation of Tumor-Initiating Cells from Cells-of-Origin of Breast, Prostate, and Ovarian Cancers. Cancers 2023, 15, 757. https://doi.org/10.3390/cancers15030757
Han S, Chen X, Li Z. Innate Immune Program in Formation of Tumor-Initiating Cells from Cells-of-Origin of Breast, Prostate, and Ovarian Cancers. Cancers. 2023; 15(3):757. https://doi.org/10.3390/cancers15030757
Chicago/Turabian StyleHan, Sen, Xueqing Chen, and Zhe Li. 2023. "Innate Immune Program in Formation of Tumor-Initiating Cells from Cells-of-Origin of Breast, Prostate, and Ovarian Cancers" Cancers 15, no. 3: 757. https://doi.org/10.3390/cancers15030757
APA StyleHan, S., Chen, X., & Li, Z. (2023). Innate Immune Program in Formation of Tumor-Initiating Cells from Cells-of-Origin of Breast, Prostate, and Ovarian Cancers. Cancers, 15(3), 757. https://doi.org/10.3390/cancers15030757




