SARS-CoV-2 Establishes a Productive Infection in Hepatoma and Glioblastoma Multiforme Cell Lines
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Plasmid Construction
2.3. Cell Culture
2.4. Infection Assays
2.5. Lentivirus Assembly and Cell Transduction
2.6. Reverse Transcription—Real-Time Polymerase Chain Reaction (RT-qPCR)
2.7. Immunofluorescence
2.8. Interferon Production
2.9. Statistical Analysis
3. Results
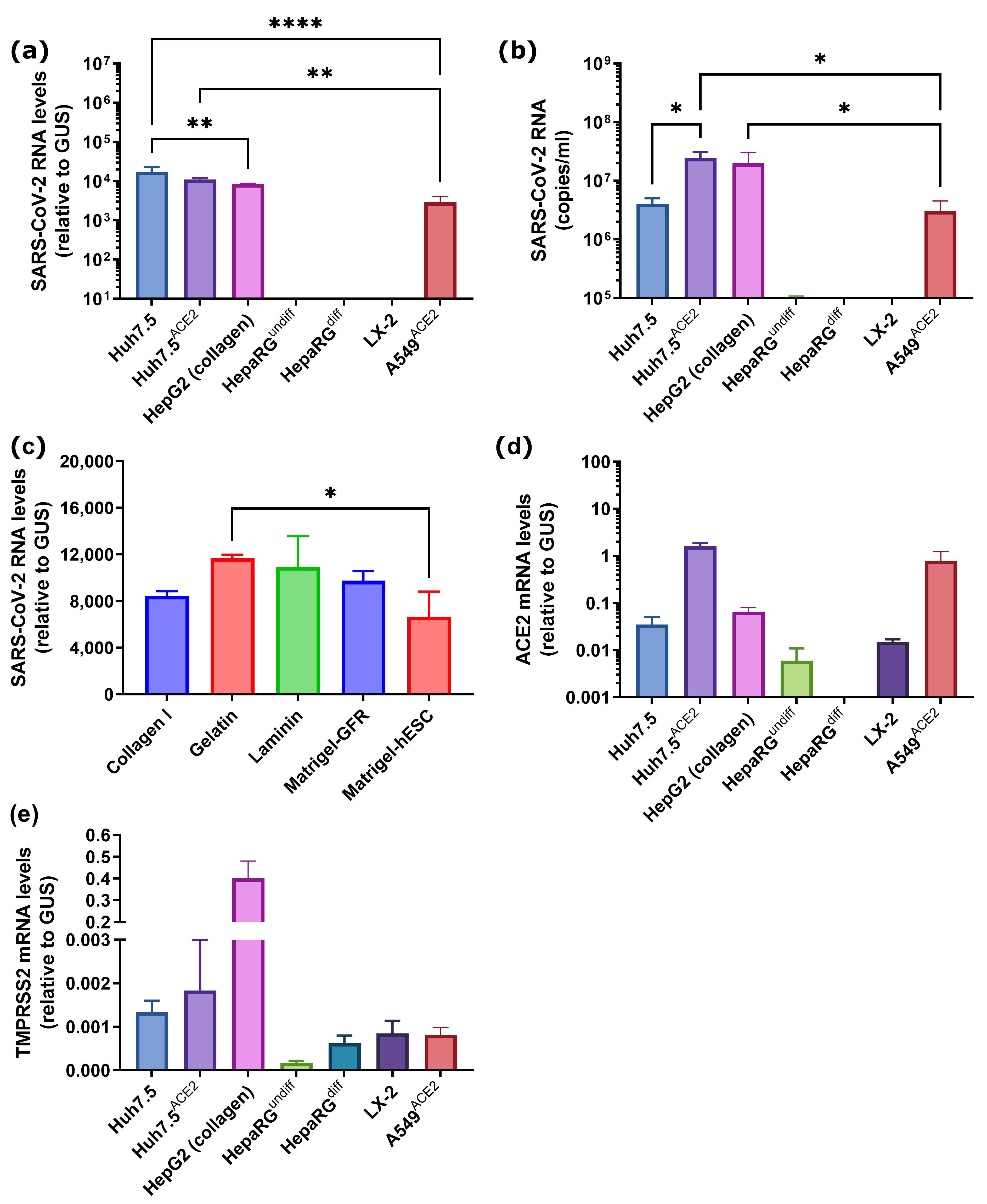
3.1. Cell Lines Overexpressing ACE2 and TMPRSS2
3.2. SARS-CoV-2 Infects Liver Hepatoma but Not Non-Transformed Hepatocyte-like Cells
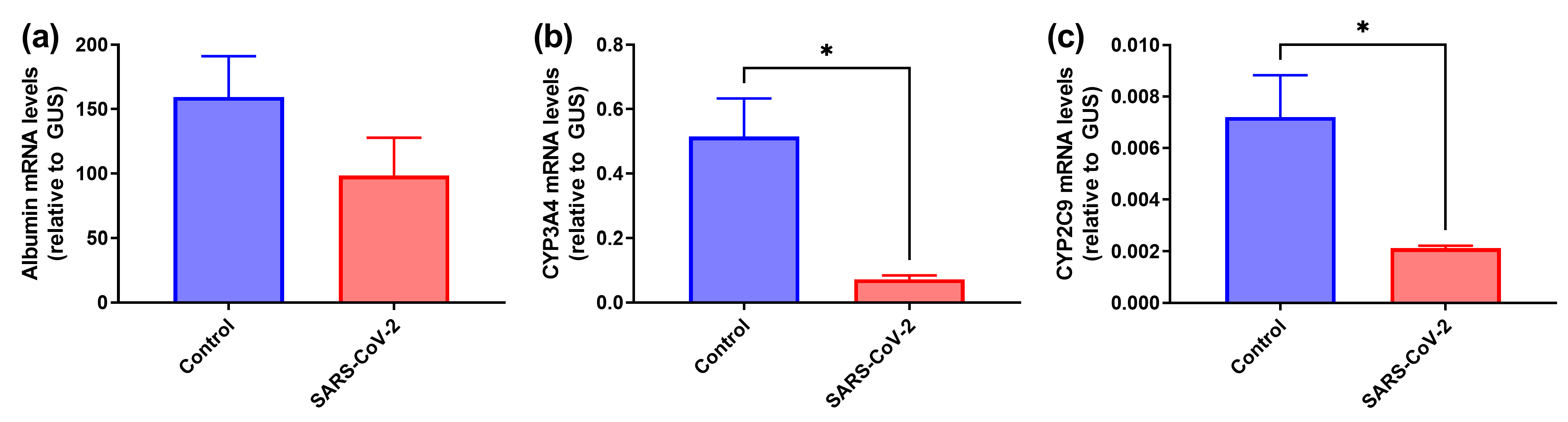
3.3. SARS-CoV-2 Induces Decrease in Expression of Hepatocyte-Specific Markers in HepaRGdiff Cells
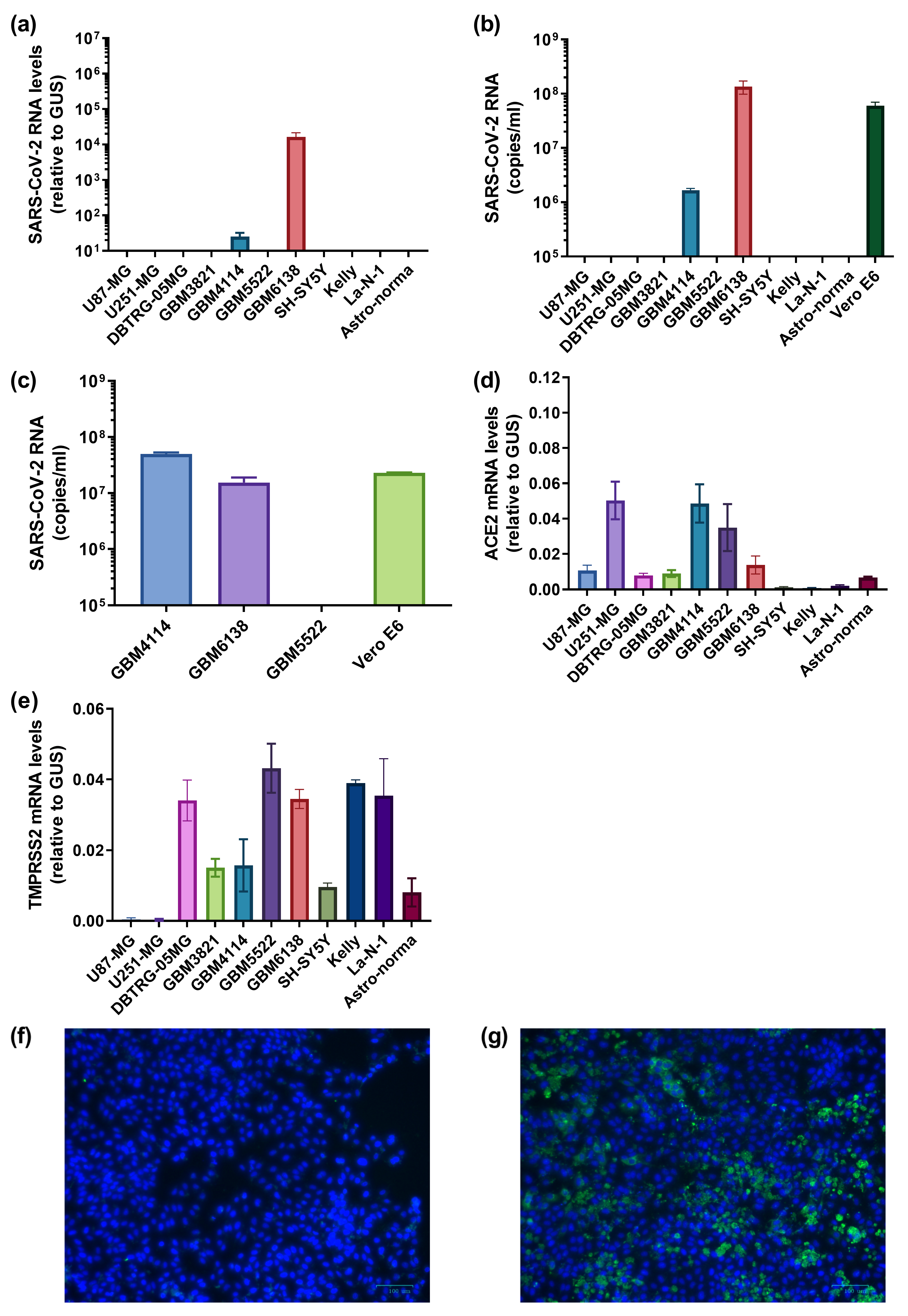
3.4. Several Primary Glioblastoma Cell Lines Support Highly Efficient SARS-CoV-2 Replication
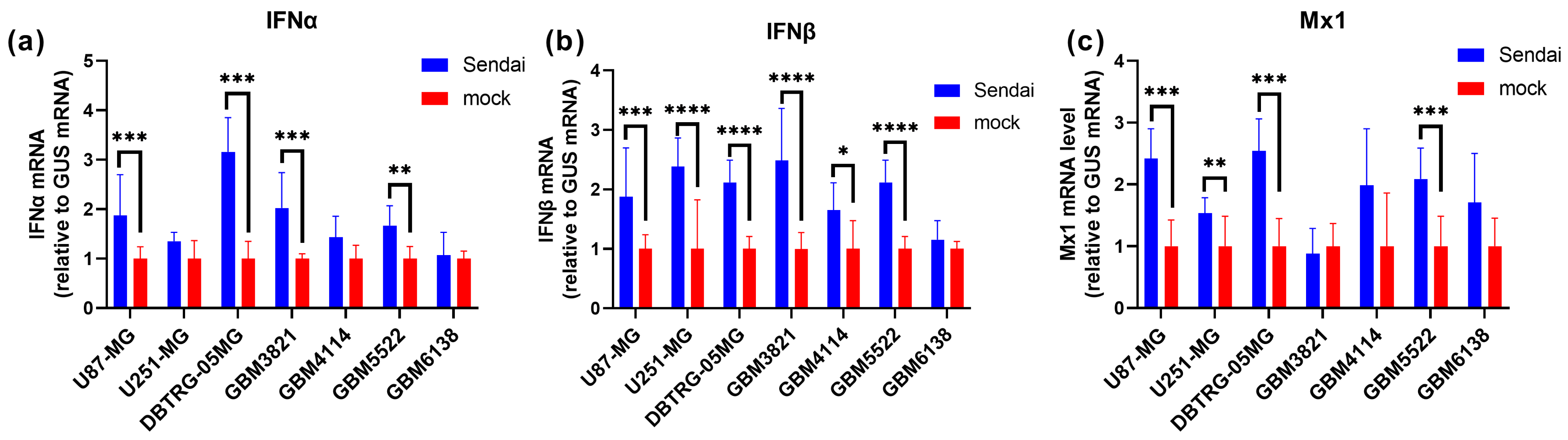
3.5. The Highly Permissive Glioblastoma GBM6138 Cell Line Has Defects in Type I Interferon Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Torres Acosta, M.A.; Singer, B.D. Pathogenesis of COVID-19-induced ARDS: Implications for an ageing population. Eur. Respir. J. 2020, 56, 2002049. [Google Scholar] [CrossRef] [PubMed]
- Acherjee, T.; Behara, A.; Saad, M.; Vittorio, T.J. Mechanisms and management of prothrombotic state in COVID-19 disease. Ther. Adv. Cardiovasc. Dis. 2021, 15, 17539447211053470. [Google Scholar] [CrossRef] [PubMed]
- Ball, L.; Silva, P.L.; Giacobbe, D.R.; Bassetti, M.; Zubieta-Calleja, G.R.; Rocco, P.R.M.; Pelosi, P. Understanding the pathophysiology of typical acute respiratory distress syndrome and severe COVID-19. Expert Rev. Respir. Med. 2022, 16, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Vanderheiden, A.; Ralfs, P.; Chirkova, T.; Upadhyay, A.A.; Zimmerman, M.G.; Bedoya, S.; Aoued, H.; Tharp, G.M.; Pellegrini, K.L.; Manfredi, C.; et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 2020, 94, e00985-20. [Google Scholar] [CrossRef]
- Schultheiss, C.; Willscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T.; et al. The IL-1beta, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep. Med. 2022, 3, 100663. [Google Scholar] [CrossRef]
- Khan, S.; Shafiei, M.S.; Longoria, C.; Schoggins, J.W.; Savani, R.C.; Zaki, H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-kappaB pathway. Elife 2021, 10, e68563. [Google Scholar] [CrossRef]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Choudhury, A.; Mukherjee, S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020, 92, 2105–2113. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Das, N.C.; Patra, R.; Mukherjee, S. In silico analyses on the comparative sensing of SARS-CoV-2 mRNA by the intracellular TLRs of humans. J. Med. Virol. 2021, 93, 2476–2486. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S. Toll-like receptor 4 in COVID-19: Friend or foe? Future Virol. 2022, 17. [Google Scholar] [CrossRef] [PubMed]
- Group, R.C.; Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; et al. Dexamethasone in Hospitalized Patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef]
- Atzeni, F.; Gerratana, E.; Giallanza, M.; La Corte, L.; Nucera, V.; Miceli, G.; Sangari, D.; Masala, I.F. The effect of drugs used in rheumatology for treating SARS-CoV2 infection. Expert Opin. Biol. Ther. 2021, 21, 219–228. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, J.Y.; Yang, J.W.; Lee, K.H.; Effenberger, M.; Szpirt, W.; Kronbichler, A.; Shin, J.I. Immunopathogenesis and treatment of cytokine storm in COVID-19. Theranostics 2021, 11, 316–329. [Google Scholar] [CrossRef]
- Kalil, A.C.; Patterson, T.F.; Mehta, A.K.; Tomashek, K.M.; Wolfe, C.R.; Ghazaryan, V.; Marconi, V.C.; Ruiz-Palacios, G.M.; Hsieh, L.; Kline, S.; et al. Baricitinib plus Remdesivir for Hospitalized Adults with COVID-19. N. Engl. J. Med. 2021, 384, 795–807. [Google Scholar] [CrossRef]
- Pruijssers, A.J.; George, A.S.; Schafer, A.; Leist, S.R.; Gralinksi, L.E.; Dinnon, K.H., III; Yount, B.L.; Agostini, M.L.; Stevens, L.J.; Chappell, J.D.; et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020, 32, 107940. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schafer, A.; Dinnon, K.H., III; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12, eabb5883. [Google Scholar] [CrossRef]
- Reis, S.; Metzendorf, M.I.; Kuehn, R.; Popp, M.; Gagyor, I.; Kranke, P.; Meybohm, P.; Skoetz, N.; Weibel, S. Nirmatrelvir combined with ritonavir for preventing and treating COVID-19. Cochrane Database Syst. Rev. 2022, 9, CD015395. [Google Scholar] [CrossRef]
- Doroftei, B.; Ciobica, A.; Ilie, O.D.; Maftei, R.; Ilea, C. Mini-Review Discussing the Reliability and Efficiency of COVID-19 Vaccines. Diagnostics 2021, 11, 579. [Google Scholar] [CrossRef] [PubMed]
- Gorchakov, A.A.; Kulemzin, S.V.; Guselnikov, S.V.; Baranov, K.O.; Belovezhets, T.N.; Mechetina, L.V.; Volkova, O.Y.; Najakshin, A.M.; Chikaev, N.A.; Chikaev, A.N.; et al. Isolation of a panel of ultra-potent human antibodies neutralizing SARS-CoV-2 and viral variants of concern. Cell Discov. 2021, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.F.; Zhao, F.; Huang, D.; Beutler, N.; Burns, A.; He, W.T.; Limbo, O.; Smith, C.; Song, G.; Woehl, J.; et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020, 369, 956–963. [Google Scholar] [CrossRef]
- Piccicacco, N.; Zeitler, K.; Ing, A.; Montero, J.; Faughn, J.; Silbert, S.; Kim, K. Real-world effectiveness of early remdesivir and sotrovimab in the highest-risk COVID-19 outpatients during the Omicron surge. J. Antimicrob. Chemother. 2022, 77, 2693–2700. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.J.; Pade, C.; Gibbons, J.M.; Otter, A.D.; Lin, K.M.; Munoz Sandoval, D.; Pieper, F.P.; Butler, D.K.; Liu, S.; Joy, G.; et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science 2022, 377, eabq1841. [Google Scholar] [CrossRef]
- Stalman, E.W.; Wieske, L.; van Dam, K.P.J.; Kummer, L.Y.; van Kempen, Z.L.E.; Killestein, J.; Volkers, A.G.; Tas, S.W.; Boekel, L.; Wolbink, G.J.; et al. Breakthrough infections with the SARS-CoV-2 omicron (B.1.1.529) variant in patients with immune-mediated inflammatory diseases. Ann. Rheum. Dis. 2022, 81, 1757–1766. [Google Scholar] [CrossRef]
- Hernandez Acosta, R.A.; Esquer Garrigos, Z.; Marcelin, J.R.; Vijayvargiya, P. COVID-19 Pathogenesis and Clinical Manifestations. Infect. Dis. Clin. North Am. 2022, 36, 231–249. [Google Scholar] [CrossRef]
- Keyhanian, K.; Umeton, R.P.; Mohit, B.; Davoudi, V.; Hajighasemi, F.; Ghasemi, M. SARS-CoV-2 and nervous system: From pathogenesis to clinical manifestation. J. Neuroimmunol. 2020, 350, 577436. [Google Scholar] [CrossRef]
- Castanares-Zapatero, D.; Chalon, P.; Kohn, L.; Dauvrin, M.; Detollenaere, J.; Maertens de Noordhout, C.; Primus-de Jong, C.; Cleemput, I.; Van den Heede, K. Pathophysiology and mechanism of long COVID: A comprehensive review. Ann. Med. 2022, 54, 1473–1487. [Google Scholar] [CrossRef]
- Marjot, T.; Webb, G.J.; Barritt, A.S.t.; Moon, A.M.; Stamataki, Z.; Wong, V.W.; Barnes, E. COVID-19 and liver disease: Mechanistic and clinical perspectives. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 348–364. [Google Scholar] [CrossRef]
- Khunti, K.; Del Prato, S.; Mathieu, C.; Kahn, S.E.; Gabbay, R.A.; Buse, J.B. COVID-19, Hyperglycemia, and New-Onset Diabetes. Diabetes Care 2021, 44, 2645–2655. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Kondo, R.; Kawaguchi, N.; Iwakiri, Y. COVID-19 and Liver Injury: Role of Inflammatory Endotheliopathy, Platelet Dysfunction, and Thrombosis. Hepatol. Commun. 2022, 6, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Phipps, M.M.; Barraza, L.H.; LaSota, E.D.; Sobieszczyk, M.E.; Pereira, M.R.; Zheng, E.X.; Fox, A.N.; Zucker, J.; Verna, E.C. Acute Liver Injury in COVID-19: Prevalence and Association with Clinical Outcomes in a Large U.S. Cohort. Hepatology 2020, 72, 807–817. [Google Scholar] [CrossRef] [PubMed]
- El Kazafy, S.A.; Fouad, Y.M.; Said, A.F.; Assal, H.H.; Ali, T.M.; Ahmed, A.E.; Elesawy, B.H.; Ahmed, O.M. Correlations between Cytokine Levels, Liver Function Markers, and Neuropilin-1 Expression in Patients with COVID-19. Vaccines 2022, 10, 1636. [Google Scholar] [CrossRef]
- Effenberger, M.; Grander, C.; Grabherr, F.; Griesmacher, A.; Ploner, T.; Hartig, F.; Bellmann-Weiler, R.; Joannidis, M.; Zoller, H.; Weiss, G.; et al. Systemic inflammation as fuel for acute liver injury in COVID-19. Dig. Liver Dis. 2021, 53, 158–165. [Google Scholar] [CrossRef]
- Sonzogni, A.; Previtali, G.; Seghezzi, M.; Grazia Alessio, M.; Gianatti, A.; Licini, L.; Morotti, D.; Zerbi, P.; Carsana, L.; Rossi, R.; et al. Liver histopathology in severe COVID 19 respiratory failure is suggestive of vascular alterations. Liver Int. 2020, 40, 2110–2116. [Google Scholar] [CrossRef]
- Oleksowicz, L.; Mrowiec, Z.; Isaacs, R.; Dutcher, J.P.; Puszkin, E. Morphologic and ultrastructural evidence of interleukin-6 induced platelet activation. Am. J. Hematol. 1995, 48, 92–99. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Wang, R.; Feng, Z.; Zhang, J.; Yang, H.; Tan, Y.; Wang, H.; Wang, C.; Liu, L.; et al. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 infection. Nat. Commun. 2021, 12, 2506. [Google Scholar] [CrossRef]
- Jansen, J.; Reimer, K.C.; Nagai, J.S.; Varghese, F.S.; Overheul, G.J.; de Beer, M.; Roverts, R.; Daviran, D.; Fermin, L.A.S.; Willemsen, B.; et al. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell 2022, 29, 217–231.e8. [Google Scholar] [CrossRef]
- Muller, J.A.; Gross, R.; Conzelmann, C.; Kruger, J.; Merle, U.; Steinhart, J.; Weil, T.; Koepke, L.; Bozzo, C.P.; Read, C.; et al. SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nat. Metab. 2021, 3, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.T.; Lidsky, P.V.; Xiao, Y.; Lee, I.T.; Cheng, R.; Nakayama, T.; Jiang, S.; Demeter, J.; Bevacqua, R.J.; Chang, C.A.; et al. SARS-CoV-2 infects human pancreatic beta cells and elicits beta cell impairment. Cell Metab. 2021, 33, 1565–1576.e5. [Google Scholar] [CrossRef] [PubMed]
- Fahmi, A.; Brugger, M.; Demoulins, T.; Zumkehr, B.; Oliveira Esteves, B.I.; Bracher, L.; Wotzkow, C.; Blank, F.; Thiel, V.; Baud, D.; et al. SARS-CoV-2 can infect and propagate in human placenta explants. Cell Rep. Med. 2021, 2, 100456. [Google Scholar] [CrossRef]
- Sharma, A.; Garcia, G., Jr.; Wang, Y.; Plummer, J.T.; Morizono, K.; Arumugaswami, V.; Svendsen, C.N. Human iPSC-Derived Cardiomyocytes Are Susceptible to SARS-CoV-2 Infection. Cell Rep. Med. 2020, 1, 100052. [Google Scholar] [CrossRef]
- Braun, F.; Lutgehetmann, M.; Pfefferle, S.; Wong, M.N.; Carsten, A.; Lindenmeyer, M.T.; Norz, D.; Heinrich, F.; Meissner, K.; Wichmann, D.; et al. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet 2020, 396, 597–598. [Google Scholar] [CrossRef] [PubMed]
- Puelles, V.G.; Lutgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Wanner, N.; Andrieux, G.; Badia, I.M.P.; Edler, C.; Pfefferle, S.; Lindenmeyer, M.T.; Schmidt-Lauber, C.; Czogalla, J.; Wong, M.N.; Okabayashi, Y.; et al. Molecular consequences of SARS-CoV-2 liver tropism. Nat. Metab. 2022, 4, 310–319. [Google Scholar] [CrossRef]
- Rebendenne, A.; Roy, P.; Bonaventure, B.; Chaves Valadão, A.L.; Desmarets, L.; Arnaud-Arnould, M.; Rouillé, Y.; Tauziet, M.; Giovannini, D.; Touhami, J.; et al. Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs. Nat. Genet. 2022, 54, 1090–1102. [Google Scholar] [CrossRef]
- Baggen, J.; Persoons, L.; Vanstreels, E.; Jansen, S.; Van Looveren, D.; Boeckx, B.; Geudens, V.; De Man, J.; Jochmans, D.; Wauters, J.; et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021, 53, 435–444. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Andrews, M.G.; Mukhtar, T.; Eze, U.C.; Simoneau, C.R.; Ross, J.; Parikshak, N.; Wang, S.; Zhou, L.; Koontz, M.; Velmeshev, D.; et al. Tropism of SARS-CoV-2 for human cortical astrocytes. Proc. Natl. Acad. Sci. USA 2022, 119, e2122236119. [Google Scholar] [CrossRef] [PubMed]
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Silva, L.S.; Nogueira, M.H.; Antunes, A.; Vendramini, P.H.; Valenca, A.G.F.; Brandao-Teles, C.; Zuccoli, G.D.S.; et al. Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc. Natl. Acad. Sci. USA 2022, 119, e2200960119. [Google Scholar] [CrossRef]
- Lipatova, A.V.; Soboleva, A.V.; Gorshkov, V.A.; Bubis, J.A.; Solovyeva, E.M.; Krasnov, G.S.; Kochetkov, D.V.; Vorobyev, P.O.; Ilina, I.Y.; Moshkovskii, S.A.; et al. Multi-Omics Analysis of Glioblastoma Cells’ Sensitivity to Oncolytic Viruses. Cancers 2021, 13, 5268. [Google Scholar] [CrossRef] [PubMed]
- Vorobyev, P.O.; Kochetkov, D.V.; Chumakov, P.M.; Zakirova, N.F.; Zotova-Nefedorova, S.I.; Vasilenko, K.V.; Alekseeva, O.N.; Kochetkov, S.N.; Bartosch, B.; Lipatova, A.V.; et al. 2-Deoxyglucose, an Inhibitor of Glycolysis, Enhances the Oncolytic Effect of Coxsackievirus. Cancers 2022, 14, 5611. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, O.N.; Snezhkina, A.V.; Krasnov, G.S.; Valuev-Elliston, V.T.; Khomich, O.A.; Khomutov, A.R.; Keinanen, T.A.; Alhonen, L.; Bartosch, B.; Kudryavtseva, A.V.; et al. Activation of Polyamine Catabolism by N(1),N(11)-Diethylnorspermine in Hepatic HepaRG Cells Induces Dedifferentiation and Mesenchymal-Like Phenotype. Cells 2018, 7, 275. [Google Scholar] [CrossRef] [PubMed]
- Golikov, M.V.; Karpenko, I.L.; Lipatova, A.V.; Ivanova, O.N.; Fedyakina, I.T.; Larichev, V.F.; Zakirova, N.F.; Leonova, O.G.; Popenko, V.I.; Bartosch, B.; et al. Cultivation of Cells in a Physiological Plasmax Medium Increases Mitochondrial Respiratory Capacity and Reduces Replication Levels of RNA Viruses. Antioxidants 2022, 11, 97. [Google Scholar] [CrossRef]
- Baklaushev, V.P.; Averyanov, A.V.; Sotnikova, A.G.; Perkina, A.S.; Ivanov, A.V.; Yusubalieva, G.M.; Novikova, O.N.; Shikina, V.E.; Dupik, N.V.; Kedrova, A.G.; et al. Safety and efficacy of convalescent plasma for COVID-19: The preliminary results of a clinical trial. J. Clin. Pract. 2020, 11, 38–50. [Google Scholar] [CrossRef]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Barcena, M.; et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef]
- Parent, R.; Marion, M.J.; Furio, L.; Trepo, C.; Petit, M.A. Origin and characterization of a human bipotent liver progenitor cell line. Gastroenterology 2004, 126, 1147–1156. [Google Scholar] [CrossRef]
- Winer, B.Y.; Huang, T.S.; Pludwinski, E.; Heller, B.; Wojcik, F.; Lipkowitz, G.E.; Parekh, A.; Cho, C.; Shrirao, A.; Muir, T.W.; et al. Long-term hepatitis B infection in a scalable hepatic co-culture system. Nat. Commun. 2017, 8, 125. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef] [PubMed]
- Mao, R.; Qiu, Y.; He, J.S.; Tan, J.Y.; Li, X.H.; Liang, J.; Shen, J.; Zhu, L.R.; Chen, Y.; Iacucci, M.; et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Hellmuth, J.C.; Scherer, C.; Muenchhoff, M.; Mayerle, J.; Gerbes, A.L. Liver function test abnormalities at hospital admission are associated with severe course of SARS-CoV-2 infection: A prospective cohort study. Gut 2021, 70, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.P.; Mishra, S.; Jha, D.K.; Shukla, J.; Choudhury, A.; Mohindra, R.; Mandavdhare, H.S.; Dutta, U.; Sharma, V. Coronavirus disease (COVID-19) and the liver: A comprehensive systematic review and meta-analysis. Hepatol. Int. 2020, 14, 711–722. [Google Scholar] [CrossRef]
- Yip, T.C.-F.; Lui, G.C.-Y.; Wong, V.W.-S.; Chow, V.C.-Y.; Ho, T.H.-Y.; Li, T.C.-M.; Tse, Y.-K.; Hui, D.S.-C.; Chan, H.L.-Y.; Wong, G.L.-H. Liver injury is independently associated with adverse clinical outcomes in patients with COVID-19. Gut 2021, 70, 733–742. [Google Scholar] [CrossRef]
- Weber, S.; Mayerle, J.; Irlbeck, M.; Gerbes, A.L. Severe liver failure during SARS-CoV-2 infection. Gut 2020, 69, 1365–1367. [Google Scholar] [CrossRef]
- Marjot, T.; Moon, A.M.; Cook, J.A.; Abd-Elsalam, S.; Aloman, C.; Armstrong, M.J.; Pose, E.; Brenner, E.J.; Cargill, T.; Catana, M.A.; et al. Outcomes following SARS-CoV-2 infection in patients with chronic liver disease: An international registry study. J. Hepatol. 2021, 74, 567–577. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Del Zompo, F.; Nesci, A.; Santopaolo, F.; Ianiro, G.; Pompili, M.; Gasbarrini, A.; Gemelli against, COVID group. Liver involvement is not associated with mortality: Results from a large cohort of SARS-CoV-2-positive patients. Aliment. Pharmacol. Ther. 2020, 52, 1060–1068. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. 2011, 6, 425–456. [Google Scholar] [CrossRef]
- Belopasov, V.V.; Yachou, Y.; Samoilova, E.M.; Baklaushev, V.P. The nervous system damage in COVID-19. J. Clin. Pract. 2020, 11, 60–80. [Google Scholar] [CrossRef]
- Bhola, S.; Trisal, J.; Thakur, V.; Kaur, P.; Kulshrestha, S.; Bhatia, S.K.; Kumar, P. Neurological toll of COVID-19. Neurol. Sci. 2022, 43, 2171–2186. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.H.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950.e9. [Google Scholar] [CrossRef] [PubMed]
- McMahon, C.L.; Staples, H.; Gazi, M.; Carrion, R.; Hsieh, J. SARS-CoV-2 targets glial cells in human cortical organoids. Stem Cell Rep. 2021, 16, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, L.; Albecka, A.; Mallery, D.L.; Kellner, M.J.; Paul, D.; Carter, A.P.; James, L.C.; Lancaster, M.A. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell 2020, 27, 951–961.e5. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, M.; Garcia, G., Jr.; Tian, E.; Cui, Q.; Chen, X.; Sun, G.; Wang, J.; Arumugaswami, V.; Shi, Y. ApoE-Isoform-Dependent SARS-CoV-2 Neurotropism and Cellular Response. Cell Stem Cell 2021, 28, 331–342.e5. [Google Scholar] [CrossRef]
- Zhang, B.Z.; Chu, H.; Han, S.; Shuai, H.; Deng, J.; Hu, Y.F.; Gong, H.R.; Lee, A.C.; Zou, Z.; Yau, T.; et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020, 30, 928–931. [Google Scholar] [CrossRef]
- Beckman, D.; Bonillas, A.; Diniz, G.B.; Ott, S.; Roh, J.W.; Elizaldi, S.R.; Schmidt, B.A.; Sammak, R.L.; Van Rompay, K.K.A.; Iyer, S.S.; et al. SARS-CoV-2 infects neurons and induces neuroinflammation in a non-human primate model of COVID-19. Cell Rep. 2022, 41, 111573. [Google Scholar] [CrossRef]
- Lei, J.; Liu, Y.; Xie, T.; Yao, G.; Wang, G.; Diao, B.; Song, J. Evidence for residual SARS-CoV-2 in glioblastoma tissue of a convalescent patient. Neuroreport 2021, 32, 771–775. [Google Scholar] [CrossRef]
- Bielarz, V.; Willemart, K.; Avalosse, N.; De Swert, K.; Lotfi, R.; Lejeune, N.; Poulain, F.; Ninanne, N.; Gilloteaux, J.; Gillet, N.; et al. Susceptibility of neuroblastoma and glioblastoma cell lines to SARS-CoV-2 infection. Brain Res. 2021, 1758, 147344. [Google Scholar] [CrossRef]
- Vanhulle, E.; Stroobants, J.; Provinciael, B.; Camps, A.; Noppen, S.; Maes, P.; Vermeire, K. SARS-CoV-2 Permissive glioblastoma cell line for high throughput antiviral screening. Antiviral Res. 2022, 203, 105342. [Google Scholar] [CrossRef]
- Itou, M.; Kawaguchi, T.; Taniguchi, E.; Sata, M. Dipeptidyl peptidase-4: A key player in chronic liver disease. World J. Gastroenterol. 2013, 19, 2298–2306. [Google Scholar] [CrossRef]
- Desmarais, F.; Herve, V.; Bergeron, K.F.; Ravaut, G.; Perrotte, M.; Fyfe-Desmarais, G.; Rassart, E.; Ramassamy, C.; Mounier, C. Cerebral Apolipoprotein D Exits the Brain and Accumulates in Peripheral Tissues. Int. J. Mol. Sci. 2021, 22, 4118. [Google Scholar] [CrossRef] [PubMed]
- Lokugamage, K.G.; Hage, A.; de Vries, M.; Valero-Jimenez, A.M.; Schindewolf, C.; Dittmann, M.; Rajsbaum, R.; Menachery, V.D. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020, 94, e01410-20. [Google Scholar] [CrossRef] [PubMed]
- Blight, K.J.; McKeating, J.A.; Rice, C.M. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002, 76, 13001–13014. [Google Scholar] [CrossRef] [PubMed]
- Sumpter, R., Jr.; Loo, Y.M.; Foy, E.; Li, K.; Yoneyama, M.; Fujita, T.; Lemon, S.M.; Gale, M., Jr. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005, 79, 2689–2699. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, O.V.; Chumakov, P.M. Defects in interferon pathways as potential biomarkers of sensitivity to oncolytic viruses. Rev. Med. Virol. 2018, 28, e2008. [Google Scholar] [CrossRef]
- Ohadi, L.; Hosseinzadeh, F.; Dadkhahfar, S.; Nasiri, S. Oncolytic effect of SARS-CoV-2 in a patient with mycosis fungoides: A case report. Clin. Case Rep. 2022, 10, e05682. [Google Scholar] [CrossRef]
- Pasin, F.; Mascalchi Calveri, M.; Calabrese, A.; Pizzarelli, G.; Bongiovanni, I.; Andreoli, M.; Cattaneo, C.; Rignanese, G. Oncolytic effect of SARS-CoV2 in a patient with NK lymphoma. Acta Bio Med. Atenei Parmensis 2020, 91, e2020047. [Google Scholar] [CrossRef]
- Kandeel, E.Z.; Refaat, L.; Abdel-Fatah, R.; Samra, M.; Bayoumi, A.; Abdellateif, M.S.; Abdel-Hady, H.; Ali, M.; Khafagy, M. Could COVID-19 induce remission of acute leukemia? Hematology 2021, 26, 870–873. [Google Scholar] [CrossRef]
- Grivas, P.; Khaki, A.R.; Wise-Draper, T.M.; French, B.; Hennessy, C.; Hsu, C.Y.; Shyr, Y.; Li, X.; Choueiri, T.K.; Painter, C.A.; et al. Association of clinical factors and recent anticancer therapy with COVID-19 severity among patients with cancer: A report from the COVID-19 and Cancer Consortium. Ann. Oncol. 2021, 32, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Parmar, H.S.; Nayak, A.; Kataria, S.; Tripathi, V.; Jaiswal, P.; Gavel, P.K.; Jha, H.C.; Bhagwat, S.; Dixit, A.K.; Lukashevich, V.; et al. Restructuring the ONYX-015 adenovirus by using spike protein genes from SARS-CoV-2 and MERS-CoV: Possible implications in breast cancer treatment. Med. Hypotheses 2022, 159, 110750. [Google Scholar] [CrossRef]
- Donia, A.; Shahid, R.; Nawaz, M.; Yaqub, T.; Bokhari, H. Can we develop oncolytic SARS-CoV-2 to specifically target cancer cells? Ther. Adv. Med. Oncol. 2021, 13, 17588359211061988. [Google Scholar] [CrossRef] [PubMed]
- Corey, L.; Beyrer, C.; Cohen, M.S.; Michael, N.L.; Bedford, T.; Rolland, M. SARS-CoV-2 Variants in Patients with Immunosuppression. N. Engl. J. Med. 2021, 385, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, S.; Fernandez-Antunez, C.; Galli, A.; Underwood, A.; Pham, L.V.; Ryberg, L.A.; Feng, S.; Pedersen, M.S.; Mikkelsen, L.S.; Belouzard, S.; et al. Overcoming Culture Restriction for SARS-CoV-2 in Human Cells Facilitates the Screening of Compounds Inhibiting Viral Replication. Antimicrob. Agents Chemother. 2021, 65, e0009721. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smirnova, O.A.; Ivanova, O.N.; Fedyakina, I.T.; Yusubalieva, G.M.; Baklaushev, V.P.; Yanvarev, D.V.; Kechko, O.I.; Mitkevich, V.A.; Vorobyev, P.O.; Fedorov, V.S.; et al. SARS-CoV-2 Establishes a Productive Infection in Hepatoma and Glioblastoma Multiforme Cell Lines. Cancers 2023, 15, 632. https://doi.org/10.3390/cancers15030632
Smirnova OA, Ivanova ON, Fedyakina IT, Yusubalieva GM, Baklaushev VP, Yanvarev DV, Kechko OI, Mitkevich VA, Vorobyev PO, Fedorov VS, et al. SARS-CoV-2 Establishes a Productive Infection in Hepatoma and Glioblastoma Multiforme Cell Lines. Cancers. 2023; 15(3):632. https://doi.org/10.3390/cancers15030632
Chicago/Turabian StyleSmirnova, Olga A., Olga N. Ivanova, Irina T. Fedyakina, Gaukhar M. Yusubalieva, Vladimir P. Baklaushev, Dmitry V. Yanvarev, Olga I. Kechko, Vladimir A. Mitkevich, Pavel O. Vorobyev, Vyacheslav S. Fedorov, and et al. 2023. "SARS-CoV-2 Establishes a Productive Infection in Hepatoma and Glioblastoma Multiforme Cell Lines" Cancers 15, no. 3: 632. https://doi.org/10.3390/cancers15030632
APA StyleSmirnova, O. A., Ivanova, O. N., Fedyakina, I. T., Yusubalieva, G. M., Baklaushev, V. P., Yanvarev, D. V., Kechko, O. I., Mitkevich, V. A., Vorobyev, P. O., Fedorov, V. S., Bartosch, B., Valuev-Elliston, V. T., Lipatova, A. L., & Ivanov, A. V. (2023). SARS-CoV-2 Establishes a Productive Infection in Hepatoma and Glioblastoma Multiforme Cell Lines. Cancers, 15(3), 632. https://doi.org/10.3390/cancers15030632







