The Genomic Landscape of Urothelial Carcinoma with High and Low ERBB2 Expression
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Cohort
2.2. Defining Primary, Metastatic, Lower, and Upper Urothelial Tract UC
2.3. DNA Next-Generation Sequencing (NGS)
2.4. Identification of Genetic Variants and Copy Number Amplification (CNA)
2.5. Tumor Mutational Burden (TMB)
2.6. MSI/MMR Status
2.7. Whole Transcriptome Sequencing
2.8. RNA Signatures
2.9. Immunohistochemistry (IHC) for PD-L1 and HER2
2.10. Clinical Outcomes
2.11. Statistics
3. Results
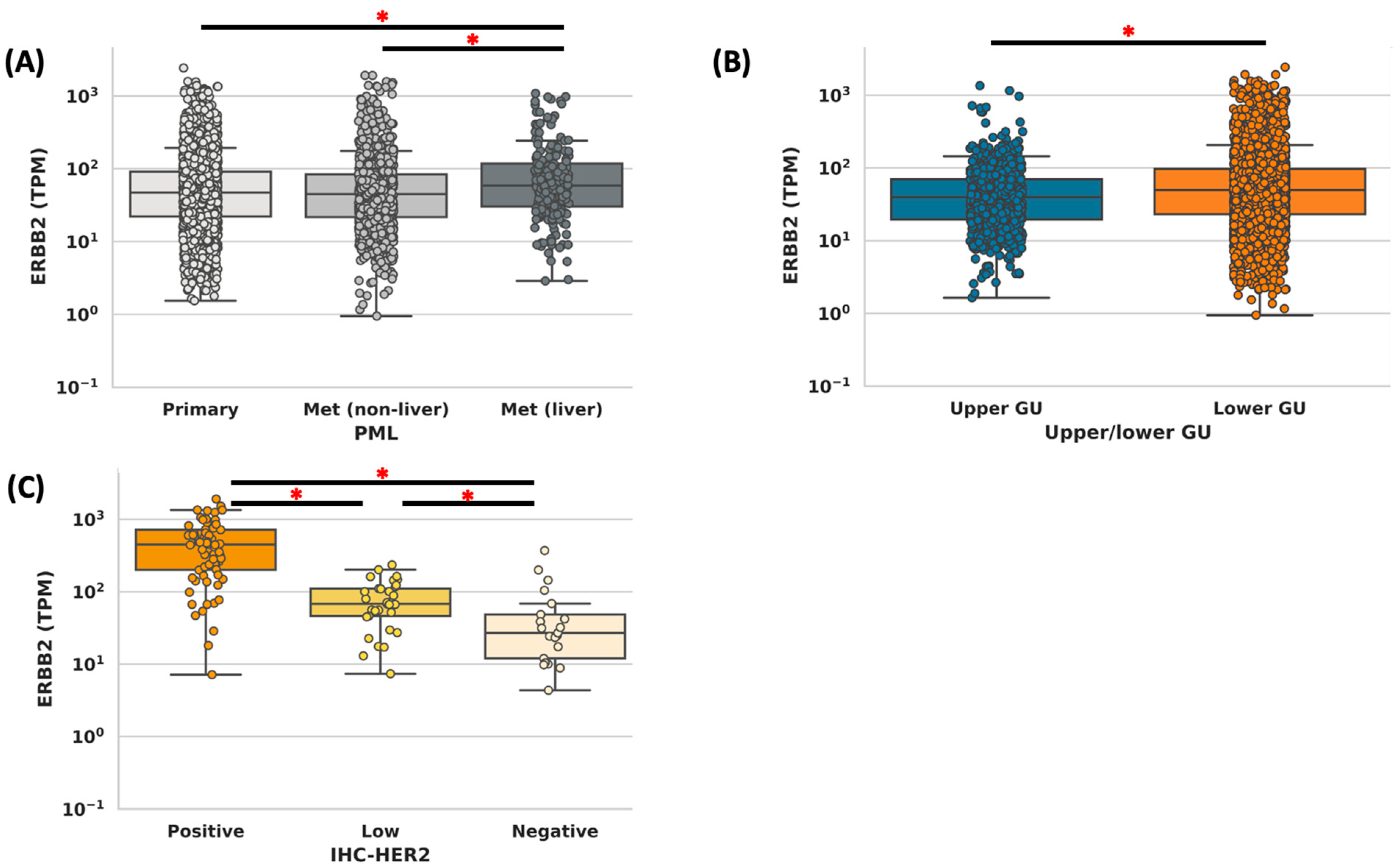
3.1. Cohort Description
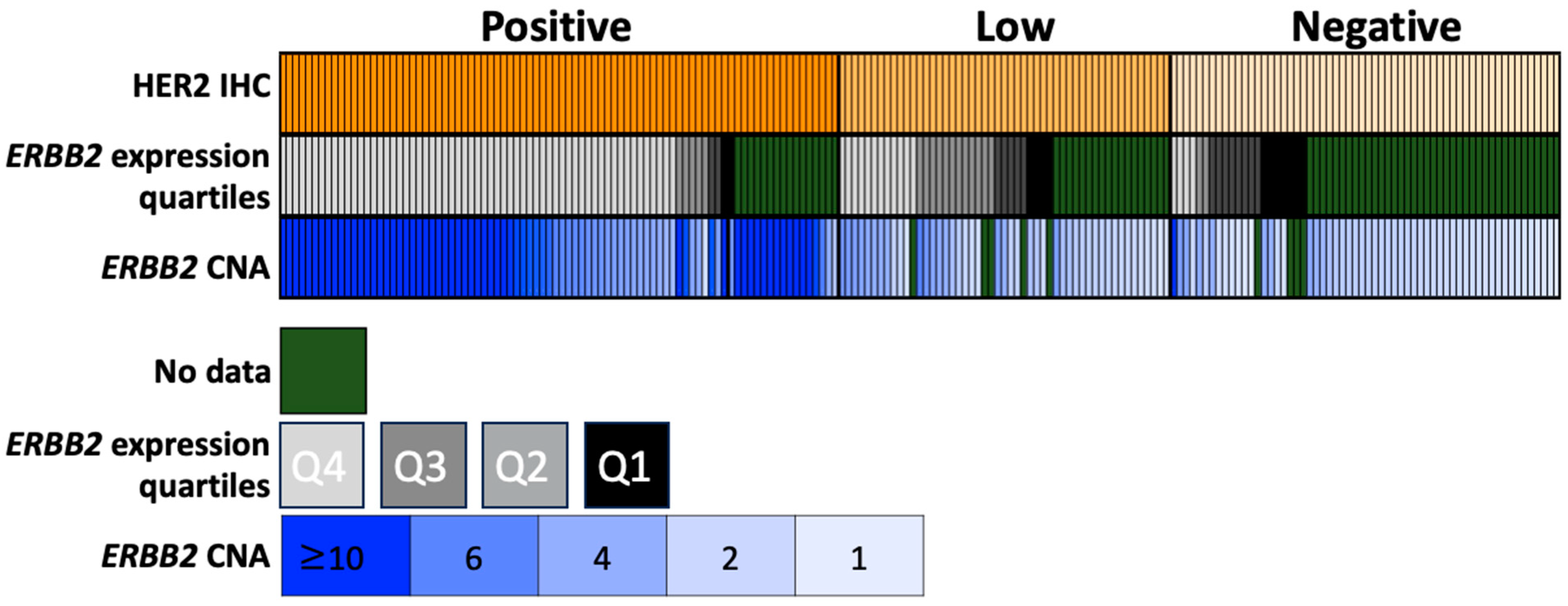
3.2. Concordance Analysis
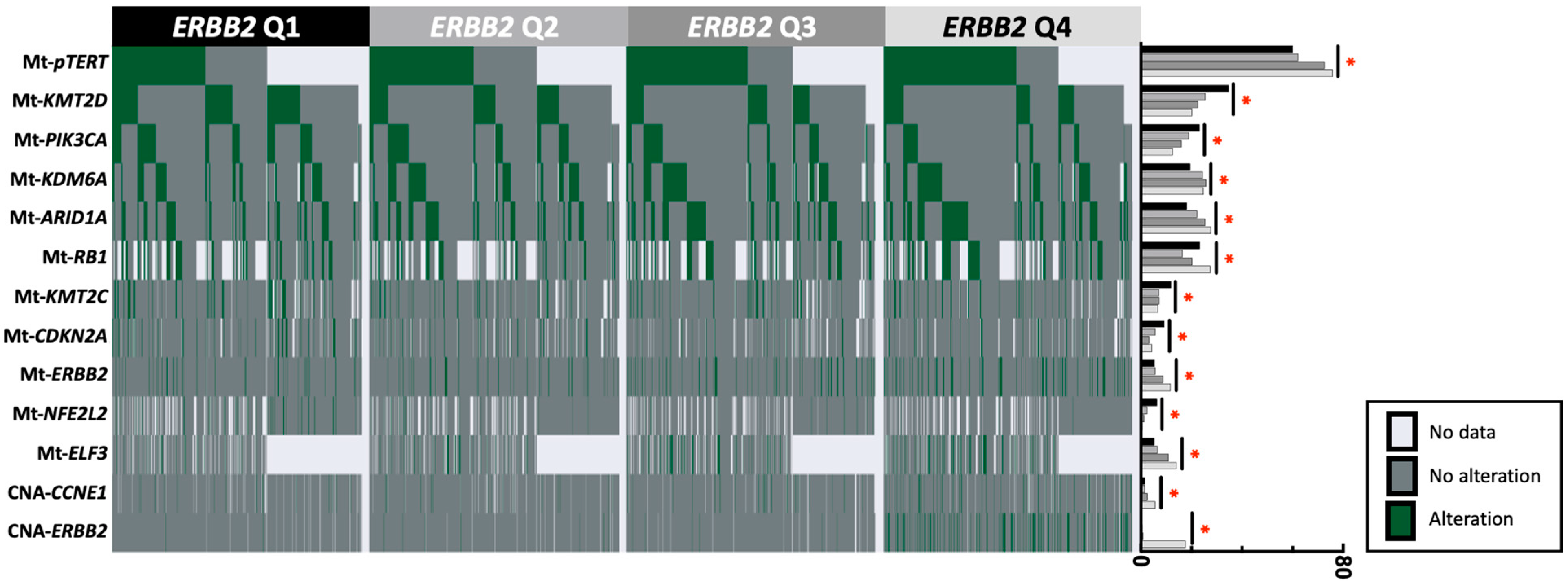
3.3. Gene Alterations in ERBB2-High vs. ERBB2-Low Tumors
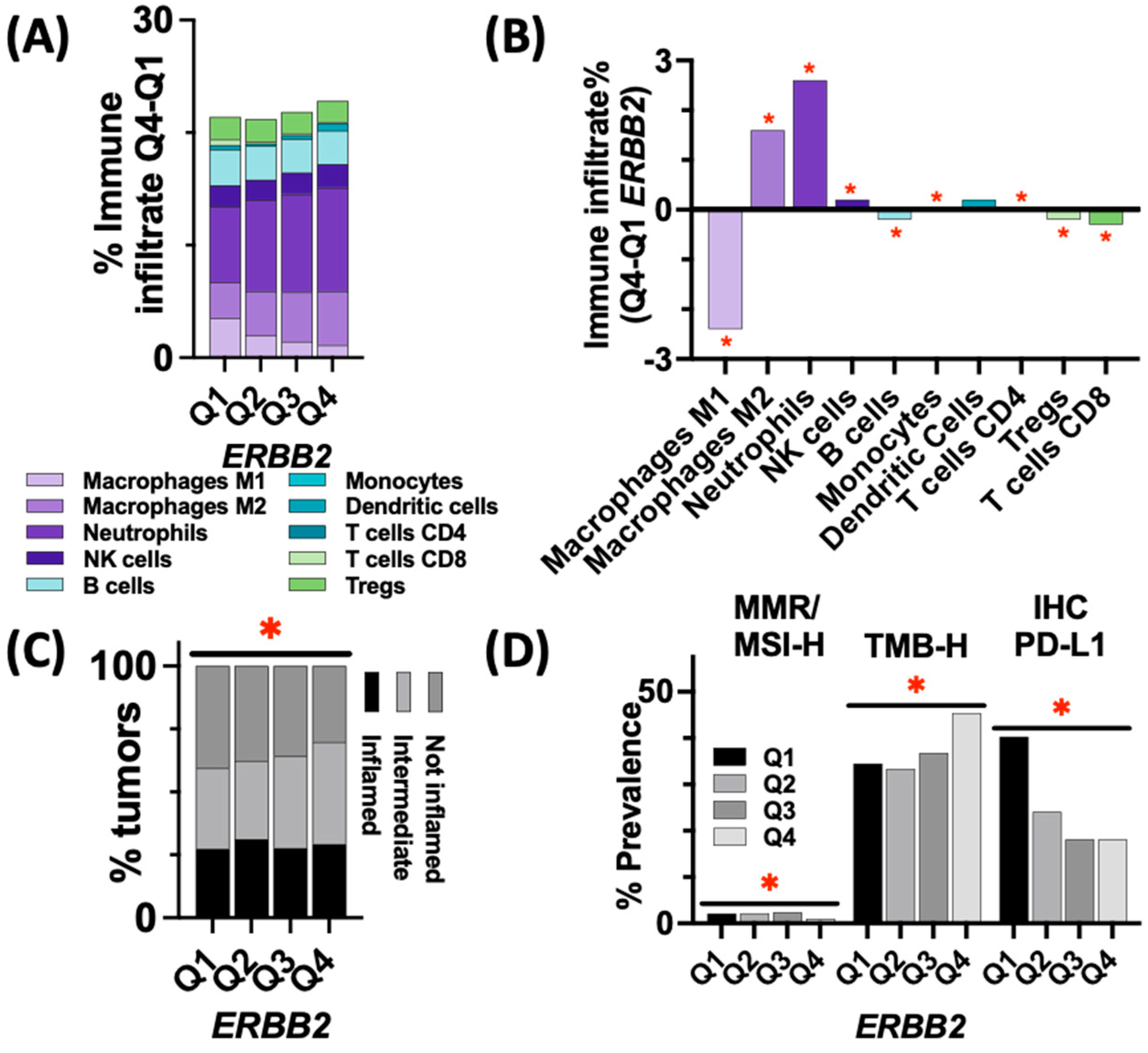
3.4. Immune Landscape and Markers of ICI Response in ERBB2-High vs. ERBB2-Low Tumors
3.5. Expression of ADC Targets in ERBB2-High vs. ERBB2-Low Tumors
3.6. Clinical Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Cancer Society. Key Statistics for Bladder Cancer; American Cancer Society: New York, NY, USA, 2023; Available online: https://www.cancer.org/cancer/bladder-cancer/about/key-statistics.html (accessed on 13 March 2023).
- Chen, D.; Ye, Y.; Guo, S.; Yao, K. Progress in the Research and Targeted Therapy of ErbB/HER Receptors in Urothelial Bladder Cancer. Front. Mol. Biosci. 2021, 8, 800945. [Google Scholar] [CrossRef] [PubMed]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Alimohamed, N.; Grewal, S.; Wirtz, H.S.; Hepp, Z.; Sauvageau, S.; Boyne, D.J.; Brenner, D.R.; Cheung, W.Y.; Jarada, T.N. Understanding Treatment Patterns and Outcomes among Patients with De Novo Unresectable Locally Advanced or Metastatic Urothelial Cancer: A Population-Level Retrospective Analysis from Alberta, Canada. Curr. Oncol. 2022, 29, 7587–7597. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, S.; Thompson, M.; Sopwith, W.; Godden, P.; Seshagiri, D.; Adedokun, L.; Zucker, K.; Jain, S.; Kotwal, S.; Prescott, S.; et al. Current Treatment and Outcomes Benchmark for Locally Advanced or Metastatic Urothelial Cancer from a Large UK-Based Single Centre. Front. Oncol. 2020, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Lattanzi, M.; Niederhausern, A.; Zheng, J.; Bahadur, N.; Nichols, C.; Barton, L.; Gandhi, F.; Chan, K.; Insinga, A.; Philip, A.; et al. Incidence and clinical outcomes of HER2-altered bladder cancer (BC) patients (pts). J. Clin. Oncol. 2022, 40 (Suppl. S6), 556. [Google Scholar] [CrossRef]
- Scherrer, E.; Kang, A.; Bloudek, L.M.; Koshkin, V.S. HER2 expression in urothelial carcinoma, a systematic literature review. Front. Oncol. 2022, 12, 1011885. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J.M.; Kim, J.-S.; Kim, S.; Kim, K.-H. Differential Expression and Clinicopathological Significance of HER2, Indoleamine 2,3-Dioxygenase and PD-L1 in Urothelial Carcinoma of the Bladder. J. Clin. Med. 2020, 9, 1265. [Google Scholar] [CrossRef] [PubMed]
- Gandour-Edwards, R.; Lara, P.N., Jr.; Folkins, A.K.; LaSalle, J.M.; Beckett, L.; Li, Y.; Meyers, F.J.; DeVere-White, R. Does HER2/neu expression provide prognostic information in patients with advanced urothelial carcinoma? Cancer 2002, 95, 1009–1015. [Google Scholar] [CrossRef]
- Gan, K.; Gao, Y.; Liu, K.; Xu, B.; Qin, W. The Clinical Significance and Prognostic Value of HER2 Expression in Bladder Cancer: A Meta-Analysis and a Bioinformatic Analysis. Front. Oncol. 2021, 11, 653491. [Google Scholar] [CrossRef]
- Albarrán, V.; Rosero, D.I.; Chamorro, J.; Pozas, J.; San Román, M.; Barrill, A.M.; Alía, V.; Sotoca, P.; Guerrero, P.; Calvo, J.C.; et al. Her-2 Targeted Therapy in Advanced Urothelial Cancer: From Monoclonal Antibodies to Antibody-Drug Conjugates. Int. J. Mol. Sci. 2022, 23, 12659. [Google Scholar] [CrossRef]
- Powles, T.; Rosenberg, J.E.; Sonpavde, G.P.; Loriot, Y.; Durán, I.; Lee, J.L.; Matsubara, N.; Vulsteke, C.; Castellano, D.; Wu, C.; et al. Enfortumab Vedotin in Previously Treated Advanced Urothelial Carcinoma. N. Engl. J. Med. 2021, 384, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Cimpean, A.M.; Tarlui, V.; Cumpănaş, A.A.; Bolintineanu, S.; Cumpănaş, A.; Raica, M. Critical Overview of HER2 Assessment in Bladder Cancer: What Is Missing for a Better Therapeutic Approach? Anticancer Res. 2017, 37, 4935–4942. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- Merino, D.M.; McShane, L.M.; Fabrizio, D.; Funari, V.; Chen, S.J.; White, J.R.; Wenz, P.; Baden, J.; Barrett, J.C.; Chaudhary, R.; et al. TMB Harmonization Consortium. Establishing guidelines to harmonize tumor mutational burden (TMB): In silico assessment of variation in TMB quantification across diagnostic platforms: Phase I of the Friends of Cancer Research TMB Harmonization Project. J. Immunother. Cancer 2020, 8, e000147. [Google Scholar] [CrossRef] [PubMed]
- Vanderwalde, A.; Spetzler, D.; Xiao, N.; Gatalica, Z.; Marshall, J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018, 7, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Bao, R.; Stapor, D.; Luke, J.J. Molecular correlates and therapeutic targets in T cell-inflamed versus non-T cell-inflamed tumors across cancer types. Genome Med. 2020, 12, 90. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Höglund, M.; Bernardo, C.; Sjödahl, G.; Eriksson, P.; Axelson, H.; Liedberg, F. The Lund taxonomy for bladder cancer classification—From gene expression clustering to cancer cell molecular phenotypes, and back again. J. Pathol. 2023, 259, 369–375. [Google Scholar] [CrossRef]
- Marzouka, N.; Eriksson, P.; Rovira, C.; Liedberg, F.; Sjodahl, G.; Hoglund, M. A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort. Sci. Rep. 2018, 8, 3737. [Google Scholar] [CrossRef]
- Chu, C.E.; Sjöström, M.; Egusa, E.A.; Gibb, E.A.; Badura, M.L.; Zhu, J.; Koshkin, V.S.; Stohr, B.A.; Meng, M.V.; Pruthi, R.S.; et al. Heterogeneity in NECTIN4 Expression Across Molecular Subtypes of Urothelial Cancer Mediates Sensitivity to Enfortumab Vedotin. Clin. Cancer Res. 2021, 27, 5123–5130. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Rong, G.; Guan, Y.; Li, J.; Chang, L.; Li, H.; Liu, B.; Wang, W.; Guan, X.; Ouyang, Q.; et al. Molecular landscape and efficacy of HER2-targeted therapy in patients with HER2-mutated metastatic breast cancer. Npj Breast Cancer 2020, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Chumsri, S.; Weidler, J.; Ali, S.; Balasubramanian, S.; Wallweber, G.; DeFazio-Eli, L.; Chenna, A.; Huang, W.; DeRidder, A.; Goicocheal, L.; et al. Prolonged Response to Trastuzumab in a Patient with HER2-Nonamplified Breast Cancer With Elevated HER2 Dimerization Harboring an ERBB2 S310F Mutation. J. Natl. Compr. Cancer Netw. JNCCN 2015, 13, 1066–1070. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Tan, J.; Shen, X.; Jiang, K.; Wang, C.; Zhu, G.; Xing, M. Therapeutic targeting of FOS in mutant TERT cancers through removing TERT suppression of apoptosis via regulating survivin and TRAIL-R2. Proc. Natl. Acad. Sci. USA 2021, 118, e2022779118. [Google Scholar] [CrossRef]
- Kamat, A.M.; Shore, N.; Hahn, N.; Alanee, S.; Nishiyama, H.; Shariat, S.; Nam, K.; Kapadia, E.; Frenkl, T.; Steinberg, G. KEYNOTE-676: Phase III study of BCG and pembrolizumab for persistent/recurrent high-risk NMIBC. Future Oncol. 2020, 16, 507–516. [Google Scholar] [CrossRef]
- De Santis, M.; Abdrashitov, R.; Hegele, A.; Kolb, M.; Parker, S.; Palou Redorta, J.; Nishiyama, H.; Xiao, F.; Gupta, A.K.; Shore, N.D. A phase III, randomized, open-label, multicenter, global study of durvalumab and bacillus calmette-guérin (BCG) versus BCG alone in high-risk, BCG-naïve non-muscle-invasive bladder cancer (NMIBC) patients (POTOMAC). J. Clin. Oncol. 2019, 37 (Suppl. S7), TPS500. [Google Scholar] [CrossRef]
- Li, S.; Wu, X.; Yan, X.; Zhou, L.; Xu, H.; Li, J.; Liu, Y.; Tang, B.; Chi, Z.; Si, L.; et al. Prognostic value of HER2 expression levels for upper tract urothelial carcinoma. J. Clin. Oncol. 2022, 40 (Suppl. S6), 557. [Google Scholar] [CrossRef]

| WTS | |||||
|---|---|---|---|---|---|
| Q1 | Q2 | Q3 | Q4 | ||
| IHC | Positive | 2 | 2 | 5 | 61 |
| 15% (2/13) | 13% (2/15) | 26% (5/19) | 79% (61/77) | ||
| Low | 4 | 5 | 12 | 12 | |
| 31% (4/13) | 33% (5/15) | 63% (12/19) | 16% (12/77) | ||
| Negative | 7 | 8 | 2 | 4 | |
| 54% (7/13) | 54% (8/15) | 11% (2/18) | 5% (4/77) | ||
| CNA | ||||
|---|---|---|---|---|
| Not Amplified | Intermediate | Amplified | ||
| (<4) | (>4) | (>6) | ||
| IHC | Positive | 4 | 21 | 61 |
| 5% (4/83) | 42% (21/42) | 97% (61/63) | ||
| Low | 30 | 15 | 1 | |
| 36% (30/83) | 42% (15/42) | 1.5% (1/63) | ||
| Negative | 49 | 6 | 1 | |
| 59% (49/83) | 16% (6/42) | 1.5% (1/63) | ||
| ERBB2 Q1 | ERBB2 Q2 | ERBB2 Q3 | ERBB2 Q4 | Statistic | q-Value | |
|---|---|---|---|---|---|---|
| Count (N) | 1032 | 1031 | 1031 | 1031 | ||
| Median Age [range] (N) | 72 [26–>89] (1032) | 72 [18–>89] (1031) | 72 [28–>89] (1031) | 72 [24–>89] (1031) | Kruskal-Wallis | 0.32 |
| Female | 34.50% | 29.00% | 25.60% | 22.70% | chi-square | <0.001 |
| (356/1032) | (299/1031) | (264/1031) | (234/1031) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadadi, A.; Krause, H.B.; Elliott, A.; Brown, J.T.; Nazha, B.; Harik, L.R.; Carthon, B.C.; Miron, B.; Nabhan, C.; Barata, P.C.; et al. The Genomic Landscape of Urothelial Carcinoma with High and Low ERBB2 Expression. Cancers 2023, 15, 5721. https://doi.org/10.3390/cancers15245721
Hadadi A, Krause HB, Elliott A, Brown JT, Nazha B, Harik LR, Carthon BC, Miron B, Nabhan C, Barata PC, et al. The Genomic Landscape of Urothelial Carcinoma with High and Low ERBB2 Expression. Cancers. 2023; 15(24):5721. https://doi.org/10.3390/cancers15245721
Chicago/Turabian StyleHadadi, Agreen, Harris B. Krause, Andrew Elliott, Jacqueline T. Brown, Bassel Nazha, Lara R. Harik, Bradley C. Carthon, Benjamin Miron, Chadi Nabhan, Pedro C. Barata, and et al. 2023. "The Genomic Landscape of Urothelial Carcinoma with High and Low ERBB2 Expression" Cancers 15, no. 24: 5721. https://doi.org/10.3390/cancers15245721
APA StyleHadadi, A., Krause, H. B., Elliott, A., Brown, J. T., Nazha, B., Harik, L. R., Carthon, B. C., Miron, B., Nabhan, C., Barata, P. C., Saleh, M., Yang, Y., McKay, R. R., & Bilen, M. A. (2023). The Genomic Landscape of Urothelial Carcinoma with High and Low ERBB2 Expression. Cancers, 15(24), 5721. https://doi.org/10.3390/cancers15245721






