The Future of Minimally Invasive Capsule Panendoscopy: Robotic Precision, Wireless Imaging and AI-Driven Insights
Abstract
:Simple Summary
Abstract
1. Introduction to Panendoscopy and Its Challenges
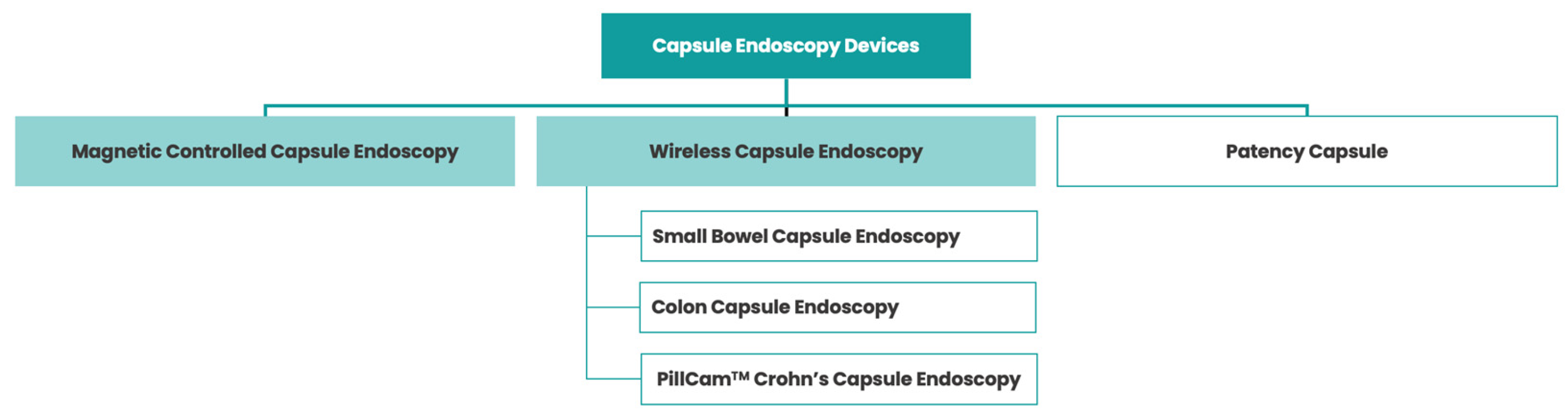
2. Wireless Capsule Endoscopy: A Pill-Sized Revolution in Gastrointestinal Imaging
3. Robotic-Assisted Panendoscopy: Advancements and Benefits
4. Artificial Intelligence in Panendoscopy: Enhancing Diagnostic Accuracy
5. Integrating Robotic Systems and Wireless Capsules: A Synergetic Approach
6. Overcoming Limitations: AI-Assisted Navigation in Panendoscopy
7. Improving Patient Experience: Wireless Capsule Endoscopy vs. Traditional Endoscopy
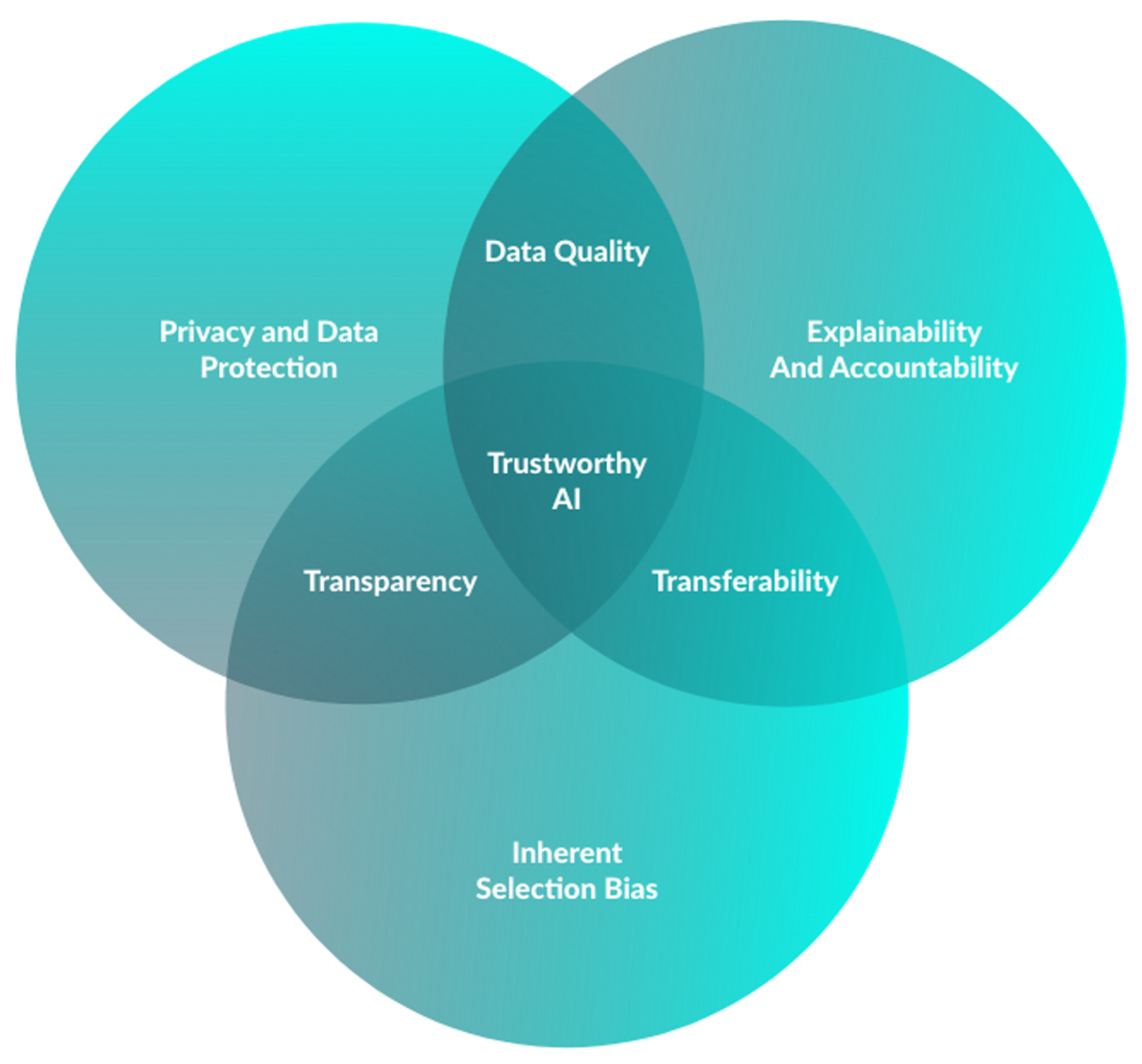
8. Ethical Considerations and Challenges in AI-Assisted Panendoscopy
9. Concluding Remarks—Enabling the Goal of Establishing the Use of Panendoscopy: Robotic and Wireless Capsule Endoscopy Assisted by Artificial Intelligence
Author Contributions
Funding
Conflicts of Interest
References
- Iddan, G.; Meron, G.; Glukhovsky, A.; Swain, P. Wireless capsule endoscopy. Nature 2000, 405, 417. [Google Scholar] [CrossRef] [PubMed]
- Eliakim, R.; Fireman, Z.; Gralnek, I.M.; Yassin, K.; Waterman, M.; Kopelman, Y.; Lachter, J.; Koslowsky, B.; Adler, S.N. Evaluation of the PillCam Colon capsule in the detection of colonic pathology: Results of the first multicenter, prospective, comparative study. Endoscopy 2006, 38, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Eliakim, R.; Adler, S.N. Colon PillCam: Why not just take a pill? Dig. Dis. Sci. 2015, 60, 660–663. [Google Scholar] [CrossRef]
- Piccirelli, S.; Mussetto, A.; Bellumat, A.; Cannizzaro, R.; Pennazio, M.; Pezzoli, A.; Bizzotto, A.; Fusetti, N.; Valiante, F.; Hassan, C.; et al. New Generation Express View: An Artificial Intelligence Software Effectively Reduces Capsule Endoscopy Reading Times. Diagnostics 2022, 12, 1783. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Cho, Y.K.; Kim, J.H. Current and Future Use of Esophageal Capsule Endoscopy. Clin. Endosc. 2018, 51, 317–322. [Google Scholar] [CrossRef]
- Kim, J.H.; Nam, S.J. Capsule Endoscopy for Gastric Evaluation. Diagnostics 2021, 11, 1792. [Google Scholar] [CrossRef]
- Spada, C.; McNamara, D.; Despott, E.J.; Adler, S.; Cash, B.D.; Fernández-Urién, I.; Ivekovic, H.; Keuchel, M.; McAlindon, M.; Saurin, J.C.; et al. Performance measures for small-bowel endoscopy: A European Society of Gastrointestinal Endoscopy (ESGE) Quality Improvement Initiative. United Eur. Gastroenterol. J. 2019, 7, 614–641. [Google Scholar] [CrossRef]
- Tabone, T.; Koulaouzidis, A.; Ellul, P. Scoring Systems for Clinical Colon Capsule Endoscopy-All You Need to Know. J. Clin. Med. 2021, 10, 2372. [Google Scholar] [CrossRef]
- Rosa, B.; Margalit-Yehuda, R.; Gatt, K.; Sciberras, M.; Girelli, C.; Saurin, J.C.; Valdivia, P.C.; Cotter, J.; Eliakim, R.; Caprioli, F.; et al. Scoring systems in clinical small-bowel capsule endoscopy: All you need to know! Endosc. Int. Open 2021, 9, E802–E823. [Google Scholar] [CrossRef]
- Cortegoso Valdivia, P.; Pennazio, M. Chapter 2—Wireless capsule endoscopy: Concept and modalities. In Artificial Intelligence in Capsule Endoscopy; Mascarenhas, M., Cardoso, H., Macedo, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 11–20. [Google Scholar] [CrossRef]
- Eliakim, R.; Yassin, K.; Niv, Y.; Metzger, Y.; Lachter, J.; Gal, E.; Sapoznikov, B.; Konikoff, F.; Leichtmann, G.; Fireman, Z.; et al. Prospective multicenter performance evaluation of the second-generation colon capsule compared with colonoscopy. Endoscopy 2009, 41, 1026–1031. [Google Scholar] [CrossRef]
- Tontini, G.E.; Rizzello, F.; Cavallaro, F.; Bonitta, G.; Gelli, D.; Pastorelli, L.; Salice, M.; Vecchi, M.; Gionchetti, P.; Calabrese, C. Usefulness of panoramic 344°-viewing in Crohn’s disease capsule endoscopy: A proof of concept pilot study with the novel PillCam™ Crohn’s system. BMC Gastroenterol. 2020, 20, 97. [Google Scholar] [CrossRef] [PubMed]
- Pennazio, M.; Rondonotti, E.; Despott, E.J.; Dray, X.; Keuchel, M.; Moreels, T.; Sanders, D.S.; Spada, C.; Carretero, C.; Cortegoso Valdivia, P.; et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Guideline—Update 2022. Endoscopy 2023, 55, 58–95. [Google Scholar] [CrossRef] [PubMed]
- Vuik, F.E.R.; Nieuwenburg, S.A.V.; Moen, S.; Spada, C.; Senore, C.; Hassan, C.; Pennazio, M.; Rondonotti, E.; Pecere, S.; Kuipers, E.J.; et al. Colon capsule endoscopy in colorectal cancer screening: A systematic review. Endoscopy 2021, 53, 815–824. [Google Scholar] [CrossRef]
- Kjølhede, T.; Ølholm, A.M.; Kaalby, L.; Kidholm, K.; Qvist, N.; Baatrup, G. Diagnostic accuracy of capsule endoscopy compared with colonoscopy for polyp detection: Systematic review and meta-analyses. Endoscopy 2021, 53, 713–721. [Google Scholar] [CrossRef]
- Möllers, T.; Schwab, M.; Gildein, L.; Hoffmeister, M.; Albert, J.; Brenner, H.; Jäger, S. Second-generation colon capsule endoscopy for detection of colorectal polyps: Systematic review and meta-analysis of clinical trials. Endosc. Int. Open 2021, 9, E562–E571. [Google Scholar] [CrossRef]
- Nakamura, M.; Kawashima, H.; Ishigami, M.; Fujishiro, M. Indications and Limitations Associated with the Patency Capsule Prior to Capsule Endoscopy. Intern. Med. 2022, 61, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Garrido, I.; Andrade, P.; Lopes, S.; Macedo, G. Chapter 5—The role of capsule endoscopy in diagnosis and clinical management of inflammatory bowel disease. In Artificial Intelligence in Capsule Endoscopy; Mascarenhas, M., Cardoso, H., Macedo, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 69–90. [Google Scholar] [CrossRef]
- Pasha, S.F.; Pennazio, M.; Rondonotti, E.; Wolf, D.; Buras, M.R.; Albert, J.G.; Cohen, S.A.; Cotter, J.; D’Haens, G.; Eliakim, R.; et al. Capsule Retention in Crohn’s Disease: A Meta-analysis. Inflamm. Bowel Dis. 2020, 26, 33–42. [Google Scholar] [CrossRef]
- Silva, M.; Cardoso, H.; Macedo, G. Patency Capsule Safety in Crohn’s Disease. J. Crohn’s Colitis 2017, 11, 1288. [Google Scholar] [CrossRef]
- Rondonotti, E.; Spada, C.; Adler, S.; May, A.; Despott, E.J.; Koulaouzidis, A.; Panter, S.; Domagk, D.; Fernandez-Urien, I.; Rahmi, G.; et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Technical Review. Endoscopy 2018, 50, 423–446. [Google Scholar] [CrossRef]
- Tabet, R.; Nassani, N.; Karam, B.; Shammaa, Y.; Akhrass, P.; Deeb, L. Pooled Analysis of the Efficacy and Safety of Video Capsule Endoscopy in Patients with Implantable Cardiac Devices. Can. J. Gastroenterol. Hepatol. 2019, 2019, 3953807. [Google Scholar] [CrossRef]
- Liao, Z.; Zou, W.; Li, Z.S. Clinical application of magnetically controlled capsule gastroscopy in gastric disease diagnosis: Recent advances. Sci. China Life Sci. 2018, 61, 1304–1309. [Google Scholar] [CrossRef]
- Shamsudhin, N.; Zverev, V.I.; Keller, H.; Pane, S.; Egolf, P.W.; Nelson, B.J.; Tishin, A.M. Magnetically guided capsule endoscopy. Med. Phys. 2017, 44, e91–e111. [Google Scholar] [CrossRef] [PubMed]
- Swain, P.; Toor, A.; Volke, F.; Keller, J.; Gerber, J.; Rabinovitz, E.; Rothstein, R.I. Remote magnetic manipulation of a wireless capsule endoscope in the esophagus and stomach of humans (with videos). Gastrointest. Endosc. 2010, 71, 1290–1293. [Google Scholar] [CrossRef] [PubMed]
- Rahman, I.; Afzal, N.A.; Patel, P. The role of magnetic assisted capsule endoscopy (MACE) to aid visualisation in the upper GI tract. Comput. Biol. Med. 2015, 65, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Hou, X.; Lin-Hu, E.Q.; Sheng, J.Q.; Ge, Z.Z.; Jiang, B.; Hou, X.H.; Liu, J.Y.; Li, Z.; Huang, Q.Y.; et al. Accuracy of Magnetically Controlled Capsule Endoscopy, Compared With Conventional Gastroscopy, in Detection of Gastric Diseases. Clin. Gastroenterol. Hepatol. 2016, 14, 1266–1273.e1. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wang, Q.; Jiang, X.; Jiang, B.; Qian, Y.-Y.; Pan, J.; Liao, Z. Chapter 13—Magnetic capsule endoscopy: Concept and application of artificial intelligence. In Artificial Intelligence in Capsule Endoscopy; Mascarenhas, M., Cardoso, H., Macedo, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 217–241. [Google Scholar] [CrossRef]
- Liao, Z.; Duan, X.D.; Xin, L.; Bo, L.M.; Wang, X.H.; Xiao, G.H.; Hu, L.H.; Zhuang, S.L.; Li, Z.S. Feasibility and safety of magnetic-controlled capsule endoscopy system in examination of human stomach: A pilot study in healthy volunteers. J. Interv. Gastroenterol. 2012, 2, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Currie, G.; Hawk, K.E.; Rohren, E.; Vial, A.; Klein, R. Machine Learning and Deep Learning in Medical Imaging: Intelligent Imaging. J. Med. Imaging Radiat. Sci. 2019, 50, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Handelman, G.S.; Kok, H.K.; Chandra, R.V.; Razavi, A.H.; Lee, M.J.; Asadi, H. eDoctor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.; Jang, G.J.; Lee, M. Fast learning method for convolutional neural networks using extreme learning machine and its application to lane detection. Neural Netw. 2017, 87, 109–121. [Google Scholar] [CrossRef]
- Afonso, J.; Martins, M.; Ferreira, J.; Mascarenhas, M. Chapter 1—Artificial intelligence: Machine learning, deep learning, and applications in gastrointestinal endoscopy. In Artificial Intelligence in Capsule Endoscopy; Mascarenhas, M., Cardoso, H., Macedo, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 1–10. [Google Scholar] [CrossRef]
- Amisha; Malik, P.; Pathania, M.; Rathaur, V.K. Overview of artificial intelligence in medicine. J. Family Med. Prim. Care 2019, 8, 2328–2331. [Google Scholar] [CrossRef]
- Li, N.; Zhao, X.; Yang, Y.; Zou, X. Objects Classification by Learning-Based Visual Saliency Model and Convolutional Neural Network. Comput. Intell. Neurosci. 2016, 2016, 7942501. [Google Scholar] [CrossRef]
- Fisher, M.; Mackiewicz, M. Colour Image Analysis of Wireless Capsule Endoscopy Video: A Review. In Color Medical Image Analysis; Celebi, M.E., Schaefer, G., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 129–144. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Kato, Y.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J. Gastroenterol. Hepatol. 2020, 35, 1196–1200. [Google Scholar] [CrossRef]
- Afonso, J.; Saraiva, M.M.; Ferreira, J.P.S.; Ribeiro, T.; Cardoso, H.; Macedo, G. Performance of a convolutional neural network for automatic detection of blood and hematic residues in small bowel lumen. Dig. Liver Dis. 2021, 53, 654–657. [Google Scholar] [CrossRef]
- Leenhardt, R.; Vasseur, P.; Li, C.; Saurin, J.C.; Rahmi, G.; Cholet, F.; Becq, A.; Marteau, P.; Histace, A.; Dray, X. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest. Endosc. 2019, 89, 189–194. [Google Scholar] [CrossRef]
- Tsuboi, A.; Oka, S.; Aoyama, K.; Saito, H.; Aoki, T.; Yamada, A.; Matsuda, T.; Fujishiro, M.; Ishihara, S.; Nakahori, M.; et al. Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images. Dig. Endosc. 2020, 32, 382–390. [Google Scholar] [CrossRef]
- Houdeville, C.; Souchaud, M.; Leenhardt, R.; Beaumont, H.; Benamouzig, R.; McAlindon, M.; Grimbert, S.; Lamarque, D.; Makins, R.; Saurin, J.C.; et al. A multisystem-compatible deep learning-based algorithm for detection and characterization of angiectasias in small-bowel capsule endoscopy. A proof-of-concept study. Dig. Liver Dis. 2021, 53, 1627–1631. [Google Scholar] [CrossRef]
- Ribeiro, T.; Saraiva, M.M.; Ferreira, J.P.S.; Cardoso, H.; Afonso, J.; Andrade, P.; Parente, M.; Jorge, R.N.; Macedo, G. Artificial intelligence and capsule endoscopy: Automatic detection of vascular lesions using a convolutional neural network. Ann. Gastroenterol. 2021, 34, 820–828. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2019, 89, 357–363.e2. [Google Scholar] [CrossRef] [PubMed]
- Klang, E.; Barash, Y.; Margalit, R.Y.; Soffer, S.; Shimon, O.; Albshesh, A.; Ben-Horin, S.; Amitai, M.M.; Eliakim, R.; Kopylov, U. Deep learning algorithms for automated detection of Crohn’s disease ulcers by video capsule endoscopy. Gastrointest. Endosc. 2020, 91, 606–613.e2. [Google Scholar] [CrossRef] [PubMed]
- Barash, Y.; Azaria, L.; Soffer, S.; Margalit Yehuda, R.; Shlomi, O.; Ben-Horin, S.; Eliakim, R.; Klang, E.; Kopylov, U. Ulcer severity grading in video capsule images of patients with Crohn’s disease: An ordinal neural network solution. Gastrointest. Endosc. 2021, 93, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Afonso, J.; Saraiva, M.J.M.; Ferreira, J.P.S.; Cardoso, H.; Ribeiro, T.; Andrade, P.; Parente, M.; Jorge, R.N.; Saraiva, M.M.; Macedo, G. Development of a Convolutional Neural Network for Detection of Erosions and Ulcers With Distinct Bleeding Potential in Capsule Endoscopy. Tech. Innov. Gastrointest. Endosc. 2021, 23, 291–296. [Google Scholar] [CrossRef]
- Saito, H.; Aoki, T.; Aoyama, K.; Kato, Y.; Tsuboi, A.; Yamada, A.; Fujishiro, M.; Oka, S.; Ishihara, S.; Matsuda, T.; et al. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2020, 92, 144–151.e1. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas Saraiva, M.; Afonso, J.; Ribeiro, T.; Ferreira, J.; Cardoso, H.; Andrade, P.; Gonçalves, R.; Cardoso, P.; Parente, M.; Jorge, R.; et al. Artificial intelligence and capsule endoscopy: Automatic detection of enteric protruding lesions using a convolutional neural network. Rev. Esp. Enferm. Dig. 2021, 115, 75–79. [Google Scholar] [CrossRef]
- Yamada, A.; Niikura, R.; Otani, K.; Aoki, T.; Koike, K. Automatic detection of colorectal neoplasia in wireless colon capsule endoscopic images using a deep convolutional neural network. Endoscopy 2021, 53, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.M.; Ferreira, J.P.S.; Cardoso, H.; Afonso, J.; Ribeiro, T.; Andrade, P.; Parente, M.P.L.; Jorge, R.N.; Macedo, G. Artificial intelligence and colon capsule endoscopy: Development of an automated diagnostic system of protruding lesions in colon capsule endoscopy. Tech. Coloproctol. 2021, 25, 1243–1248. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.; Mascarenhas, M.; Afonso, J.; Cardoso, H.; Andrade, P.; Lopes, S.; Ferreira, J.; Mascarenhas Saraiva, M.; Macedo, G. Artificial intelligence and colon capsule endoscopy: Automatic detection of ulcers and erosions using a convolutional neural network. J. Gastroenterol. Hepatol. 2022, 37, 2282–2288. [Google Scholar] [CrossRef] [PubMed]
- Majtner, T.; Brodersen, J.B.; Herp, J.; Kjeldsen, J.; Halling, M.L.; Jensen, M.D. A deep learning framework for autonomous detection and classification of Crohn’s disease lesions in the small bowel and colon with capsule endoscopy. Endosc. Int. Open 2021, 9, E1361–E1370. [Google Scholar] [CrossRef]
- Ferreira, J.P.S.; de Mascarenhas Saraiva, M.; Afonso, J.P.L.; Ribeiro, T.F.C.; Cardoso, H.M.C.; Ribeiro Andrade, A.P.; de Mascarenhas Saraiva, M.N.G.; Parente, M.P.L.; Natal Jorge, R.; Lopes, S.I.O.; et al. Identification of Ulcers and Erosions by the Novel Pillcam™ Crohn’s Capsule Using a Convolutional Neural Network: A Multicentre Pilot Study. J. Crohn’s Colitis 2022, 16, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas Saraiva, M.; Ferreira, J.P.S.; Cardoso, H.; Afonso, J.; Ribeiro, T.; Andrade, P.; Parente, M.P.L.; Jorge, R.N.; Macedo, G. Artificial intelligence and colon capsule endoscopy: Automatic detection of blood in colon capsule endoscopy using a convolutional neural network. Endosc. Int. Open 2021, 9, E1264–E1268. [Google Scholar] [CrossRef]
- Ding, Z.; Shi, H.; Zhang, H.; Meng, L.; Fan, M.; Han, C.; Zhang, K.; Ming, F.; Xie, X.; Liu, H.; et al. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology 2019, 157, 1044–1054.e5. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Kato, Y.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of various abnormalities in capsule endoscopy videos by a deep learning-based system: A multicenter study. Gastrointest. Endosc. 2021, 93, 165–173.e1. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas Saraiva, M.J.; Afonso, J.; Ribeiro, T.; Ferreira, J.; Cardoso, H.; Andrade, A.P.; Parente, M.; Natal, R.; Mascarenhas Saraiva, M.; Macedo, G. Deep learning and capsule endoscopy: Automatic identification and differentiation of small bowel lesions with distinct haemorrhagic potential using a convolutional neural network. BMJ Open Gastroenterol. 2021, 8, e000753. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, M.; Ribeiro, T.; Afonso, J.; Ferreira, J.P.S.; Cardoso, H.; Andrade, P.; Parente, M.P.L.; Jorge, R.N.; Mascarenhas Saraiva, M.; Macedo, G. Deep learning and colon capsule endoscopy: Automatic detection of blood and colonic mucosal lesions using a convolutional neural network. Endosc. Int. Open 2022, 10, E171–E177. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Xiao, Y.F.; Zhao, X.Y.; Li, J.J.; Yang, Q.Q.; Peng, X.; Nie, X.B.; Zhou, J.Y.; Zhao, Y.B.; Yang, H.; et al. Development and Validation of an Artificial Intelligence Model for Small Bowel Capsule Endoscopy Video Review. JAMA Netw. Open 2022, 5, e2221992. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Xia, T.; Pan, J.; Gao, F.; Wang, S.; Qian, Y.Y.; Wang, H.; Zhao, J.; Jiang, X.; Zou, W.B.; et al. Use of artificial intelligence for detection of gastric lesions by magnetically controlled capsule endoscopy. Gastrointest. Endosc. 2021, 93, 133–139.e4. [Google Scholar] [CrossRef]
- Pan, J.; Xia, J.; Jiang, B.; Zhang, H.; Zhang, H.; Li, Z.S.; Liao, Z. Real-time identification of gastric lesions and anatomical landmarks by artificial intelligence during magnetically controlled capsule endoscopy. Endoscopy 2022, 54, E622–E623. [Google Scholar] [CrossRef]
- Mascarenhas, M.; Mendes, F.; Ribeiro, T.; Afonso, J.; Cardoso, P.; Martins, M.; Cardoso, H.; Andrade, P.; Ferreira, J.; Saraiva, M.M.; et al. Deep Learning and Minimally Invasive Endoscopy: Automatic Classification of Pleomorphic Gastric Lesions in Capsule Endoscopy. Clin. Transl. Gastroenterol. 2023, 14, e00609. [Google Scholar] [CrossRef]
- Eliakim, R. The impact of panenteric capsule endoscopy on the management of Crohn’s disease. Therap. Adv. Gastroenterol. 2017, 10, 737–744. [Google Scholar] [CrossRef]
- Eliakim, R.; Yablecovitch, D.; Lahat, A.; Ungar, B.; Shachar, E.; Carter, D.; Selinger, L.; Neuman, S.; Ben-Horin, S.; Kopylov, U. A novel PillCam Crohn’s capsule score (Eliakim score) for quantification of mucosal inflammation in Crohn’s disease. United Eur. Gastroenterol. J. 2020, 8, 544–551. [Google Scholar] [CrossRef]
- Mussetto, A.; Arena, R.; Fuccio, L.; Trebbi, M.; Tina Garribba, A.; Gasperoni, S.; Manzi, I.; Triossi, O.; Rondonotti, E. A new panenteric capsule endoscopy-based strategy in patients with melena and a negative upper gastrointestinal endoscopy: A prospective feasibility study. Eur. J. Gastroenterol. Hepatol. 2021, 33, 686–690. [Google Scholar] [CrossRef]
- Kim, S.H.; Chun, H.J. Capsule Endoscopy: Pitfalls and Approaches to Overcome. Diagnostics 2021, 11, 1765. [Google Scholar] [CrossRef]
- Cancer Today IARC. Global Cancer Observatory: Cancer Today. Available online: https://gco.iarc.fr/today (accessed on 30 August 2023).
- Ribeiro, T.; Fernández-Urien, I.; Cardoso, H. Chapter 15—Colon capsule endoscopy and artificial intelligence: A perfect match for panendoscopy. In Artificial Intelligence in Capsule Endoscopy; Mascarenhas, M., Cardoso, H., Macedo, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 255–269. [Google Scholar] [CrossRef]
- Baddeley, R.; Aabakken, L.; Veitch, A.; Hayee, B. Green Endoscopy: Counting the Carbon Cost of Our Practice. Gastroenterology 2022, 162, 1556–1560. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, S.; Dhar, A.; Baddeley, R.; Donnelly, L.; Haddock, R.; Arasaradnam, R.; Coulter, A.; Disney, B.R.; Griffiths, H.; Healey, C.; et al. Green endoscopy: British Society of Gastroenterology (BSG), Joint Accreditation Group (JAG) and Centre for Sustainable Health (CSH) joint consensus on practical measures for environmental sustainability in endoscopy. Gut 2023, 72, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Levy, I.; Gralnek, I.M. Complications of diagnostic colonoscopy, upper endoscopy, and enteroscopy. Best. Pract. Res. Clin. Gastroenterol. 2016, 30, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Helmers, R.A.; Dilling, J.A.; Chaffee, C.R.; Larson, M.V.; Narr, B.J.; Haas, D.A.; Kaplan, R.S. Overall Cost Comparison of Gastrointestinal Endoscopic Procedures With Endoscopist- or Anesthesia-Supported Sedation by Activity-Based Costing Techniques. Mayo Clin. Proc. Innov. Qual. Outcomes 2017, 1, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.M.; Rosa, B.; Mendes, F.; Mascarenhas, M.; Saraiva, M.M.; Cotter, J. Chapter 11—Small bowel and colon cleansing in capsule endoscopy. In Artificial Intelligence in Capsule Endoscopy; Mascarenhas, M., Cardoso, H., Macedo, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 181–197. [Google Scholar] [CrossRef]
- Mascarenhas, M.; Afonso, J.; Ribeiro, T.; Andrade, P.; Cardoso, H.; Macedo, G. The Promise of Artificial Intelligence in Digestive Healthcare and the Bioethics Challenges It Presents. Medicina 2023, 59, 790. [Google Scholar] [CrossRef] [PubMed]
- Kruse, C.S.; Frederick, B.; Jacobson, T.; Monticone, D.K. Cybersecurity in healthcare: A systematic review of modern threats and trends. Technol. Health Care 2017, 25, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Regulation (EU). Regulation (EU) 2016/679 of the European Parliament and of the Council of 27 April 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing Directive 95/46/EC (General Data Protection Regulation) (Text with EEA relevance); Publications Office of the European Union: Luxembourg, 2016; pp. 1–88. [Google Scholar]
- Mascarenhas, M.; Santos, A.; Macedo, G. Chapter 12—Introducing blockchain technology in data storage to foster big data and artificial intelligence applications in healthcare systems. In Artificial Intelligence in Capsule Endoscopy; Mascarenhas, M., Cardoso, H., Macedo, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 199–216. [Google Scholar] [CrossRef]
- Suresh, H.; Guttag, J.V. A Framework for Understanding Sources of Harm throughout the Machine Learning Life Cycle. In Proceedings of the EAAMO’21: Proceedings of the 1st ACM Conference on Equity and Access in Algorithms, Mechanisms, and Optimization, New York, NY, USA, 5–9 October 2021. [Google Scholar] [CrossRef]
- Ying, X. An Overview of Overfitting and its Solutions. J. Phys. Conf. Ser. 2019, 1168, 022022. [Google Scholar] [CrossRef]
- Price, W.N., II. Black-Box Medicine. Harv. J. Law. Technol. 2014, 28, 419. [Google Scholar]
- Messmann, H.; Bisschops, R.; Antonelli, G.; Libânio, D.; Sinonquel, P.; Abdelrahim, M.; Ahmad, O.F.; Areia, M.; Bergman, J.; Bhandari, P.; et al. Expected value of artificial intelligence in gastrointestinal endoscopy: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy 2022, 54, 1211–1231. [Google Scholar] [CrossRef]
- FDA. Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD) Action Plan; FDA Statement: Silver Spring, MD, USA, 2021.


| Publication, Author, Year | Study Aim | Capsule Types | Centers n | Exams n | Frames n | Types of CNN | Dataset Methods | Analysis Methods | Classification Categories | Performance Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Lesion | SEN | SPE | AUC | ||||||||||
| Specific CNN for Small Bowel Lesions | Aoki, 2020 [37] | Detection of blood | SB2 SB3 | 1 | 66 | 38,055 | 6711 | ResNet | Frame labeling of all datasets (normal vs. blood content) | Train–test split (73–26%) | Blood | 97 | 100 | 100 |
| Afonso, 2021 [38] | Detection of blood | SB3 | 2 | 1483 | 23,190 | 13,515 | Xception | Frame labeling of all datasets (normal vs. blood or hematic residues) | Staged incremental frame with train–test split (80–20%) | Blood/hematic residues | 98 | 98 | 100 | |
| Leenhardt, 2019 [39] | Detection of angiectasia | SB3 | French national of still frames database (from 13 centers) | NA | 1200 | 600 | YOLO | Previous manual annotation of all angiectasias for the French national database | Deep feature extraction of dataset already manually annotated (300 lesions and 300 normal frames) Validation with classification of new dataset (300 lesions and 300 normal frames) | Angiectasia | 100 | 96 | NK | |
| Tsuboi, 2020 [40] | Detection of angiectasia | SB2 SB3 | 2 | 169 | 12,725 | 2725 | SSD | Manual annotation of all angiectasias | CNN is trained exclusively in positive frames (2237 with angiectasias) Validation on mixed data with positive and negative frames (488 angiectasias and 10,000 normal frames) | Angiectasia | 99 | 98 | 100 | |
| Houdeville, 2021 [41] | Detection of angiectasia | SB3 Mirocam | NA | NA | 12,255 | 613 | YOLO | Previous trained on SB3 devices | Validation with 626 new SB3 still frames and 621 new Mirocam still frames | Angiectasia (SB3) | 97 | 99 | NK | |
| Angiectasia (Mirocam) | 96 | 98 | NK | |||||||||||
| Ribeiro, 2021 [42] | Detection of vascular lesions + categorization of bleeding potential | SB3 | 2 | 1483 | 11,588 | 2063 | Xception | Frame labeling of all datasets (normal (N) vs. red spots (P1V) vs. angiectasia or varices (P2V)) | Train–test split (80–20%) with 3 × 3 confusion matrix | N vs. all | 90 | 97 | 98 | |
| P1V vs. all | 92 | 95 | 97 | |||||||||||
| P2V vs. all | 94 | 95 | 98 | |||||||||||
| Aoki, 2019 [43] | Detection of ulcerative lesions | SB2 SB3 | 1 | 180 | 15,800 | 5800 | SSD | Manual annotation of all ulcers or erosions | CNN is trained exclusively in positive frames (5630 with ulcers) Validation on mixed data with positive and negative frames (440 lesions and 10,000 normal frames) | Ulcers or erosions | 88 | 91 | 96 | |
| Klang, 2020 [44] | Detection of ulcers + differentiation from normal mucosa | SB3 | 1 | 49 | 17,640 | 7391 | Xception | Frame labeling of all datasets (normal vs. ulcer) | 5-fold cross-validation with train–test split (80 vs. 20%) | Ulcers (mean of cross-validation) | 95 | 97 | 99 | |
| Barash, 2021 [45] | Categorization of severity grade of ulcers | SB3 | 1 | NK | Random selection of 1546 ulcer frames from Klang dataset | ResNet | Frame labeling of all datasets (mild ulceration (1) vs. moderate ulceration (2) vs. severe ulceration (3)) | Train–test split (80–20%) with 3 × 3 confusion matrix | 1 vs. 2 | 34 | 71 | 57 | ||
| 2 vs. 3 | 73 | 91 | 93 | |||||||||||
| 1 vs. 3 | 91 | 91 | 96 | |||||||||||
| Afonso, 2021 [46] | Detection of ulcerative lesions + categorization of bleeding potential | SB3 | 2 | 2565 | 23,720 | 5675 | Xception | Frame labeling of all datasets (normal (N) vs. erosions (P1E) vs. ulcers with uncertain/intermediate bleeding potential (P1U) vs. ulcers with high bleeding potential (P2U)) | Train–test split (80–20%) with 4 × 4 confusion matrix | N vs. all | 94 | 91 | 98 | |
| P1E vs. all | 73 | 96 | 95 | |||||||||||
| P1U vs. all | 72 | 96 | 96 | |||||||||||
| P2U vs. all | 91 | 99 | 100 | |||||||||||
| Saito, 2020 [47] | Detection of protruding lesions | SB2 SB3 | 3 | 385 | 48,091 | 38091 | SSD | Manual annotation of all protruding lesions (polyps, nodules, epithelial tumors, submucosal tumors, venous structures) | CNN is trained exclusively in positive frames (30,584 with protruding lesions) Validation on mixed data with positive and negative frames (7507 lesions and 10,000 normal frames) | Protruding lesions | 91 | 80 | 91 | |
| Saraiva, 2021 [48] | Detection of protruding lesions + categorization of bleeding potential | SB3 | 1 | 1483 | 18,625 | 2830 | Xception | Frame labeling of all data (normal (N) vs. protruding lesions with uncertain/intermediate bleeding potential (P1PR) vs. protruding lesions with high bleeding potential (P2PR)) | Train–test split (80–20%) with 3 × 3 confusion matrix | N vs. all | 92 | 99 | 99 | |
| P1PR vs. all | 96 | 94 | 99 | |||||||||||
| P2PR vs. all | 97 | 98 | 100 | |||||||||||
| Specific CNN for Colonic Lesions | Yamada, 2021 [49] | Detection of colorectal neoplasias | COLON2 | 1 | 184 | 20,717 | 17,783 | SSD | Manual annotation of all colorectal neoplasias (polyps and cancers) | CNN is trained exclusively in positive frames (15,933 with colorectal neoplasias) Validation on mixed data with positive and negative frames (1805 lesions and 2934 normal frames) | Colorectal neoplasias | 79 | 87 | 90 |
| Saraiva, 2021 [50] | Detection of protruding lesions | COLON2 | 1 | 24 | 3640 | 860 | Xception | Frame labeling of all datasets (normal vs. protruding lesions: polyps, epithelial tumors, subepithelial lesions) | Train–test split (80–20%) | Protruding lesions | 91 | 93 | 97 | |
| Ribeiro, 2022 [51] | Detection of ulcerative lesions | COLON2 | 2 | 124 | 37,319 | 3570 | Xception | Frame labeling of all datasets (normal vs. ulcer or erosions) | train–validation (for hyperparameter tuning)–test split (70–20–10%) | Ulcers or erosions | 97 | 100 | 100 | |
| Majtner, 2021 [52] | Panenteric (small bowel and colon) detection of ulcerative lesions | CROHN | 1 | 38 | 77,744 | 2748 | ResNet | Frame labeling of all datasets (normal vs. ulcer or erosions) | Train–validation–test (70–20–10%) with patient split | Ulcers or erosions | 96 | 100 | NK | |
| Ferreira, 2022 [53] | Panenteric (small bowel and colon) detection of ulcerative lesions | CROHN | 2 | 59 | 24,675 | 5300 | Xception | Frame labeling of all datasets (normal vs. ulcer or erosions) | Train–test split (80–20%) | Ulcers or erosions | 98 | 99 | 100 | |
| Saraiva, 2021 [54] | Detection of blood | COLON2 | 1 | 24 | 5825 | 2975 | Xception | Frame labeling of all datasets (normal vs. blood or hematic residues) | Train–test split (80–0%) | Blood or hematic residues | 100 | 93 | 100 | |
| Complex CNN for Enteric and Colonic Lesions | Ding, 2019 [55] | Detection of abnormal findings in the small bowel without discrimination capacity | NaviCam | 77 | 1970 | 158,235 + validation set | NK | ResNet | Frame labeling of training set (inflammation, ulcer, polyps, lymphangiectasia, bleeding, vascular disease, protruding lesion, lymphatic follicular hyperplasia, diverticulum, parasite, normal) | Testing with 5000 independent CE videos | Abnormal findings | 100 | 100 | NK |
| Aoki, 2021 [56] | Detection of multiple types of lesions in the small bowel | SB3 | 3 | NK | 66,028 + validation set | 44,684 | Combined 3 SSD + 1 ResNet | Manual annotation of all mucosa breaks, angiectasias, protruding lesions and blood contents | CNN is trained on mixed data with positive and negative frames (44,684 lesions and 21,344 normal frames) Validation on 379 full videos | Mucosal brakes vs. other lesions | 96 | 99 | NK | |
| Angiectasias vs. other lesions | 79 | 99 | NK | |||||||||||
| Protruding lesions vs. other lesions | 100 | 95 | NK | |||||||||||
| Blood content vs. other lesions | 100 | 100 | NK | |||||||||||
| Saraiva, 2021 [57] | Detection of multiple types of lesions in the small bowel + categorization of bleeding potential | SB3 OMON | 2 | 5793 | 53,555 | 35,545 | Xception | Frame labeling of all data (normal (N) vs. lymphangiectasias (P0L) vs. xanthomas (P0X) vs. erosions (P1E) vs. ulcers with uncertain/intermediate bleeding potential (P1U) vs. ulcers with high bleeding potential (P2U) vs. red spots (P1RS) vs. vascular lesions (angiectasias or varices) (P2V) vs. protruding lesions with uncertain/intermediate bleeding potential (P1P) vs. protruding lesions with high bleeding potential (P2P) vs. blood or hematic residues) | Train–test split (80–20%) with 11 × 11 confusion matrix | N vs. all | 92 | 96 | 99 | |
| P0L vs. all | 88 | 99 | 99 | |||||||||||
| P0X vs. all | 85 | 98 | 99 | |||||||||||
| P1E vs. all | 73 | 99 | 97 | |||||||||||
| P1U vs. all | 81 | 99 | 99 | |||||||||||
| P2U vs. all | 94 | 98 | 100 | |||||||||||
| P1RS vs. all | 80 | 99 | 98 | |||||||||||
| P2V vs. all | 91 | 99 | 100 | |||||||||||
| P1P vs. all | 93 | 99 | 99 | |||||||||||
| P2P vs. all | 94 | 100 | 99 | |||||||||||
| Blood vs. all | 99 | 100 | 100 | |||||||||||
| Saraiva, 2022 [58] | Detection of pleomorphic lesions or blood in the colon | COLON2 | 2 | 124 | 9005 | pl5,930 | Xception | Frame labeling of all datasets (normal (N) vs. blood or hematic residues (B) vs. mucosal lesions (ML), including ulcers, erosions, vascular lesions (red spots, angiectasia and varices) and protruding lesions (polyps, epithelial tumors, submucosal tumors and nodes)) | Train–test split (80–20%) with 3 × 3 confusion matrix | N vs. all | 97 | 96 | 100 | |
| Blood vs. all | 100 | 100 | 100 | |||||||||||
| ML vs. all | 92 | 99 | 90 | |||||||||||
| Xie, 2022 [59] | Detection of multiple types of lesions in the small bowel + differentiation from normal mucosa | OMON | 51 | 5825 | 757,770 | NK | EfficientNet + Yolo | Frame labeling of all datasets Protruding lesions (venous structure, nodule, mass/tumor, polyp(s)), flat lesions (angiectasia, plaque (red), plaque (white), red spot, abnormal villi), mucosa (lymphangiectasia, erythematous, edematous), excavated lesion (erosion, ulcer, aphtha) and content (blood, parasite) | CNN is trained on mixed data with positive and negative frames Validation on 2898 full videos | Venous structure vs. all | 98 | 100 | NK | |
| Nodule vs. all | 97 | 100 | NK | |||||||||||
| Mass or tumor vs. all | 95 | 100 | NK | |||||||||||
| Polyp vs. all | 95 | 100 | NK | |||||||||||
| Angiectasia vs. all | 96 | 100 | NK | |||||||||||
| Plaque (red) vs. all | 94 | 100 | NK | |||||||||||
| Plaque (white) vs. all | 95 | 100 | NK | |||||||||||
| Red spot vs. all | 96 | 100 | NK | |||||||||||
| Abnormal villi vs. all | 95 | 100 | NK | |||||||||||
| Lymphangiectasia vs. all | 98 | 100 | NK | |||||||||||
| Erythematous mucosa vs. all | 95 | 100 | NK | |||||||||||
| Edematous mucosa vs. all | NK | NK | NK | |||||||||||
| Erosion vs. all | NK | NK | NK | |||||||||||
| Ulcer vs. all | NK | NK | NK | |||||||||||
| Aphtha vs. all | NK | NK | NK | |||||||||||
| Blood vs. all | NK | NK | NK | |||||||||||
| Parasite vs. all | NK | NK | NK | |||||||||||
| Complex CNN for Gastric Lesions | Xia, 2021 [60] | Detection of multiple types of lesions + differentiation from normal mucosa | NaviCam MCE | 1 | 797 | 1,023,955 | NK | ResNet | Frame labeling of training set (erosions, polyps, ulcers, submucosal tumors, xanthomas, normal) | testing with 100 independent CE videos | Pleomorphic lesions | 96 | 76 | 84 |
| Pan, 2022 [61] | Detect in real time of both gastric anatomic landmarks and different types of lesions | NaviCam MCE | 1 | 906 | 34,062 + validation set | NK | ResNet | Frame labeling of all datasets (ulcerative (ulcer and erosions), protruding lesions (polyps and submucosal tumors), xanthomas, normal mucosa) | Prospective validation on 50 CE exams | Gastric lesions | 99 | NK | NK | |
| Anatomic landmarks | 94 | NK | NK | |||||||||||
| Saraiva, 2023 [62] | Detection of pleomorphic gastric lesions | SB3 CROHN OMON | 2 | 107 | 12,918 | 6074 | Xception | Frame labeling of all datasets (normal vs. pleomorphic lesion (vascular, ulcerative or protruding lesion or blood/hematic residues)) | Train–test split (80–20%) with patient split design and 3-fold cross-validation during training set | Pleomorphic lesions (mean of cross-validation) | 88 | 92 | 96 | |
| Pleomorphic lesions (test set) | 97 | 96 | 100 | |||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mascarenhas, M.; Martins, M.; Afonso, J.; Ribeiro, T.; Cardoso, P.; Mendes, F.; Andrade, P.; Cardoso, H.; Ferreira, J.; Macedo, G. The Future of Minimally Invasive Capsule Panendoscopy: Robotic Precision, Wireless Imaging and AI-Driven Insights. Cancers 2023, 15, 5861. https://doi.org/10.3390/cancers15245861
Mascarenhas M, Martins M, Afonso J, Ribeiro T, Cardoso P, Mendes F, Andrade P, Cardoso H, Ferreira J, Macedo G. The Future of Minimally Invasive Capsule Panendoscopy: Robotic Precision, Wireless Imaging and AI-Driven Insights. Cancers. 2023; 15(24):5861. https://doi.org/10.3390/cancers15245861
Chicago/Turabian StyleMascarenhas, Miguel, Miguel Martins, João Afonso, Tiago Ribeiro, Pedro Cardoso, Francisco Mendes, Patrícia Andrade, Helder Cardoso, João Ferreira, and Guilherme Macedo. 2023. "The Future of Minimally Invasive Capsule Panendoscopy: Robotic Precision, Wireless Imaging and AI-Driven Insights" Cancers 15, no. 24: 5861. https://doi.org/10.3390/cancers15245861
APA StyleMascarenhas, M., Martins, M., Afonso, J., Ribeiro, T., Cardoso, P., Mendes, F., Andrade, P., Cardoso, H., Ferreira, J., & Macedo, G. (2023). The Future of Minimally Invasive Capsule Panendoscopy: Robotic Precision, Wireless Imaging and AI-Driven Insights. Cancers, 15(24), 5861. https://doi.org/10.3390/cancers15245861









