Clinical Use of a Commercial Artificial Intelligence-Based Software for Autocontouring in Radiation Therapy: Geometric Performance and Dosimetric Impact
Abstract
:Simple Summary
Abstract
1. Introduction
Research Objectives
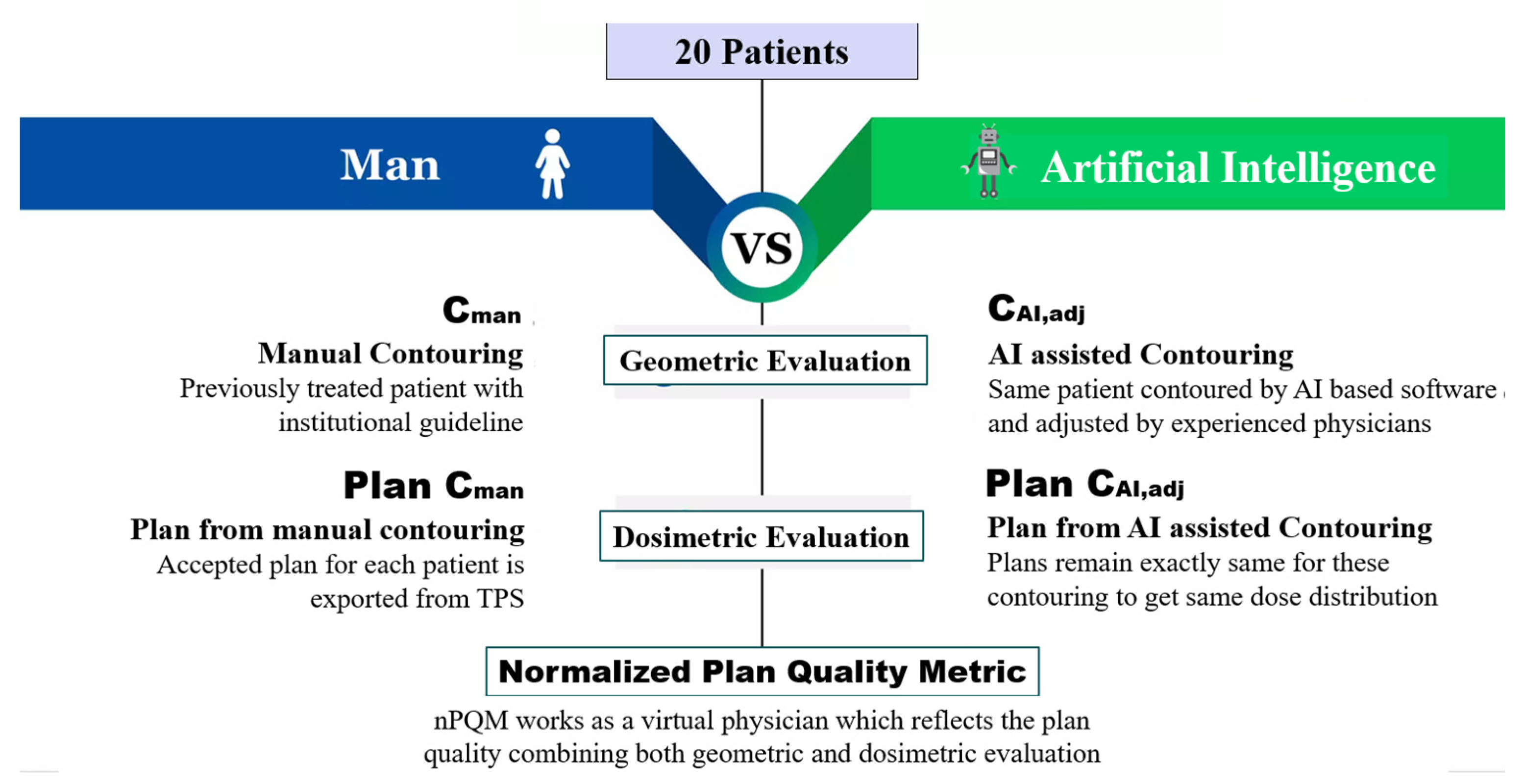
2. Materials and Methods
2.1. Patient Data
2.2. Contouring Workflows
- -
- Manual contouring (Cman). Contours were delineated by a radiation oncologist with at least ten years of experience, also using semiautomated tools like flood fill and interpolation, within the integrated ARIA and Eclipse TPS systems (version: 16.1; Varian Medical Systems, Inc., NewYork, CA, USA) [18] and following the institutional guidelines [19,20,21]. These contours were assumed as the ground truth structures.
- -
- Fully automated contouring based on artificial intelligence (CAI). These were automatically created using a research version of Limbus Contour (version: 1.0.18; Limbus AI Inc., Regina, SK, Canada) [22] software. Limbus Contour (LC) employs organ-specific deep convolutional neural network models on the basis of a U-net architecture [23], which were trained on CT images from the Cancer Imaging Archive public database [24]. Following the creation of contours, LC applies a number of postprocessing techniques including as outlier removal, slice interpolation, z-plane cutoffs, and contour smoothing [23]. Contouring the structure set on a patient required up to 7 min on 3.2 GHz Intel Pentium CPU G3420; 4 GB RAM; 64-bit Operating System. The Limbus-generated RT structure sets were exported as DICOM files into the Varian Eclipse workstation for revision and validation.
- -
- automatically generated contours reviewed and eventually adjusted by radiation oncologist (CAI,adj). Expert ROs reviewed and, if necessary, modified the CAI using the Eclipse contouring application in accordance with institutional consensus guidelines for target volume and OAR contours. In order to keep ROs blinded to the original contours’ creation, only CAI contours were visible to them during revision time.
2.3. Treatment Planning and Delivery
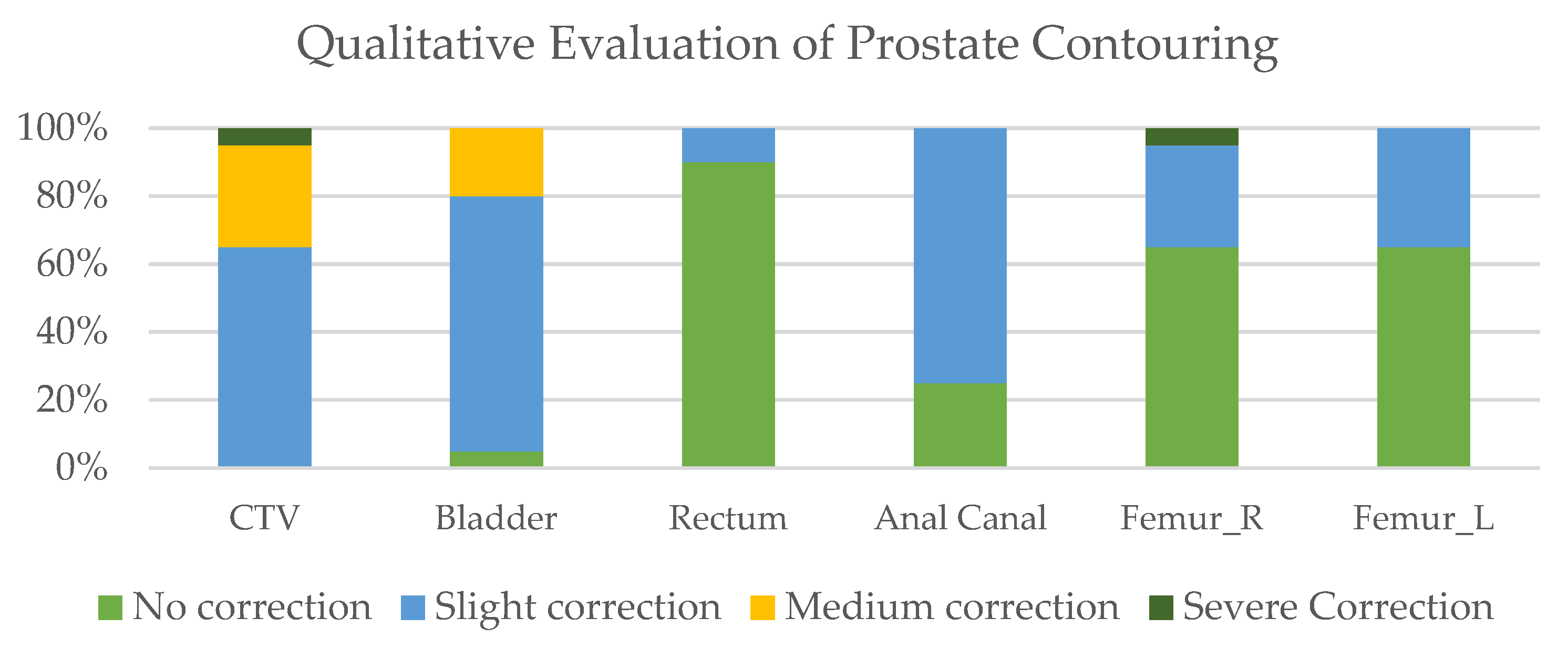
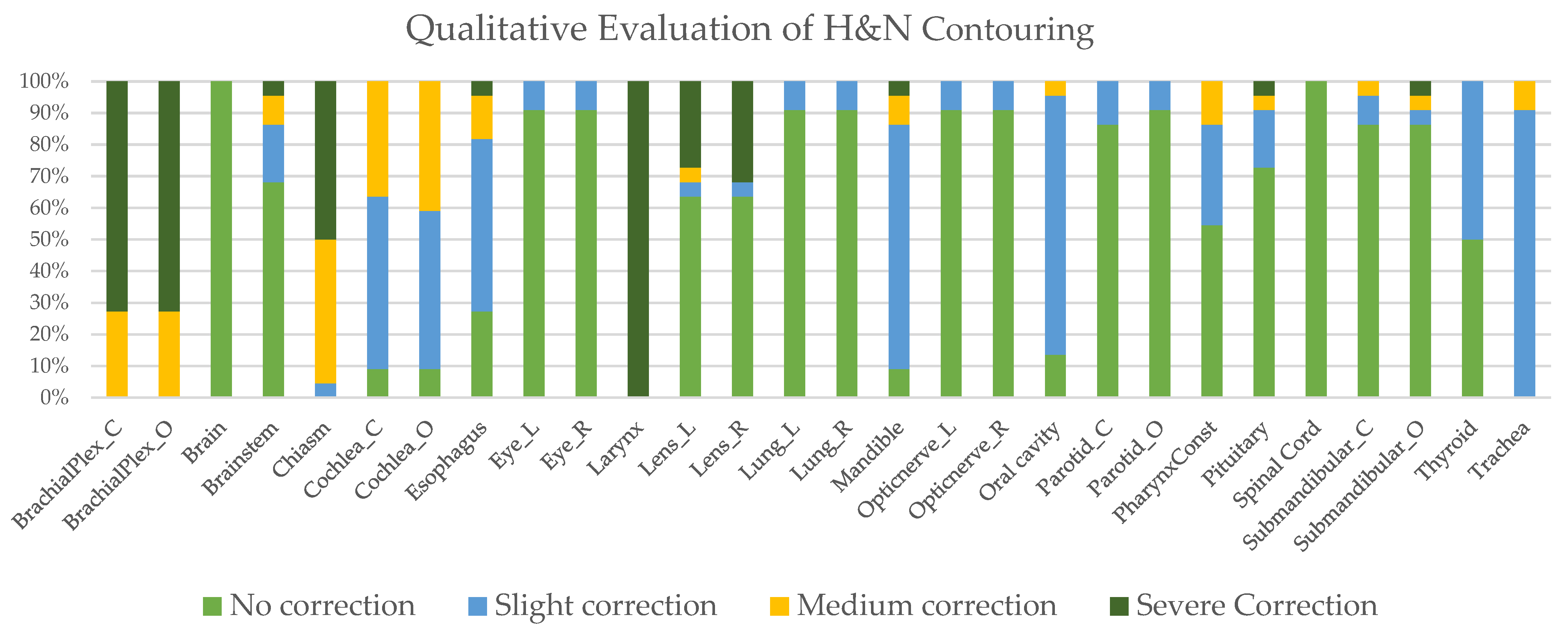
2.4. Qualitative Assessment of Automated Contouring
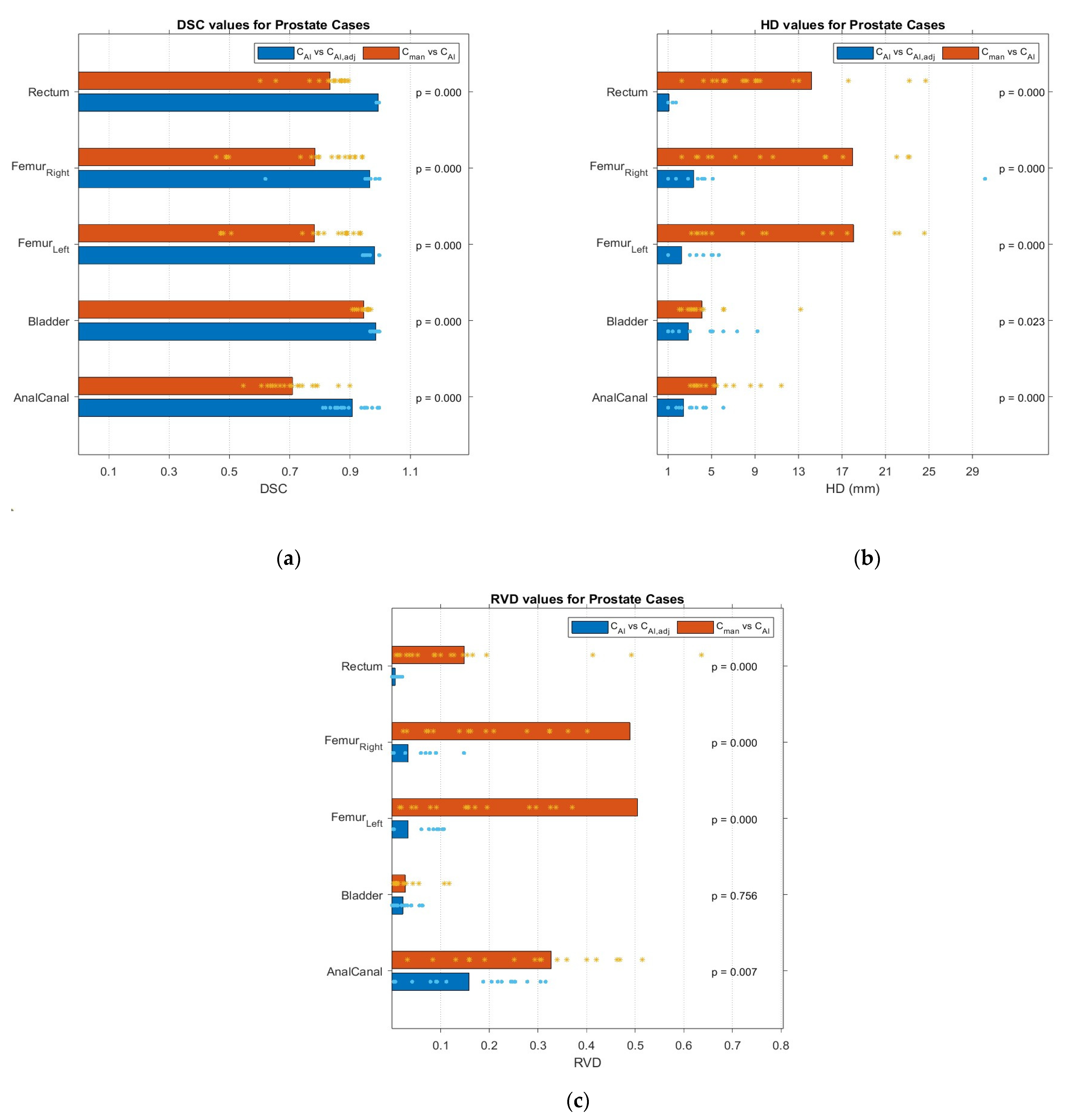
2.5. Geometric Evaluation
2.6. Evaluation of Dose Differences
2.7. Normalized Plan Quality Metric
2.8. Evaluation of Contouring Time
2.9. Interobserver Variability
2.10. Data Analysis
3. Results
3.1. Qualitative Assessment of Automated Contouring
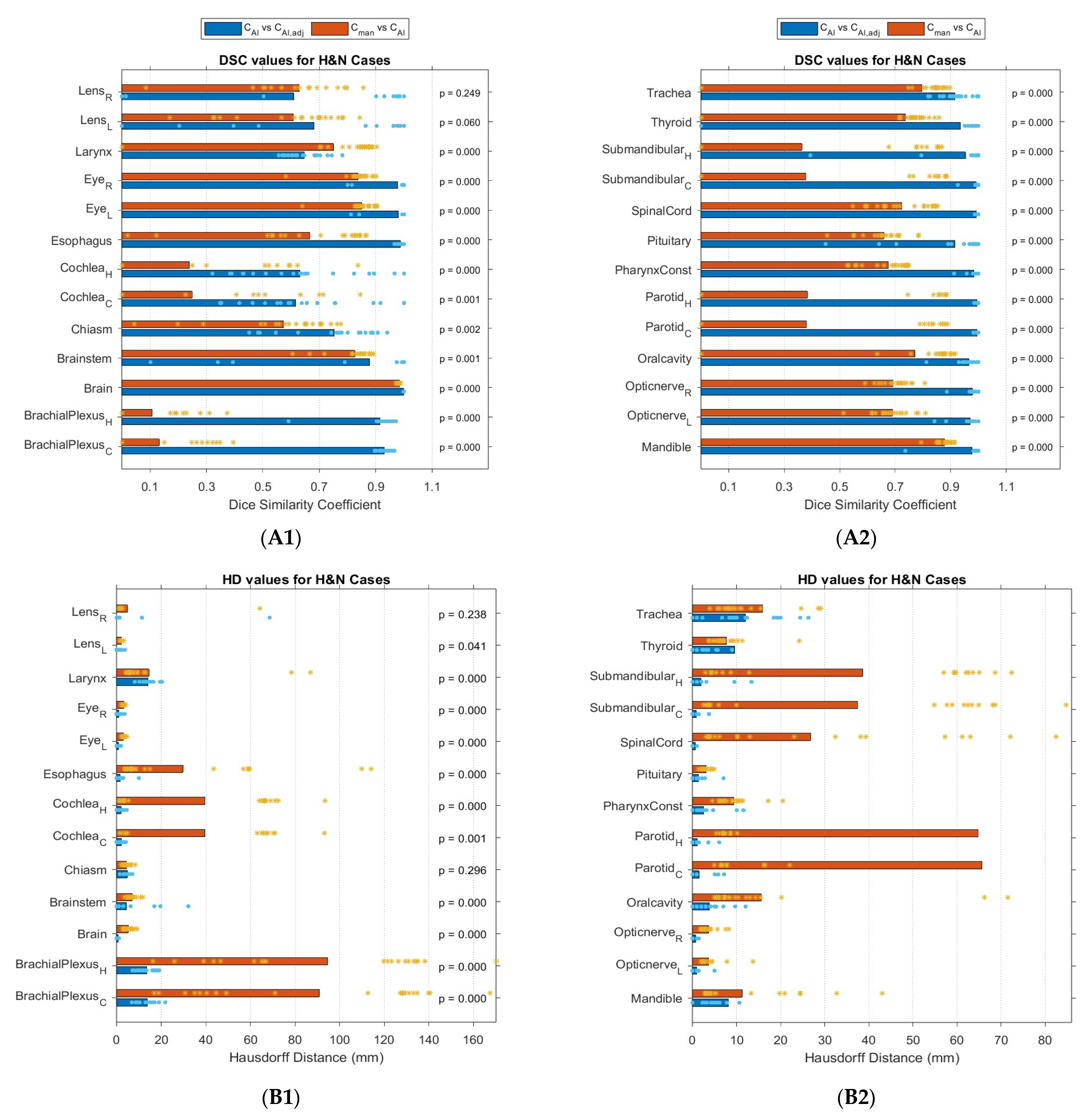
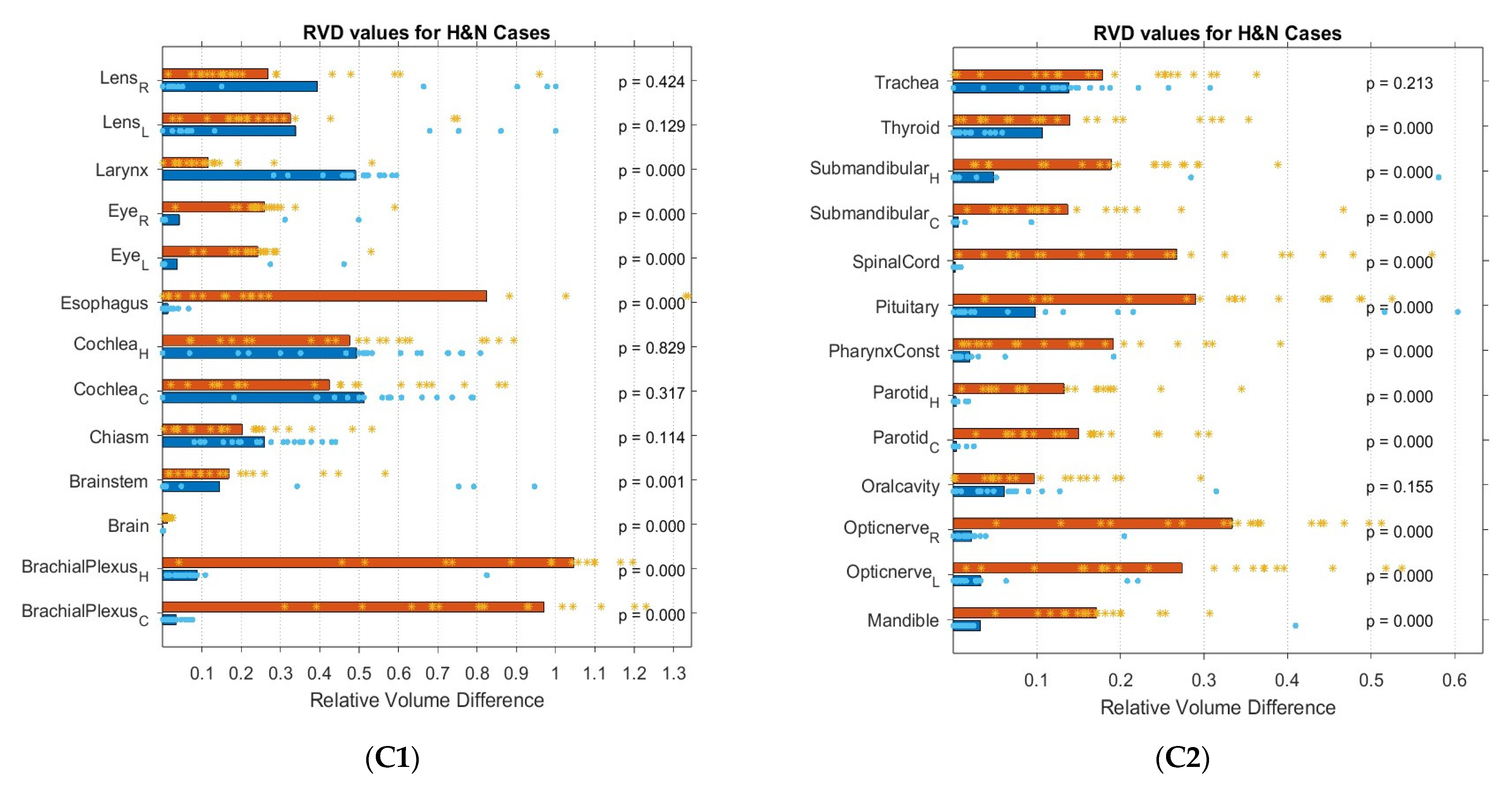
3.2. Geometric Comparison
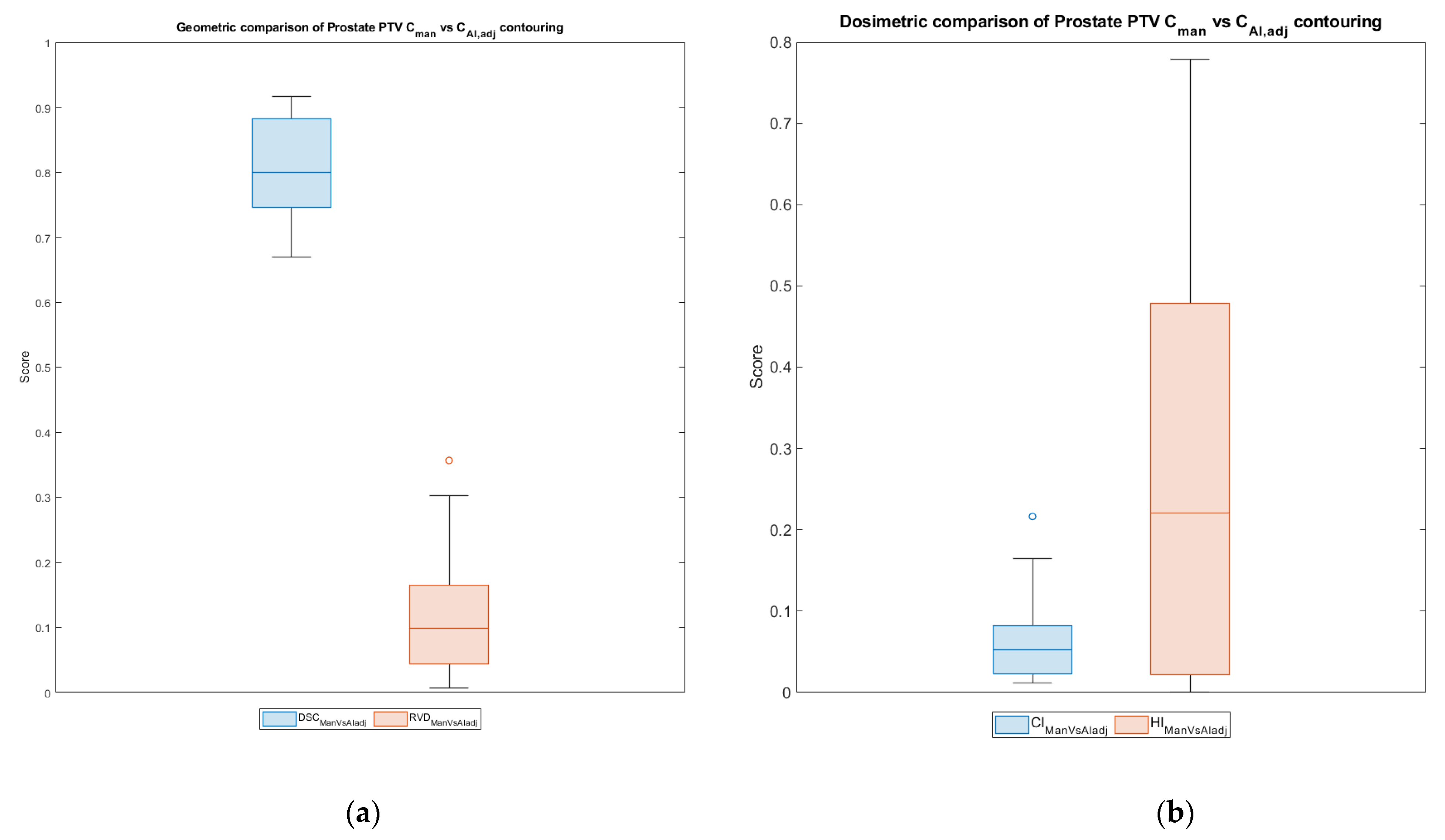
3.3. PTV Evaluation
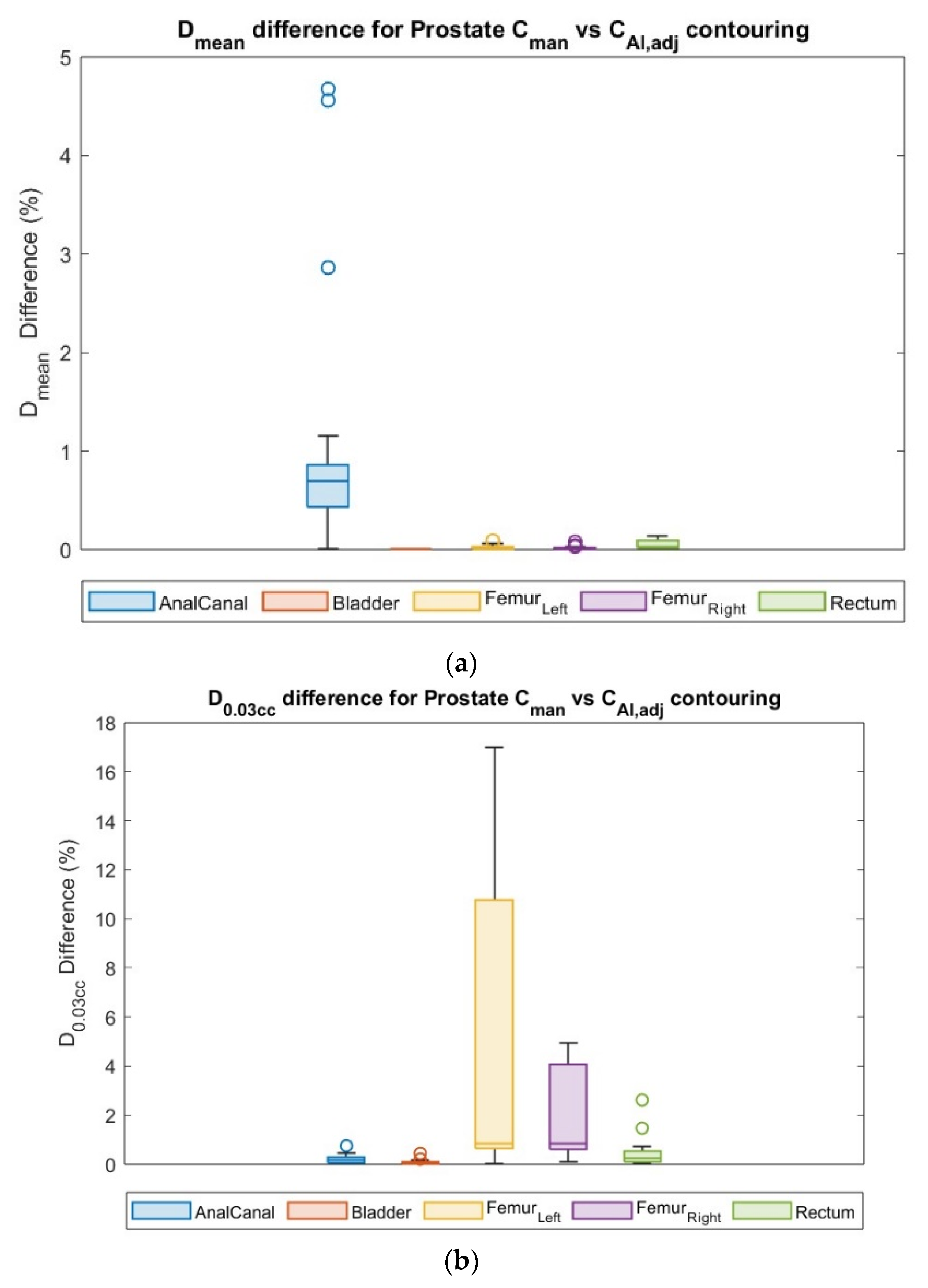
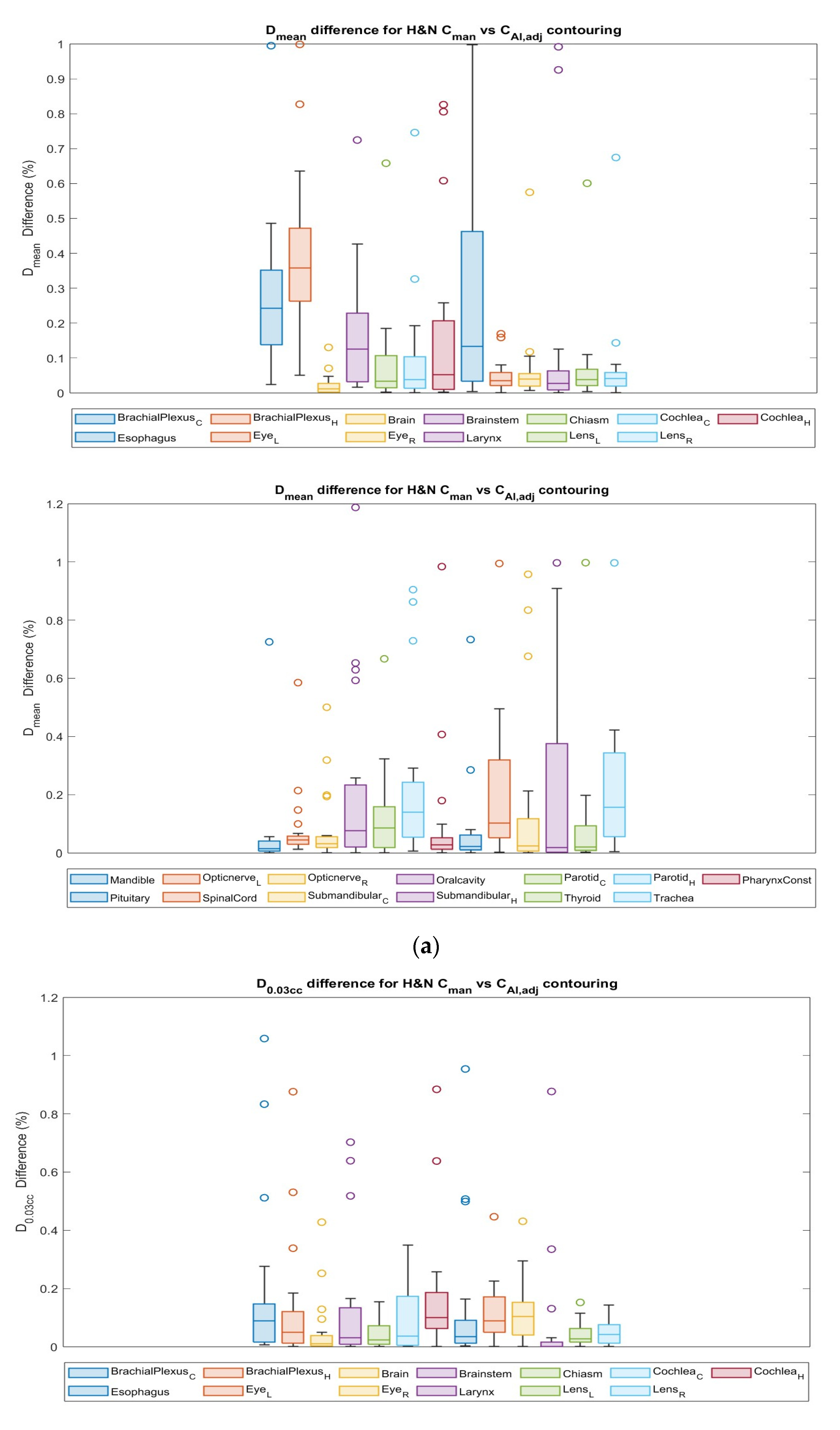
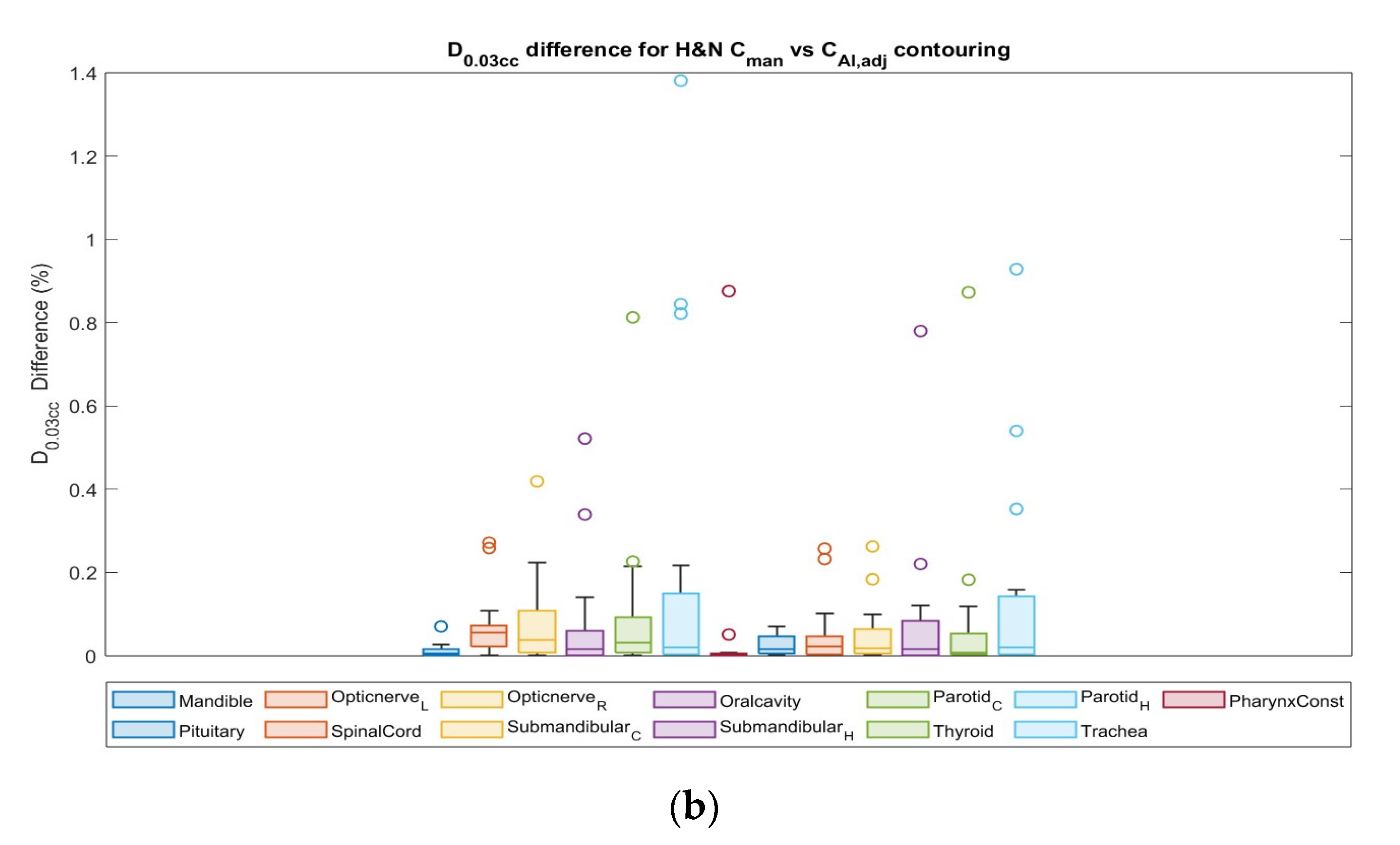
3.4. Dosimetric Comparison
3.5. nPQM Comparison
3.6. Time Savings
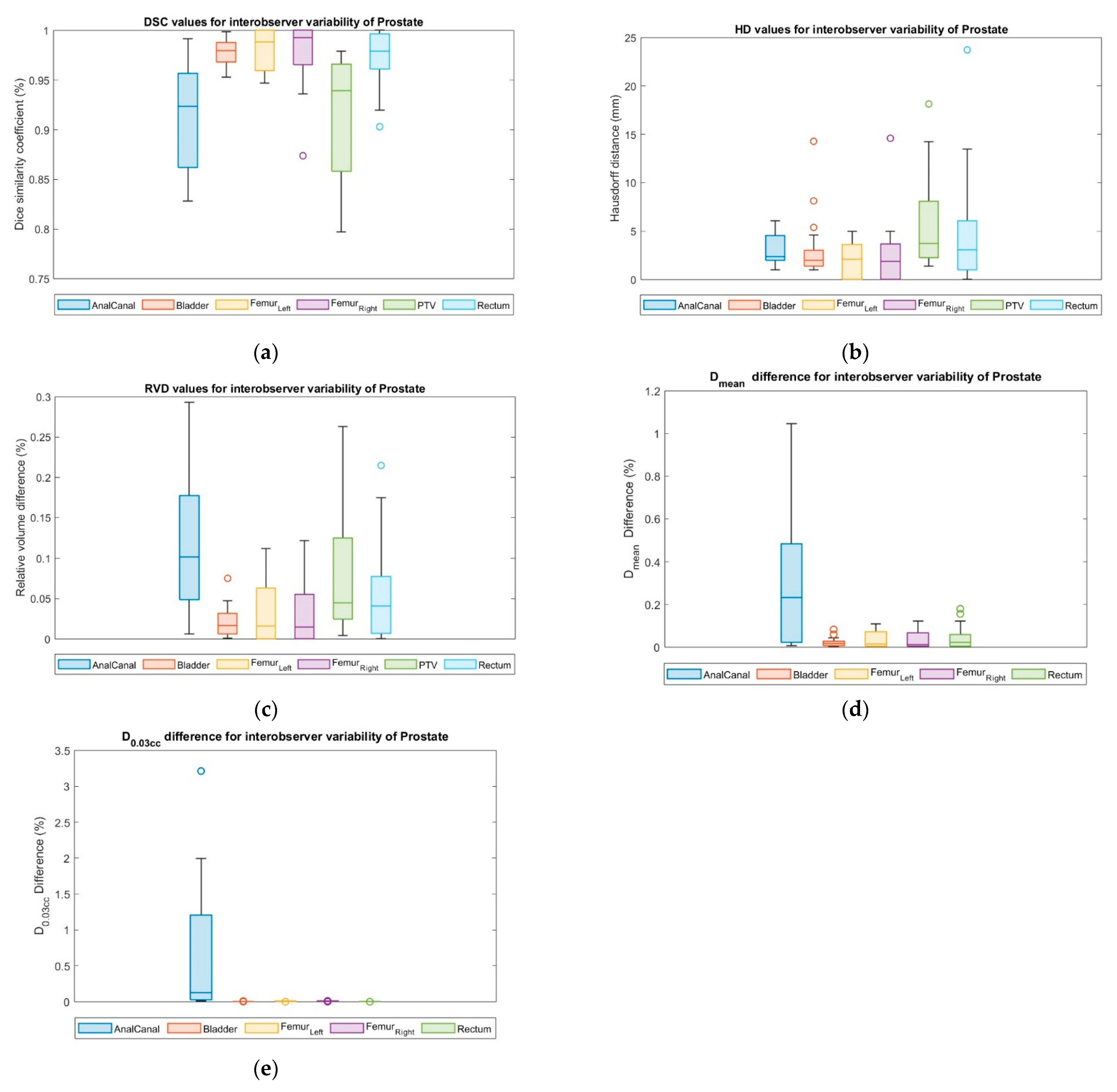
3.7. Interobserver Variability
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barton, M.B.; Jacob, S.; Shafiq, J.; Wong, K.; Thompson, S.R.; Hanna, T.P.; Delaney, G.P. Estimating the demand for radiotherapy from the evidence: A review of changes from 2003 to 2012. Radiother. Oncol. 2014, 112, 140–144. [Google Scholar] [CrossRef]
- Barton, M.B.; Frommer, M.; Shafiq, J. Role of radiotherapy in cancer control in low-income and middle-income countries. Lancet Oncol. 2006, 7, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Cancer Today. Available online: http://gco.iarc.fr/today/home (accessed on 5 November 2022).
- Cao, M.; Stiehl, B.; Yu, V.Y.; Sheng, K.; Kishan, A.U.; Chin, R.K.; Yang, Y.; Ruan, D. Analysis of Geometric Performance and Dosimetric Impact of Using Automatic Contour Segmentation for Radiotherapy Planning. Front. Oncol. 2020, 10, 1762. Available online: https://www.frontiersin.org/articles/10.3389/fonc.2020.01762 (accessed on 30 November 2023). [CrossRef] [PubMed]
- Jameson, M.G.; Holloway, L.C.; Vial, P.J.; Vinod, S.K.; Metcalfe, P.E. A review of methods of analysis in contouring studies for radiation oncology. J. Med. Imaging Radiat. Oncol. 2010, 54, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Chang, J.S.; Kim, Y.B.; Kim, J.S. Atlas-based auto-segmentation for postoperative radiotherapy planning in endometrial and cervical cancers. Radiat. Oncol. 2020, 15, 106. [Google Scholar] [CrossRef] [PubMed]
- Tong, N.; Gou, S.; Yang, S.; Cao, M.; Sheng, K. Shape constrained fully convolutional DenseNet with adversarial training for multiorgan segmentation on head and neck CT and low-field MR images. Med. Phys. 2019, 46, 2669–2682. [Google Scholar] [CrossRef] [PubMed]
- Jackson, P.; Kron, T.; Hardcastle, N. A future of automated image contouring with machine learning in radiation therapy. J. Med. Radiat. Sci. 2019, 66, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Huang, V.; Wells, D.; Giambattista, J.; Giambattista, J.; Kolbeck, C.; Otto, K.; Saibishkumar, E.P.; Alexander, A. Implementation of deep learning-based auto-segmentation for radiotherapy planning structures: A workflow study at two cancer centers. Radiat. Oncol. 2021, 16, 101. [Google Scholar] [CrossRef]
- Brouwer, C.L.; Dinkla, A.M.; Vandewinckele, L.; Crijns, W.; Claessens, M.; Verellen, D.; van Elmpt, W. Machine learning applications in radiation oncology: Current use and needs to support clinical implementation. Phys. Imaging Radiat. Oncol. 2020, 16, 144–148. [Google Scholar] [CrossRef]
- Casati, M.; Piffer, S.; Calusi, S.; Marrazzo, L.; Simontacchi, G.; Di Cataldo, V.; Greto, D.; Desideri, I.; Vernaleone, M.; Francolini, G.; et al. Clinical validation of an automatic atlas-based segmentation tool for male pelvis CT images. J. Appl. Clin. Med. Phys. 2022, 23, e13507. [Google Scholar] [CrossRef]
- D’aviero, A.; Re, A.; Catucci, F.; Piccari, D.; Votta, C.; Piro, D.; Piras, A.; Di Dio, C.; Iezzi, M.; Preziosi, F.; et al. Clinical Validation of a Deep-Learning Segmentation Software in Head and Neck: An Early Analysis in a Developing Radiation Oncology Center. Int. J. Environ. Res. Public Health 2022, 19, 9057. [Google Scholar] [CrossRef] [PubMed]
- Zabel, W.J.; Conway, J.L.; Gladwish, A.; Skliarenko, J.; Didiodato, G.; Goorts-Matthews, L.; Michalak, A.; Reistetter, S.; King, J.; Nakonechny, K.; et al. Clinical Evaluation of Deep Learning and Atlas-Based Auto-Contouring of Bladder and Rectum for Prostate Radiation Therapy. Pract. Radiat. Oncol. 2021, 11, e80–e89. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Voet, P.W.; Dirkx, M.L.; Teguh, D.N.; Hoogeman, M.S.; Levendag, P.C.; Heijmen, B.J. Does atlas-based autosegmentation of neck levels require subsequent manual contour editing to avoid risk of severe target underdosage? A dosimetric analysis. Radiother. Oncol. 2011, 98, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Vinod, S.K.; Jameson, M.G.; Min, M.; Holloway, L.C. Uncertainties in volume delineation in radiation oncology: A systematic review and recommendations for future studies. Radiother. Oncol. 2016, 121, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Sharp, G.; Fritscher, K.D.; Pekar, V.; Peroni, M.; Shusharina, N.; Veeraraghavan, H.; Yang, J. Vision 20/20: Perspectives on automated image segmentation for radiotherapy. Med. Phys. 2014, 41, 050902. [Google Scholar] [CrossRef]
- Eclipse|Varian. Available online: https://www.varian.com/products/radiotherapy/treatment-planning/eclipse (accessed on 12 November 2022).
- Offersen, B.V.; Boersma, L.J.; Kirkove, C.; Hol, S.; Aznar, M.C.; Sola, A.B.; Kirova, Y.M.; Pignol, J.-P.; Remouchamps, V.; Verhoeven, K.; et al. ESTRO consensus guideline on target volume delineation for elective radiation therapy of early stage breast cancer. Radiother. Oncol. 2015, 114, 3–10. [Google Scholar] [CrossRef]
- Salembier, C.; Villeirs, G.; De Bari, B.; Hoskin, P.; Pieters, B.R.; Van Vulpen, M.; Khoo, V.; Henry, A.; Bossi, A.; De Meerleer, G.; et al. ESTRO ACROP consensus guideline on CT- and MRI-based target volume delineation for primary radiation therapy of localized prostate cancer. Radiother. Oncol. 2018, 127, 49–61. [Google Scholar] [CrossRef]
- Grégoire, V.; Evans, M.; Le, Q.-T.; Bourhis, J.; Budach, V.; Chen, A.; Eisbruch, A.; Feng, M.; Giralt, J.; Gupta, T.; et al. Delineation of the primary tumour Clinical Target Volumes (CTV-P) in laryngeal, hypopharyngeal, oropharyngeal and oral cavity squamous cell carcinoma: AIRO, CACA, DAHANCA, EORTC, GEORCC, GORTEC, HKNPCSG, HNCIG, IAG-KHT, LPRHHT, NCIC CTG, NCRI, NRG Oncology, PHNS, SBRT, SOMERA, SRO, SSHNO, TROG consensus guidelines. Radiother. Oncol. 2018, 126, 3–24. [Google Scholar] [CrossRef]
- Limbus AI-Automatic Contouring for Radiation Therapy, Limbus AI. Available online: https://limbus.ai/ (accessed on 12 November 2022).
- Wong, J.; Fong, A.; McVicar, N.; Smith, S.; Giambattista, J.; Wells, D.; Kolbeck, C.; Giambattista, J.; Gondara, L.; Alexander, A. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother. Oncol. 2020, 144, 152–158. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Avanzo, M.; Pirrone, G.; Vinante, L.; Caroli, A.; Stancanello, J.; Drigo, A.; Massarut, S.; Mileto, M.; Urbani, M.; Trovo, M.; et al. Electron Density and Biologically Effective Dose (BED) Radiomics-Based Machine Learning Models to Predict Late Radiation-Induced Subcutaneous Fibrosis. Front. Oncol. 2020, 10, 490. [Google Scholar] [CrossRef]
- Avanzo, M.; Chiovati, P.; Boz, G.; Sartor, G.; Dozza, F.; Capra, E. Image-guided volumetric arc radiotherapy of pancreatic cancer with simultaneous integrated boost: Optimization strategies and dosimetric results. Phys. Medica 2016, 32, 169–175. [Google Scholar] [CrossRef]
- Baroudi, H.; Brock, K.K.; Cao, W.; Chen, X.; Chung, C.; Court, L.E.; El Basha, M.D.; Farhat, M.; Gay, S.; Gronberg, M.P.; et al. Automated Contouring and Planning in Radiation Therapy: What Is ‘Clinically Acceptable’? Diagnostics 2023, 13, 667. [Google Scholar] [CrossRef]
- Gooding, M.J.; Smith, A.J.; Tariq, M.; Aljabar, P.; Peressutti, D.; van der Stoep, J.; Reymen, B.; Emans, D.; Hattu, D.; van Loon, J.; et al. Comparative evaluation of autocontouring in clinical practice: A practical method using the Turing test. Med. Phys. 2018, 45, 5105–5115. [Google Scholar] [CrossRef]
- Yeghiazaryan, V.; Voiculescu, I. Family of boundary overlap metrics for the evaluation of medical image segmentation. J. Med. Imaging 2018, 5, 015006. [Google Scholar] [CrossRef] [PubMed]
- StructSeg2019-Grand Challenge, Grand-Challenge.org. Available online: https://structseg2019.grand-challenge.org/Evaluation/ (accessed on 30 November 2022).
- Mayo, C.S.; Moran, J.M.; Bosch, W.; Xiao, Y.; McNutt, T.; Popple, R.; Michalski, J.; Feng, M.; Marks, L.B.; Fuller, C.D.; et al. American Association of Physicists in Medicine Task Group 263: Standardizing Nomenclatures in Radiation Oncology. Int. J. Radiat. Oncol. 2017, 100, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Doses, A. 3. Special Considerations Regarding Absorbed-Dose and Dose–Volume Prescribing and Reporting in IMRT. J. ICRU 2010, 10, 27–40. [Google Scholar] [CrossRef]
- Feuvret, L.; Noël, G.; Mazeron, J.-J.; Bey, P. Conformity index: A review. Int. J. Radiat. Oncol. 2006, 64, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Nelms, B.E.; Robinson, G.; Markham, J.; Velasco, K.; Boyd, S.; Narayan, S.; Wheeler, J.; Sobczak, M.L. Variation in external beam treatment plan quality: An inter-institutional study of planners and planning systems. Pract. Radiat. Oncol. 2012, 2, 296–305. [Google Scholar] [CrossRef] [PubMed]
- MATLAB-MathWorks. Available online: https://ww2.mathworks.cn/en/products/matlab.html (accessed on 30 November 2022).
- Avanzo, M.; Trianni, A.; Botta, F.; Talamonti, C.; Stasi, M.; Iori, M. Artificial Intelligence and the Medical Physicist: Welcome to the Machine. Appl. Sci. 2021, 11, 1691. [Google Scholar] [CrossRef]
- Zanca, F.; Brusasco, C.; Pesapane, F.; Kwade, Z.; Beckers, R.; Avanzo, M. Regulatory Aspects of the Use of Artificial Intelligence Medical Software. Semin. Radiat. Oncol. 2022, 32, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Mackay, K.; Bernstein, D.; Glocker, B.; Kamnitsas, K.; Taylor, A. A Review of the Metrics Used to Assess Auto-Contouring Systems in Radiotherapy. Clin. Oncol. 2023, 35, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Hsu, I.J.; Lessard, E.; Pouliot, J.; Vujic, J. Dose uncertainty due to computed tomography (CT) slice thickness in CT-based high dose rate brachytherapy of the prostate cancer. Med. Phys. 2004, 31, 2543–2548. [Google Scholar] [CrossRef] [PubMed]
- Berthelet, E.; Liu, M.; Truong, P.; Czaykowski, P.; Kalach, N.; Yu, C.; Patterson, K.; Currie, T.; Kristensen, S.; Kwan, W.; et al. CT slice index and thickness: Impact on organ contouring in radiation treatment planning for prostate cancer. J. Appl. Clin. Med. Phys. 2003, 4, 365–373. [Google Scholar] [CrossRef]
| Prostate (6000 cGy × 20 fx) | |
|---|---|
| PTV | Dmin > 5700 cGy |
| Dmax < 6420 cGy | |
| Rectum | V4500 cGy < 15% |
| V2800 cGy < 35% | |
| V900 cGy < 80% | |
| Bladder | V4500 cGy < 25% |
| V2800 cGy < 50% | |
| Anal canal | V4500 cGy < 15% |
| V2800 cGy < 35% | |
| V900 cGy < 80% | |
| Femoral head | V3100 cGy < 1% |
| H&N (7095 cGy × 33 fx) | |
|---|---|
| PTV7095 | Dmin > 6953 cGy |
| Dmax < 7591 cGy | |
| PTV6270 | Dmin > 5956 cGy |
| PTV5610 | Dmin > 5329 cGy |
| Brain | V6000 cGy < 3% |
| Brainstem | Dmax < 5000 cGy |
| Cochlea | Dmean < 4500 cGy |
| Spinal cord | Dmax < 3600 cGy |
| PRV_Spinal cord | Dmax < 4000 cGy |
| Oral cavity | Dmean < 3000 cGy |
| V3000 cGy < 73% | |
| V4000 cGy < 20% | |
| Ipsilateral parotid | Dmean < 2600 cGy |
| Contralateral parotid | Dmean < 2000 cGy |
| Ipsilateral submandibular gland | Dmean < 5000 cGy |
| Contralateral submandibular gland | Dmean < 3900 cGy |
| Mandible | V5000 cGy < 31% |
| Arytenoid cartilage | V5000 cGy < 50% |
| Constrictor muscle | V5000 cGy < 70% |
| Constrictor muscle-PTV | Dmean < 5000 cGy |
| V5000 cGy < 31% | |
| V5000 cGy < 31 cc | |
| Thyroid | Dmean < 4500 cGy |
| V4000 cGy < 50% | |
| V3000 cGy < 60% | |
| Brachial plexus | V6000 cGy < 0.1 cc |
| Esophagus | V3500 cGy < 50% |
| V5000 cGy < 40% | |
| V7000 cGy < 20% | |
| Likert Scale for Each Patient | ||
|---|---|---|
| : Require correction | → Large and obvious errors |
| : Require correction | → Minor errors that need a small amount of editing |
| : Accepted | → Minor errors, but these are clinically not significant |
| : Accepted | → Contour is very precise |
| Structure | Constraint | Function | Thresholds | Max Score |
|---|---|---|---|---|
| PTV | D99% | Linear | >6000 cGY (100%) | 5 |
| >5700 cGy (95%) | 4 | |||
| D0.1 cc | Linear | <6300 cGy(105%) | 5 | |
| <6420 cGy(107%) | 4 | |||
| Rectum | V4500 cGy | Threshold | <15% | 3 |
| V2800 cGy | Threshold | <35% | 3 | |
| V900 cGy | Threshold | <80% | 3 | |
| Bladder | V4500 cGy | Threshold | <25% | 3 |
| V2800 cGy | Threshold | <50% | 3 | |
| Anal canal | V4500 cGy | Threshold | <15% | 3 |
| V2800 cGy | Threshold | <35% | 3 | |
| V900 cGy | Threshold | <80% | 3 | |
| Femur Right | V3500 cGy | Threshold | <1% | 2 |
| Femur Left | V3500 cGy | Threshold | <1% | 2 |
| Structure | Constraint | Function | Thresholds | Max Score |
|---|---|---|---|---|
| BrachialPlexus_C | Dmax | Threshold | <6000 cGy | 3 |
| BrachialPlexus_O | Dmax | Threshold | <6270 cGy | 3 |
| Brain | V6000 cGy | Threshold | <3% | 3 |
| Brainstem | Dmax | Threshold | <5000 cGy | 5 |
| Chiasm | Dmax | Threshold | <5400 cGy | 5 |
| Cochlea_Contralateral | Dmax | Threshold | <1000 cGy | 2 |
| Cochlea_Ipsilateral | Dmax | Threshold | <3500 cGy | 2 |
| Esophagus | V3500 cGy | Threshold | <50% | 1 |
| V5000 cGy | Threshold | <40% | 1 | |
| V7000 cGy | Threshold | <20% | 1 | |
| Eye_L | Dmax | Threshold | <4500 cGy | 5 |
| Eye_R | Dmax | Threshold | <4500 cGy | 5 |
| Larynx | Dmean | Threshold | <4000 cGy | 2 |
| V5000 cGy | Threshold | <27% | 2 | |
| Lens_L | Dmax | Threshold | <400 cGy | 4 |
| Lens_R | Dmax | Threshold | <400 cGy | 4 |
| Mandible | V5000 cGy | Threshold | <31% | 3 |
| OpticNerve_L | Dmax | Threshold | <5400 cGy | 5 |
| OpticNerve_R | Dmax | Threshold | <5400 cGy | 5 |
| Oral Cavity | Dmean | Threshold | <3000 cGy | 3 |
| V3000 cGy | Threshold | <73% | 2 | |
| V4000 cGy | Threshold | <20% | 2 | |
| Parotid_Contralateral | Dmean | Threshold | <2000 cGy | 2 |
| Dmedian | Threshold | <2000 cGy | 4 | |
| Parotid_Ipsilatateral | Dmean | Threshold | <2600 cGy | 2 |
| Dmedian | Threshold | <2600 cGy | 4 | |
| PharynxConst | V5000 cGy | Threshold | <70% | 2 |
| Pituitary | Dmax | Threshold | <5000 cGy | 5 |
| SpinalCord | Dmax | Threshold | <4000 cGy | 5 |
| Submandibular_Co | Dmean | Threshold | <3900 cGy | 3 |
| Submandibular_Ho | Dmean | Threshold | <5000 cGy | 3 |
| Thyroid | Dmean | Threshold | <4500 cGy | 1 |
| V4000 cGy | Threshold | <50% | 1 | |
| V3000 cGy | Threshold | <60% | 1 |
| DSC | HD (mm) | RVD | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | |
| CTV | 0.93 (0.07) | 0.96 (0.68–0.99) | 4.19 (3.31) | 3.00 (1.0–11.09) | 0.08 (0.11) | 0.03 (0.0–0.46) |
| Rectum | 0.99 (0.00) | 0.99 (0.99–1.0) | 1.08 (0.20) | 1.00 (1.0–1.73) | 0.01 (0.0) | 0.01 (0.0–0.02) |
| Bladder | 0.99 (0.01) | 0.99 (0.97–1.0) | 2.85 (2.54) | 1.21 (1.0–9.22) | 0.02 (0.02) | 0.02 (0.0–0.06) |
| Anal Canal | 0.91 (0.06) | 0.89 (0.81–1.0) | 2.44 (1.36) | 2.00 (1.0–6.08) | 0.16 (0.11) | 0.20 (0.0–0.32) |
| Left Femur | 0.98 (0.02) | 1.00 (0.94–1.0) | 2.23 (1.80) | 1.00 (1.0–5.66) | 0.03 (0.04) | 0.00 (0.0–0.11) |
| Right Femur | 0.97 (0.08) | 1.00 (0.62–1.0) | 3.34 (6.46) | 1.00 (1.0–30.15) | 0.03 (0.04) | 0.00 (0.0–0.15) |
| DSC | HD (mm) | RVD | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | |
| PTV | 0.80 (0.07) | 0.80 (0.67–0.91) | 15.38 (9.20) | 15.39 (3.16–37.7) | 0.12 (0.10) | 0.10 (0.01–0.35) |
| Rectum | 0.83 (0.07) | 0.85 (0.60–0.89) | 14.20 (20.2) | 8.62 (2.24–96.30) | 0.15 (0.17) | 0.09 (0.01–0.64) |
| Bladder | 0.94 (0.01) | 0.95 (0.90–0.97) | 4.11 (2.49) | 3.39 (2.0–13.19) | 0.03 (0.03) | 0.01 (0.0–0.12) |
| Anal Canal | 0.70 (0.08) | 0.70 (0.54–0.90) | 5.43 (2.26) | 4.47 (3.0–11.40) | 0.33 (0.27) | 0.30 (0.03–1.29) |
| Left Femur | 0.78 (0.16) | 0.83 (0.47–0.93) | 18.06 (16.4) | 12.64 (3.16–54.0) | 0.50 (0.69) | 0.18 (0.02–1.98) |
| Right Femur | 0.78 (0.16) | 0.84 (0.45–0.94) | 17.98 (16.8) | 13.01 (2.24–54.3) | 0.49 (0.65) | 0.20 (0.02–1.78) |
| DSC | HD (mm) | RVD | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | |
| Contralateral Brachial Plexus | 0.93 (0.02) | 0.93 (0.90–0.97) | 13.87 (3.80) | 14.59 (6.78–21.8) | 0.03 (0.02) | 0.03 (0.00–0.08) |
| Ipsilateral Brachial Plexus | 0.92 (0.08) | 0.93 (0.59–0.97) | 13.52 (4.12) | 13.28 (7.07–19.1) | 0.09 (0.18) | 0.05 (0.01–0.82) |
| Brain | 1.00 (0.00) | 1.00 (1.00–1.00) | 00.65 (0.49) | 01.00 (0.00–1.00) | 0.00 (0.00) | 0.00 (0.00–0.00) |
| Brainstem | 0.88 (0.27) | 1.00 (0.10–1.00) | 04.35 (8.54) | 1.00 (0.00–32.16) | 0.15 (0.31) | 0.00 (0.00–0.95) |
| Chiasm | 0.75 (0.15) | 0.78 (0.45–0.94) | 04.82 (1.40) | 05.10 (1.41–7.00) | 0.26 (0.12) | 0.26 (0.08–0.44) |
| Contralateral Cochlea | 0.62 (0.19) | 0.60 (0.35–1.00) | 02.21 (1.06) | 02.24 (0.00–4.12) | 0.51 (0.23) | 0.57 (0.00–0.79) |
| Ipsilateral Cochlea | 0.63 (0.20) | 0.63 (0.32–1.00) | 02.03 (0.96) | 01.87 (0.00–4.58) | 0.49 (0.24) | 0.52 (0.00–0.81) |
| Esophagus | 0.99 (0.01) | 0.99 (0.97–1.00) | 01.67 (2.12) | 01.00 (0.00–10.0) | 0.01 (0.02) | 0.01 (0.00–0.07) |
| Left Eye | 0.98 (0.05) | 0.99 (0.81–1.00) | 00.85 (0.59) | 01.00 (0.00–2.00) | 0.04 (0.12) | 0.00 (0.00–0.46) |
| Right Eye | 0.98 (0.06) | 0.99 (0.80–1.00) | 00.93 (0.82) | 01.00 (0.00–3.61) | 0.04 (0.13) | 0.00 (0.00–0.50) |
| Larynx | 0.65 (0.06) | 0.63 (0.56–0.78) | 14.11 (2.79) | 13.83 (8.12–20.3) | 0.49 (0.08) | 0.51 (0.28–0.60) |
| Left Lens | 0.68 (0.42) | 0.93 (0.00–1.00) | 01.00 (1.00) | 01.00 (0.00–3.74) | 0.34 (0.43) | 0.06 (0.00–1.00) |
| Right Lens | 0.61 (0.47) | 0.95 (0.00–1.00) | 5.96 (17.54) | 01.00 (0.00–68.6) | 0.39 (0.47) | 0.05 (0.00–1.00) |
| Mandible | 0.98 (0.06) | 0.99 (0.74–1.00) | 8.13 (19.10) | 3.32 (0.00–88.21) | 0.03 (0.09) | 0.01 (0.00–0.41) |
| Left Optic Nerve | 0.97 (0.04) | 0.98 (0.84–1.00) | 00.99 (1.06) | 01.00 (0.00–5.00) | 0.03 (0.06) | 0.01 (0.00–0.22) |
| Right Optic Nerve | 0.98 (0.03) | 0.98 (0.89–1.00) | 00.72 (0.49) | 01.00 (0.00–1.41) | 0.02 (0.04) | 0.01 (0.00–0.20) |
| Oral Cavity | 0.97 (0.04) | 0.98 (0.81–1.00) | 03.82 (3.07) | 3.50 (0.00–12.04) | 0.06 (0.07) | 0.04 (0.00–0.31) |
| Contralateral Parotid | 0.99 (0.00) | 0.99 (0.98–1.00) | 01.50 (2.02) | 01.00 (0.00–7.14) | 0.00 (0.01) | 0.00 (0.00–0.02) |
| Ipsilateral Parotid | 0.99 (0.00) | 0.99 (0.98–1.00) | 01.13 (1.43) | 01.00 (0.00–6.08) | 0.00 (0.00) | 0.00 (0.00–0.02) |
| Pharynx Constrictor Muscle | 0.98 (0.02) | 0.99 (0.91–1.00) | 02.57 (3.03) | 1.21 (0.00–11.58) | 0.02 (0.04) | 0.01 (0.00–0.19) |
| Pituitary | 0.92 (0.15) | 0.97 (0.45–1.00) | 01.33 (1.50) | 01.00 (0.00–7.07) | 0.10 (0.17) | 0.01 (0.00–0.60) |
| Spinal Cord | 0.99 (0.01) | 0.99 (0.99–1.00) | 00.65 (0.49) | 01.00 (0.00–1.00) | 0.00 (0.00) | 0.00 (0.00–0.01) |
| Contralateral Submandibular | 0.99 (0.02) | 0.99 (0.93–1.00) | 00.86 (0.84) | 01.00 (0.00–3.74) | 0.01 (0.02) | 0.00 (0.00–0.09) |
| Ipsilateral Submandibular | 0.95 (0.14) | 0.99 (0.39–1.00) | 01.91 (3.42) | 1.00 (0.00–13.42) | 0.05 (0.14) | 0.00 (0.00–0.58) |
| Thyroid | 0.94 (0.22) | 0.99 (0.00–1.00) | 9.58 (31.67) | 01.62 (0.0–143.8) | 0.11 (0.41) | 0.01 (0.00–1.84) |
| Trachea | 0.92 (0.05) | 0.92 (0.82–1.00) | 12.04 (7.25) | 11.05 (0.0–26.29) | 0.14 (0.08) | 0.14 (0.00–0.31) |
| DSC | HD (mm) | RVD | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | |
| Contralateral Brachial Plexus | 0.13 (0.16) | 0.00 (0.00–0.39) | 90.86 (50.80) | 119.9 (16.9–167) | 0.97 (0.46) | 0.87 (0.31–2.08) |
| Ipsilateral Brachial Plexus | 0.11 (0.13) | 0.00 (0.00–0.37) | 94.65 (46.10) | 120.6 (16.4–170) | 1.05 (0.43) | 1.07 (0.04–1.80) |
| Brain | 0.98 (0.00) | 0.99 (0.97–0.99) | 05.20 (01.90) | 04.79 (2.80–9.22) | 0.01 (0.01) | 00.01 (0.0–0.03) |
| Brainstem | 0.83 (0.08) | 0.85 (0.60–0.89) | 06.99 (02.30) | 6.44 (3.50–11.31) | 0.17 (0.15) | 0.12 (0.01–0.57) |
| Chiasm | 0.57 (0.19) | 0.63 (0.04–0.78) | 04.50 (01.70) | 04.12 (2.20–8.31) | 0.20 (0.15) | 00.20 (0.0–0.53) |
| Contralateral Cochlea | 0.25 (0.31) | 0.00 (0.00–0.85) | 39.55 (34.70) | 64.07 (1.4–93.09) | 0.42 (0.27) | 0.45 (0.02–0.87) |
| Ipsilateral Cochlea | 0.24 (0.29) | 0.00 (0.00–0.84) | 39.66 (34.90) | 64.55 (1.00–93.3) | 0.48 (0.26) | 0.51 (0.07–0.89) |
| Esophagus | 0.67 (0.24) | 0.79 (0.02–0.87) | 29.76 (35.40) | 8.33 (3.60–114.2) | 5.62 (21.33) | 0.22 (0.02–95.9) |
| Left Eye | 0.85 (0.06) | 0.85 (0.64–0.91) | 02.97 (00.60) | 03.00 (2.00–4.58) | 0.24 (0.09) | 0.25 (0.08–0.53) |
| Right Eye | 0.84 (0.07) | 0.85 (0.58–0.90) | 03.17 (00.60) | 03.08 (2.40–4.24) | 0.26 (0.10) | 0.25 (0.03–0.59) |
| Larynx | 0.75 (0.26) | 0.85 (0.00–0.90) | 14.50 (23.50) | 06.20 (4.0–86.98) | 11.93 (36.3) | 0.11 (0.03–121) |
| Lens_L | 0.61 (0.19) | 0.69 (0.17–0.84) | 02.11 (00.60) | 02.00 (1.40–3.16) | 0.33 (0.32) | 0.22 (0.03–1.41) |
| Lens_R | 0.63 (0.17) | 0.65 (0.08–0.86) | 04.87 (14.00) | 01.73 (1.0–64.33) | 0.27 (0.23) | 0.18 (0.02–0.96) |
| Mandible | 0.88 (0.03) | 0.88 (0.79–0.91) | 11.29 (12.10) | 04.12 (2.8–43.12) | 0.17 (0.06) | 0.17 (0.05–0.31) |
| Left Optic Nerve | 0.69 (0.07) | 0.69 (0.51–0.81) | 03.68 (0270) | 02.83 (2.0–13.78) | 0.27 (0.15) | 0.27 (0.02–0.54) |
| Right Optic Nerve | 0.69 (0.06) | 0.70 (0.59–0.81) | 03.73 (01.90) | 03.08 (1.70–8.31) | 0.33 (0.12) | 0.36 (0.05–0.51) |
| Oral Cavity | 0.77 (0.27) | 0.87 (0.00–0.92) | 15.56 (18.70) | 9.09 (5.0–71.510) | 4.48 (15.36) | 0.12 (0.02–66.7) |
| Contralateral Parotid | 0.38 (0.43) | 0.00 (0.00–0.89) | 65.68 (52.30) | 94.79 (4.9–147.8) | 0.15 (0.08) | 0.15 (0.03–0.31) |
| Ipsilateral Parotid | 0.38 (0.44) | 0.00 (0.00–0.89) | 64.74 (54.10) | 96.12 (5.4–148.1) | 0.13 (0.08) | 0.14 (0.01–0.35) |
| Pharynx Constrictor Muscle | 0.67 (0.08) | 0.71 (0.53–0.75) | 09.34 (03.80) | 9.06 (4.60–20.45) | 0.19 (0.24) | 00.14 (0.01–1.1) |
| Pituitary | 0.66 (0.08) | 0.67 (0.45–0.79) | 03.08 (00.90) | 03.00 (1.40–5.00) | 0.29 (0.17) | 0.33 (0.04–0.52) |
| Spinal Cord | 0.72 (0.10) | 0.71 (0.55–0.85) | 26.80 (26.90) | 11.63 (3.2–82.49) | 0.27 (0.22) | 0.23 (0.01–0.91) |
| Contralateral Submandibular | 0.38 (0.43) | 0.00 (0.00–0.89) | 37.42 (31.20) | 56.35 (2.5–84.73) | 0.14 (0.10) | 0.11 (0.02–0.47) |
| Ipsilateral Submandibular | 0.36 (0.41) | 0.00 (0.00–0.87) | 38.61 (31.00) | 58.19 (3.0–88.42) | 0.19 (0.10) | 0.20 (0.02–0.39) |
| Thyroid | 0.74 (0.18) | 0.77 (0.00–0.86) | 07.68 (04.47) | 06.63 (3.6–24.19) | 0.14 (0.11) | 0.11 (0.01–0.35) |
| Trachea | 0.80 (0.19) | 0.84 (0.00–0.90) | 15.95 (18.90) | 09.43 (4.0–90.63) | 7.20 (31.34) | 0.22 (0.0–140.3) |
| ∆Dmean | ∆D0.03cc | |||
|---|---|---|---|---|
| Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | |
| Rectum | 0.13 (0.17) | 0.07 (0.0–0.73) | 0.05 (0.05) | 0.03 (0.0–0.13) |
| Bladder | 0.03 (0.02) | 0.02 (0.0–0.07) | 0.00 (0.00) | 0.00 (0.0–0.01) |
| Anal canal | 0.49 (0.28) | 0.48 (0.03–1.22) | 1.11 (1.33) | 0.69 (0.0–4.67) |
| Femur Left | 0.21 (0.16) | 0.19 (0.0–0.53) | 0.02 (0.02) | 0.01 (0.0–0.09) |
| Femur Right | 0.21 (0.16) | 0.21 (0.0–0.54) | 0.01 (0.02) | 0.01 (0.0–0.08) |
| ∆Dmean | ∆D0.03cc | |||
|---|---|---|---|---|
| Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | |
| Contralateral Brachial Plexus | 0.25 (0.13) | 0.24 (0.02–0.49) | 0.12 (0.19) | 0.08 (0.01–0.83) |
| Ipsilateral Brachial Plexus | 0.35 (0.15) | 0.35 (0.05–0.64) | 0.07 (0.09) | 0.02 (0.00–0.34) |
| Brain | 0.02 (0.03) | 0.01 (0.00–0.13) | 0.06 (0.11) | 0.02 (0.00–0.43) |
| Brainstem | 0.14 (0.11) | 0.13 (0.02–0.43) | 0.14 (0.23) | 0.04 (0.00–0.70) |
| Chiasm | 0.05 (0.05) | 0.03 (0.00–0.13) | 0.05 (0.05) | 0.04 (0.00–0.15) |
| Contralateral Cochlea | 0.08 (0.09) | 0.04 (0.00–0.33) | 0.11 (0.11) | 0.08 (0.00–0.35) |
| Ipsilateral Cochlea | 0.11 (0.20) | 0.04 (0.00–0.83) | 0.16 (0.19) | 0.10 (0.02–0.88) |
| Esophagus | 0.18 (0.21) | 0.08 (0.00–0.64) | 0.09 (0.16) | 0.03 (0.00–0.51) |
| Left Eye | 0.04 (0.04) | 0.03 (0.00–0.16) | 0.13 (0.11) | 0.09 (0.02–0.45) |
| Right Eye | 0.04 (0.03) | 0.03 (0.01–0.12) | 0.13 (0.11) | 0.10 (0.03–0.43) |
| Larynx | 0.09 (0.23) | 0.03 (0.00–0.93) | 0.07 (0.23) | 0.00 (0.00–0.88) |
| Left Lens | 0.04 (0.03) | 0.04 (0.00–0.11) | 0.05 (0.04) | 0.04 (0.00–0.15) |
| Right Lens | 0.04 (0.03) | 0.04 (0.00–0.14) | 0.05 (0.04) | 0.05 (0.00–0.14) |
| Mandible | 0.02 (0.02) | 0.01 (0.00–0.06) | 0.01 (0.02) | 0.00 (0.00–0.17) |
| Left Optic Nerve | 0.05 (0.03) | 0.04 (0.01–0.15) | 0.07 (0.08) | 0.06 (0.01–0.27) |
| Right Optic Nerve | 0.05 (0.08) | 0.03 (0.00–0.32) | 0.07 (0.10) | 0.04 (0.00–0.42) |
| Oral Cavity | 0.11 (0.15) | 0.04 (0.00–0.59) | 0.05 (0.09) | 0.02 (0.00–0.34) |
| Contralateral Parotid | 0.10 (0.09) | 0.10 (0.01–0.32) | 0.11 (0.19) | 0.05 (0.00–0.81) |
| Ipsilateral Parotid | 0.15 (0.17) | 0.09 (0.01–0.73) | 0.10 (0.20) | 0.02 (0.00–0.82) |
| Pharynx Constrictor Muscle | 0.06 (0.10) | 0.03 (0.00–0.41) | 0.00 (0.01) | 0.00 (0.00–0.05) |
| Pituitary | 0.04 (0.07) | 0.02 (0.00–0.28) | 0.02 (0.02) | 0.02 (0.00–0.07) |
| Spinal Cord | 0.17 (0.17) | 0.10 (0.00–0.50) | 0.04 (0.06) | 0.02 (0.00–0.26) |
| Contralateral Submandibular Gland | 0.13 (0.24) | 0.03 (0.00–0.83) | 0.05 (0.07) | 0.02 (0.00–0.26) |
| Ipsilateral Submandibular Gland | 0.14 (0.20) | 0.01 (0.00–0.64) | 0.04 (0.06) | 0.01 (0.00–0.22) |
| Thyroid | 0.05 (0.06) | 0.02 (0.00–0.20) | 0.03 (0.05) | 0.01 (0.00–0.18) |
| Trachea | 0.17 (0.15) | 0.13 (0.00–0.42) | 0.05 (0.09) | 0.02 (0.00–0.35) |
| Study Site | OAR | ∆Dmean | ∆D0.03cc |
|---|---|---|---|
| Prostate | Rectum | 0.64 | 0.35 |
| Bladder | 0.90 | 0.37 | |
| Anal Canal | 0.11 | 0.04 | |
| Left Femur | 0.47 | 0.93 | |
| Right Femur_R | 0.23 | 0.82 | |
| Head and Neck | Contralateral Brachial Plexus | 0.02 | 0.22 |
| Ipsilateral Brachial Plexus | 0.00 | 0.41 | |
| Brain | 0.95 | 0.79 | |
| Brainstem | 0.92 | 0.84 | |
| Chiasm | 0.90 | 0.74 | |
| Contralateral Cochlea | 0.82 | 0.58 | |
| Ipsilateral Cochlea | 0.74 | 0.97 | |
| Esophagus | 0.34 | 0.66 | |
| Left Eye | 0.71 | 0.90 | |
| Right Eye | 0.86 | 1.00 | |
| Larynx | 0.51 | 0.47 | |
| Left Lens | 0.90 | 0.51 | |
| Right Lens | 0.95 | 0.52 | |
| Mandible | 0.92 | 0.94 | |
| Left Optic Nerve | 0.84 | 0.88 | |
| Right Optic Nerve | 0.80 | 0.79 | |
| Oralcavity | 0.52 | 0.39 | |
| Parotid_C | 0.86 | 0.42 | |
| Parotid_H | 0.30 | 0.44 | |
| PharynxConst | 0.99 | 0.92 | |
| Pituitary | 0.84 | 0.82 | |
| SpinalCord | 0.34 | 0.78 | |
| Contralateral Submandibular gland | 0.12 | 0.22 | |
| Ipsilateral Submandibular gland | 0.17 | 0.37 | |
| Thyroid | 0.95 | 0.66 | |
| Trachea | 0.92 | 1.00 |
| Study Site | Mean (SD) | Median (Range) |
|---|---|---|
| Prostate | 0.080 (0.097) | 0.032 (0.00–0.276) |
| Head and Neck | 0.067 (0.057) | 0.054 (0.00–0.173) |
| Study Site | Tman | TAI,adj | Time Savings | Saved Time (%) |
|---|---|---|---|---|
| Prostate | 23 min | 6 min 25 s | 16 min 35 s | 72% |
| Head and Neck | 2 h 30 min | 23 min 35 s | 2 h 6 min 25 s | 84% |
| DSC | HD (mm) | RVD | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | |
| PTV | 0.91 (0.06) | 0.94 (0.80–0.98) | 5.87 (4.70) | 3.74 (1.41–18.14) | 0.09 (0.09) | 0.04 (0.0–0.26) |
| Rectum | 0.97 (0.03) | 0.98 (0.90–1.00) | 4.76 (5.79) | 3.08 (0.0–23.73) | 0.06 (0.06) | 0.04 (0.0–0.21) |
| Bladder | 0.98 (0.01) | 0.98 (0.95–1.00) | 3.10 (3.17) | 2.00 (1.0–14.28) | 0.02 (0.02) | 0.02 (0.0–0.08) |
| Anal Canal | 0.91 (0.05) | 0.92 (0.83–0.99) | 3.11 (1.53) | 2.34 (1.0–6.08) | 0.12 (0.09) | 0.10 (0.01–0.29) |
| Femur Left | 0.98 (0.02) | 0.99 (0.95–1.00) | 2.02 (1.71) | 02.12 (0.0–5.0) | 0.03 (0.04) | 0.02 (0.0–0.11) |
| Femur Right | 0.98 (0.03) | 0.99 (0.87–1.00) | 2.53 (3.29) | 1.87 (0.0–14.59) | 0.03 (0.04) | 0.02 (0.0–0.12) |
| ∆Dmean | ∆D0.03cc | |||
|---|---|---|---|---|
| Mean (SD) | Median (Range) | Mean (SD) | Median (Range) | |
| Rectum | 0.04 (0.05) | 0.02 (0.0–0.18) | 0.00 (0.00) | 0.00 (0.0–0.00) |
| Bladder | 0.02 (0.02) | 0.02 (0.0–0.08) | 0.00 (0.00) | 0.00 (0.0–0.01) |
| Anal Canal | 0.28 (0.31) | 0.23 (0.0–1.05) | 0.65 (0.91) | 0.12 (0.0–3.21) |
| Femur Left | 0.04 (0.04) | 0.01 (0.0–0.11) | 0.00 (0.00) | 0.00 (0.0–0.00) |
| Femur Right | 0.04 (0.04) | 0.01 (0.0–0.12) | 0.00 (0.00) | 0.00 (0.0–0.01) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoque, S.M.H.; Pirrone, G.; Matrone, F.; Donofrio, A.; Fanetti, G.; Caroli, A.; Rista, R.S.; Bortolus, R.; Avanzo, M.; Drigo, A.; et al. Clinical Use of a Commercial Artificial Intelligence-Based Software for Autocontouring in Radiation Therapy: Geometric Performance and Dosimetric Impact. Cancers 2023, 15, 5735. https://doi.org/10.3390/cancers15245735
Hoque SMH, Pirrone G, Matrone F, Donofrio A, Fanetti G, Caroli A, Rista RS, Bortolus R, Avanzo M, Drigo A, et al. Clinical Use of a Commercial Artificial Intelligence-Based Software for Autocontouring in Radiation Therapy: Geometric Performance and Dosimetric Impact. Cancers. 2023; 15(24):5735. https://doi.org/10.3390/cancers15245735
Chicago/Turabian StyleHoque, S M Hasibul, Giovanni Pirrone, Fabio Matrone, Alessandra Donofrio, Giuseppe Fanetti, Angela Caroli, Rahnuma Shahrin Rista, Roberto Bortolus, Michele Avanzo, Annalisa Drigo, and et al. 2023. "Clinical Use of a Commercial Artificial Intelligence-Based Software for Autocontouring in Radiation Therapy: Geometric Performance and Dosimetric Impact" Cancers 15, no. 24: 5735. https://doi.org/10.3390/cancers15245735
APA StyleHoque, S. M. H., Pirrone, G., Matrone, F., Donofrio, A., Fanetti, G., Caroli, A., Rista, R. S., Bortolus, R., Avanzo, M., Drigo, A., & Chiovati, P. (2023). Clinical Use of a Commercial Artificial Intelligence-Based Software for Autocontouring in Radiation Therapy: Geometric Performance and Dosimetric Impact. Cancers, 15(24), 5735. https://doi.org/10.3390/cancers15245735






