Inflammation and Immunity Gene Expression Patterns and Machine Learning Approaches in Association with Response to Immune-Checkpoint Inhibitors-Based Treatments in Clear-Cell Renal Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Differential Gene Expression Analysis (DGEA)
2.3. Machine Learning (ML)
3. Results
3.1. Prediction of Response to Therapy
3.2. Transcriptomic Changes in CB after Therapy
3.3. Transcriptomic Changes in PD after Therapy
3.4. Transcriptomic Changes Affected by Therapy in PD and CB Patients
3.5. Transcriptomic Changes Affected by Different Therapeutic Combinations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding

Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arora, R.D.; Limaiem, F. Renal Clear Cell Cancer. In StatPearls; StatPearls: Tampa, FL, USA, 2023. [Google Scholar]
- Cheaib, J.G.; Patel, H.D.; Johnson, M.H.; Gorin, M.A.; Haut, E.R.; Canner, J.K.; Allaf, M.E.; Pierorazio, P.M. Stage-Specific Conditional Survival in Renal Cell Carcinoma after Nephrectomy. Urol. Oncol. 2020, 38, 6.e1–6.e7. [Google Scholar] [CrossRef]
- Osawa, T.; Takeuchi, A.; Kojima, T.; Shinohara, N.; Eto, M.; Nishiyama, H. Overview of Current and Future Systemic Therapy for Metastatic Renal Cell Carcinoma. Jpn. J. Clin. Oncol. 2019, 49, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.T.; McGovern, F.J. Renal-cell carcinoma. N. Engl. J. Med. 2005, 353, 2477–2490. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, Z.; Li, Y.; Zhao, W.; Wu, J.; Zhang, Z. PD-1/PD-L1 Checkpoint Inhibitors in Tumor Immunotherapy. Front. Pharmacol. 2021, 12, 731798. [Google Scholar] [CrossRef]
- Tarhini, A.A.; Iqbal, F. CTLA-4 blockade: Therapeutic Potential in Cancer Treatments. OncoTargets Ther. 2010, 3, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Rasti, A.; Abolhasani, M.; Zanjani, L.S.; Asgari, M.; Mehrazma, M.; Madjd, Z. Reduced Expression of CXCR4, a Novel Renal Cancer Stem Cell Marker, Is Associated with High-Grade Renal Cell Carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Jiang, R.; Meng, E.; Wu, H. CXCL5: A Coachman to Drive Cancer Progression. Front. Oncol. 2022, 12, 944494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, M.; Wang, L.; Li, J.; Yang, T.; Shao, Q.; Liang, X.; Ma, M.; Zhang, N.; Jing, M.; et al. Identification of CCL4 as an Immune-Related Prognostic Biomarker Associated with Tumor Proliferation and the Tumor Microenvironment in Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 694664. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Zhang, F.; Feng, D.; Li, D.; Xia, Y.; Gan, M.F. An Inflammation-Related Signature Could Predict the Prognosis of Patients with Kidney Renal Clear Cell Carcinoma. Front. Genet. 2022, 13, 866696. [Google Scholar] [CrossRef]
- Jin, S.; Liu, C.; Shi, G.; Mu, Y.; Zhang, H.; Zhu, Y.; Su, H.; Ye, D. IL-1A Is Associated with Postoperative Survival and Immune Contexture in Clear Cell Renal Cell Carcinoma. Urol. Oncol. 2022, 40, 111.e111–111.e119. [Google Scholar] [CrossRef]
- Seeber, A.; Klinglmair, G.; Fritz, J.; Steinkohl, F.; Zimmer, K.C.; Aigner, F.; Horninger, W.; Gastl, G.; Zelger, B.; Brunner, A.; et al. High IDO-1 Expression in Tumor Endothelial Cells Is Associated with Response to Immunotherapy in Metastatic Renal Cell Carcinoma. Cancer Sci. 2018, 109, 1583–1591. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Y.; Wang, X.; Kang, J.; Liu, X. Prognostic Value of Performance Status in Metastatic Renal Cell Carcinoma Patients Receiving Tyrosine Kinase Inhibitors: A Systematic Review and Meta-Analysis. BMC Cancer 2019, 19, 168. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Huang, W.; Yin, C.; Cheung, Y.H.; Abrams, D.; Fallon, J.T.; Dimitrova, N.; Fanucchi, M.P. Blood-Based Gene Expression Profiling to Reveal Potential Response Biomarkers for Immunotherapy in Advanced Lung Cancer. J. Clin. Oncol. 2020, 38, e15155. [Google Scholar] [CrossRef]
- Friedlander, P.; Wassmann, K.; Christenfeld, A.M.; Fisher, D.; Kyi, C.; Kirkwood, J.M.; Bhardwaj, N.; Oh, W.K. Whole-Blood RNA Transcript-Based Models Can Predict Clinical Response in Two Large Independent Clinical Studies of Patients with Advanced Melanoma Treated with the Checkpoint Inhibitor, Tremelimumab. J. Immunother. Cancer 2017, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, K.M.; Ross-Macdonald, P.; Yuan, L.; Song, L.; Veras, E.; Wind-Rotolo, M.; McDermott, D.F.; Stephen Hodi, F.; Choueiri, T.K.; Freeman, G.J. Soluble PD-L1 as An Early Marker of Progressive Disease on Nivolumab. J. Immunother. Cancer 2022, 10, e003527. [Google Scholar] [CrossRef] [PubMed]
- Nagumo, Y.; Kandori, S.; Kojima, T.; Hamada, K.; Nitta, S.; Chihara, I.; Shiga, M.; Negoro, H.; Mathis, B.J.; Nishiyama, H. Whole-Blood Gene Expression Profiles Correlate with Response to Immune Checkpoint Inhibitors in Patients with Metastatic Renal Cell Carcinoma. Cancers 2022, 14, 6207. [Google Scholar] [CrossRef]
- Ged, Y.; Voss, M.H. Novel Emerging Biomarkers to Immunotherapy in Kidney Cancer. Ther. Adv. Med. Oncol. 2021, 13, 17588359211059367. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Lin, J.; Cai, Y.; Ma, Y.; Pan, J.; Wang, Z.; Zhang, J.; Liu, Y.; Zhao, Z. A New Signature That Predicts Progression-Free Survival of Clear Cell Renal Cell Carcinoma with Anti-PD-1 Therapy. Int. J. Mol. Sci. 2023, 24, 5332. [Google Scholar] [CrossRef]
- Antonello, P.; Pizzagalli, D.U.; Foglierini, M.; Melgrati, S.; Radice, E.; Thelen, S.; Thelen, M. ACKR3 Promotes CXCL12/CXCR4-Mediated Cell-to-Cell-Induced Lymphoma Migration through LTB4 Production. Front. Immunol. 2022, 13, 1067885. [Google Scholar] [CrossRef]
- Tang, C.; Li, L.; Xu, Q.; Xu, S.; Lin, C.; Cao, B. ACKR3 Orchestrates Hedgehog Signaling to Promote Renal Cell Carcinoma Progression. Mol. Carcinog. 2023, 62, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Mollica Poeta, V.; Massara, M.; Capucetti, A.; Bonecchi, R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front. Immunol. 2019, 10, 379. [Google Scholar] [CrossRef] [PubMed]
- Vasavada, S.P.; Novick, A.C.; Williams, B.R. P53, bcl-2, and Bax Expression in Renal Cell Carcinoma. Urology 1998, 51, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Itoi, T.; Yamana, K.; Bilim, V.; Takahashi, K.; Tomita, F. Impact of Frequent Bcl-2 Expression on Better Prognosis in Renal Cell Carcinoma Patients. Br. J. Cancer 2004, 90, 200–205. [Google Scholar] [CrossRef]
- Kausch, I.; Jiang, H.; Thode, B.; Doehn, C.; Kruger, S.; Jocham, D. Inhibition of bcl-2 Enhances the Efficacy of Chemotherapy in Renal Cell Carcinoma. Eur. Urol. 2005, 47, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Aranda, M.; Perez-Ruiz, E.; Redondo, M. Bcl-2 Inhibition to Overcome Resistance to Chemo- and Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3950. [Google Scholar] [CrossRef]
- Zeng, P.; Zhang, X.; Xiang, T.; Ling, Z.; Lin, C.; Diao, H. Secreted Phosphoprotein 1 as a Potential Prognostic and Immunotherapy Biomarker in Multiple Human Cancers. Bioengineered 2022, 13, 3221–3239. [Google Scholar] [CrossRef]
- Cebrian, A.; Gomez Del Pulgar, T.; Mendez-Vidal, M.J.; Gonzalvez, M.L.; Lainez, N.; Castellano, D.; Garcia-Carbonero, I.; Esteban, E.; Saez, M.I.; Villatoro, R.; et al. Functional PTGS2 Polymorphism-Based Models as Novel Predictive Markers in Metastatic Renal Cell Carcinoma Patients Receiving First-Line Sunitinib. Sci. Rep. 2017, 7, 41371. [Google Scholar] [CrossRef]
- Zhu, J.; Powis de Tenbossche, C.G.; Cane, S.; Colau, D.; van Baren, N.; Lurquin, C.; Schmitt-Verhulst, A.M.; Liljestrom, P.; Uyttenhove, C.; Van den Eynde, B.J. Resistance to Cancer Immunotherapy Mediated by Apoptosis of Tumor-Infiltrating Lymphocytes. Nat. Commun. 2017, 8, 1404. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, J.; Li, L.; Zhang, H.; Zhang, H.; Li, J.; Zhong, H.; Lei, T.; Jin, Y.; Xu, B.; et al. Guanylate-Binding Protein 1 as a Potential Predictor of Immunotherapy: A Pan-Cancer Analysis. Front. Genet. 2022, 13, 820135. [Google Scholar] [CrossRef]
- Ye, S.; Li, S.; Qin, L.; Zheng, W.; Liu, B.; Li, X.; Ren, Z.; Zhao, H.; Hu, X.; Ye, N. GBP2 Promotes Clear Cell Renal Cell Carcinoma Progression through Immune Infiltration and Regulation of PD-L1 Expression via STAT1 Signaling. Oncol. Rep. 2023, 49, 49. [Google Scholar] [CrossRef]
- Raulet, D.H.; Gasser, S.; Gowen, B.G.; Deng, W.; Jung, H. Regulation of Ligands for the NKG2D Activating Receptor. Annu. Rev. Immunol. 2013, 31, 413–441. [Google Scholar] [CrossRef] [PubMed]
- Torres, N.; Regge, M.V.; Secchiari, F.; Friedrich, A.D.; Spallanzani, R.G.; Raffo Iraolagoitia, X.L.; Nunez, S.Y.; Sierra, J.M.; Ziblat, A.; Santilli, M.C.; et al. Restoration of Antitumor Immunity through Anti-MICA Antibodies Elicited with a Chimeric Protein. J. Immunother. Cancer 2020, 8, e000233. [Google Scholar] [CrossRef] [PubMed]
- Secchiari, F.; Nunez, S.Y.; Sierra, J.M.; Ziblat, A.; Regge, M.V.; Raffo Iraolagoitia, X.L.; Rovegno, A.; Ameri, C.; Secin, F.P.; Richards, N.; et al. The MICA-NKG2D Axis in Clear Cell Renal Cell Carcinoma Bolsters MICA as Target in Immuno-Oncology. Oncoimmunology 2022, 11, 2104991. [Google Scholar] [CrossRef] [PubMed]
- Kahlmeyer, A.; Stohr, C.G.; Hartmann, A.; Goebell, P.J.; Wullich, B.; Wach, S.; Taubert, H.; Erlmeier, F. Expression of PD-1 and CTLA-4 Are Negative Prognostic Markers in Renal Cell Carcinoma. J. Clin. Med. 2019, 8, 743. [Google Scholar] [CrossRef] [PubMed]
- Klumper, N.; Ralser, D.J.; Zarbl, R.; Schlack, K.; Schrader, A.J.; Rehlinghaus, M.; Hoffmann, M.J.; Niegisch, G.; Uhlig, A.; Trojan, L.; et al. CTLA4 Promoter Hypomethylation Is a Negative Prognostic Biomarker at Initial Diagnosis but Predicts Response and Favorable Outcome to Anti-PD-1 Based Immunotherapy in Clear Cell Renal Cell Carcinoma. J. Immunother. Cancer 2021, 9, e002949. [Google Scholar] [CrossRef] [PubMed]
- Ross-Macdonald, P.; Walsh, A.M.; Chasalow, S.D.; Ammar, R.; Papillon-Cavanagh, S.; Szabo, P.M.; Choueiri, T.K.; Sznol, M.; Wind-Rotolo, M. Molecular Correlates of Response to Nivolumab at Baseline and on Treatment in Patients with RCC. J. Immunother. Cancer 2021, 9, e001506. [Google Scholar] [CrossRef]
- Lecoq, I.; Kopp, K.L.; Chapellier, M.; Mantas, P.; Martinenaite, E.; Perez-Penco, M.; Ronn Olsen, L.; Zocca, M.B.; Wakatsuki Pedersen, A.; Andersen, M.H. CCL22-Based Peptide Vaccines Induce Anti-Cancer Immunity by Modulating Tumor Microenvironment. Oncoimmunology 2022, 11, 2115655. [Google Scholar] [CrossRef]
- Jin, C.; Shi, L.; Li, Z.; Liu, W.; Zhao, B.; Qiu, Y.; Zhao, Y.; Li, K.; Li, Y.; Zhu, Q. Circ_0039569 Promotes Renal Cell Carcinoma Growth and Metastasis by Regulating miR-34a-5p/CCL22. Am. J. Transl. Res. 2019, 11, 4935–4945. [Google Scholar]
- Chehrazi-Raffle, A.; Meza, L.; Alcantara, M.; Dizman, N.; Bergerot, P.; Salgia, N.; Hsu, J.; Ruel, N.; Salgia, S.; Malhotra, J.; et al. Circulating Cytokines Associated with Clinical Response to Systemic Therapy in Metastatic Renal Cell Carcinoma. J. Immunother. Cancer 2021, 9, e002009. [Google Scholar] [CrossRef]
- Chen, C.; Chen, L.Y.; Yang, R.X.; Zhang, J.X.; Shao, P.F.; Xu, H.G. Identification of IRF-Associated Molecular Subtypes in Clear Cell Renal Cell Carcinoma to Characterize Immunological Characteristics and Guide Therapy. Front. Oncol. 2022, 12, 1118472. [Google Scholar] [CrossRef] [PubMed]
- Corro, C.; Healy, M.E.; Engler, S.; Bodenmiller, B.; Li, Z.; Schraml, P.; Weber, A.; Frew, I.J.; Rechsteiner, M.; Moch, H. IL-8 and CXCR1 Expression Is Associated with Cancer Stem Cell-Like Properties of Clear Cell Renal Cancer. J. Pathol. 2019, 248, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Sridhar Panaiyadiyan, B.N.; Singh, P.; Kaushal, S.; Karmakar, S.; Seth, A. Prognostic and Predictive Role of Intra-Tumoral CXCR1 Expression in Patients Receiving Tyrosine Kinase Inhibitors for Metastatic Clear-Cell Renal Cell Carcinoma. J. Clin. Urol. 2023, 16, 7. [Google Scholar] [CrossRef]
- Kohli, K.; Pillarisetty, V.G.; Kim, T.S. Key Chemokines Direct Migration of Immune Cells In Solid Tumors. Cancer Gene Ther. 2022, 29, 10–21. [Google Scholar] [CrossRef]
- Raimondi, A.; Sepe, P.; Zattarin, E.; Mennitto, A.; Stellato, M.; Claps, M.; Guadalupi, V.; Verzoni, E.; de Braud, F.; Procopio, G. Predictive Biomarkers of Response to Immunotherapy in Metastatic Renal Cell Cancer. Front. Oncol. 2020, 10, 1644. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.Y.; Heo, Y.J.; Kwon, G.Y.; Kim, K.M. Expression of CD274 mRNA Measured by qRT-PCR Correlates with PD-L1 Immunohistochemistry in Gastric and Urothelial Carcinoma. Front. Oncol. 2022, 12, 856444. [Google Scholar] [CrossRef]
- Koh, J.; Hur, J.Y.; Lee, K.Y.; Kim, M.S.; Heo, J.Y.; Ku, B.M.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Park, K.; et al. Regulatory (FoxP3+) T cells and TGF-Beta Predict the Response to Anti-PD-1 Immunotherapy in Patients with Non-Small Cell Lung Cancer. Sci. Rep. 2020, 10, 18994. [Google Scholar] [CrossRef]
- Kadam, P.; Sharma, S. PD-1 Immune Checkpoint Blockade Promotes Therapeutic Cancer Vaccine to Eradicate Lung Cancer. Vaccines 2020, 8, 317. [Google Scholar] [CrossRef]
- Cao, Y.; Jiao, N.; Sun, T.; Ma, Y.; Zhang, X.; Chen, H.; Hong, J.; Zhang, Y. CXCL11 Correlates with Antitumor Immunity and an Improved Prognosis in Colon Cancer. Front. Cell Dev. Biol. 2021, 9, 646252. [Google Scholar] [CrossRef]
- Jonas, B.A. Combination of an Oncolytic Virus with PD-L1 Blockade Keeps Cancer in Check. Sci. Transl. Med. 2017, 9, eaan2781. [Google Scholar] [CrossRef]
- Shi, J.; Wang, K.; Xiong, Z.; Yuan, C.; Wang, C.; Cao, Q.; Yu, H.; Meng, X.; Xie, K.; Cheng, Z.; et al. Impact of Inflammation and Immunotherapy in Renal Cell Carcinoma. Oncol. Lett. 2020, 20, 272. [Google Scholar] [CrossRef]
- Zhu, Y.; Knolhoff, B.L.; Meyer, M.A.; Nywening, T.M.; West, B.L.; Luo, J.; Wang-Gillam, A.; Goedegebuure, S.P.; Linehan, D.C.; DeNardo, D.G. CSF1/CSF1R Blockade Reprograms Tumor-Infiltrating Macrophages and Improves Response to T-Cell Checkpoint Immunotherapy in Pancreatic Cancer Models. Cancer Res. 2014, 74, 5057–5069. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, Y.; An, H.; Chang, Y.; Zhang, W.; Zhu, Y.; Xu, L.; Xu, J. High Expression of Colony-Stimulating Factor 1 Receptor Associates with Unfavorable Cancer-Specific Survival of Patients with Clear Cell Renal Cell Carcinoma. Ann. Surg. Oncol. 2016, 23, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Shigematsu, Y.; Ninomiya, H.; Nakashima, Y.; Kobayashi, M.; Saito, H.; Takahashi, K.; Futaya, E.; Okumura, S.; Ishikawa, Y.; et al. CSF1R-Expressing Tumor-Associated Macrophages, Smoking and Survival in Lung Adenocarcinoma: Analyses Using Quantitative Phosphor-Integrated Dot Staining. Cancers 2018, 10, 252. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Beuselinck, B.; Job, S.; Marisa, L.; Vano, Y.; Oudard, S.; Zucman-Rossi, J.; Laurent-Puig, P.; Sautes-Fridman, C.; et al. Prognostic and Theranostic Impact of Molecular Subtypes and Immune Classifications in Renal Cell Cancer (RCC) and Colorectal Cancer (CRC). Oncoimmunology 2015, 4, e1049804. [Google Scholar] [CrossRef] [PubMed]
- Firdaus, F.; Kuchakulla, M.; Qureshi, R.; Dulce, R.A.; Soni, Y.; Van Booven, D.J.; Shah, K.; Masterson, T.; Rosete, O.J.; Punnen, S. S-Nitrosylation of CSF1 Receptor Increases the Efficacy of CSF1R Blockage against Prostate Cancer. Cell Death Dis. 2022, 13, 859. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Marchesi, F.; Garlanda, C. Macrophages as Tools and Targets in Cancer Therapy. Nat. Rev. Drug Discov. 2022, 21, 799–820. [Google Scholar] [CrossRef]
- Bi, K.; He, M.X.; Bakouny, Z.; Kanodia, A.; Napolitano, S.; Wu, J.; Grimaldi, G.; Braun, D.A.; Cuoco, M.S.; Mayorga, A. Tumor and Immune Reprogramming during Immunotherapy in Advanced Renal Cell Carcinoma. Cancer Cell 2021, 39, 649–661.e645. [Google Scholar] [CrossRef]
- Beltraminelli, T.; De Palma, M. Biology and Therapeutic Targeting of Tumour-Associated Macrophages. J. Pathol. 2020, 250, 573–592. [Google Scholar] [CrossRef]
- Bertrand, F.; Montfort, A.; Marcheteau, E.; Imbert, C.; Gilhodes, J.; Filleron, T.; Rochaix, P.; Andrieu-Abadie, N.; Levade, T.; Meyer, N. TNFα Blockade Overcomes Resistance to Anti-PD-1 in Experimental Melanoma. Nat. Commun. 2017, 8, 2256. [Google Scholar] [CrossRef]
- Sun, J.-Y.; Zhang, D.; Wu, S.; Xu, M.; Zhou, X.; Lu, X.-J.; Ji, J. Resistance to PD-1/PD-L1 Blockade Cancer Immunotherapy: Mechanisms, Predictive Factors, and Future Perspectives. Biomark. Res. 2020, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Fishman, M.N.; Escudier, B.; McDermott, D.F.; Drake, C.G.; Kluger, H.; Stadler, W.M.; Perez-Gracia, J.L.; McNeel, D.G.; Curti, B. Immunomodulatory Activity of Nivolumab in Metastatic Renal Cell Carcinoma. Clin. Cancer Res. 2016, 22, 5461–5471. [Google Scholar] [CrossRef] [PubMed]
- Wightman, S.; Uppal, A.; Pitroda, S.; Ganai, S.; Burnette, B.; Stack, M.; Oshima, G.; Khan, S.; Huang, X.; Posner, M. Oncogenic CXCL10 Signalling Drives Metastasis Development and Poor Clinical Outcome. Br. J. Cancer 2015, 113, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Au, L.; Hatipoglu, E.; de Massy, M.R.; Litchfield, K.; Beattie, G.; Rowan, A.; Schnidrig, D.; Thompson, R.; Byrne, F.; Horswell, S. Determinants of Anti-PD-1 Response and Resistance in Clear Cell Renal Cell Carcinoma. Cancer Cell 2021, 39, 1497–1518.e1411. [Google Scholar] [CrossRef] [PubMed]
- Bou-Dargham, M.J.; Sang, Q.-X.A. Secretome Analysis Reveals Upregulated Granzyme B in Human Androgen-Repressed Prostate Cancer Cells with Mesenchymal and Invasive Phenotype. PLoS ONE 2020, 15, e0237222. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.K.; Donskov, F.; Nordsmark, M.; Marcussen, N.; von der Maase, H. Increased Intratumoral FOXP3-Positive Regulatory Immune Cells during Interleukin-2 Treatment in Metastatic Renal Cell Carcinoma. Clin. Cancer Res. 2009, 15, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Macher-Goeppinger, S.; Aulmann, S.; Tagscherer, K.E.; Wagener, N.; Haferkamp, A.; Penzel, R.; Brauckhoff, A.; Hohenfellner, M.; Sykora, J.; Walczak, H. Prognostic Value of Tumor Necrosis Factor–Related Apoptosis-Inducing Ligand (TRAIL) and TRAIL Receptors in Renal Cell Cancer. Clin. Cancer Res. 2009, 15, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Peng, Z.; Qin, M.; Liu, Y.; Wang, J.; Zhang, C.; Lin, J.; Dong, T.; Wang, L.; Li, S. Interferon-γ Induces Tumor Resistance to Anti-PD-1 Immunotherapy by Promoting YAP Phase Separation. Mol. Cell 2021, 81, 1216–1230.e1219. [Google Scholar] [CrossRef]
- Kim, B.-G.; Malek, E.; Choi, S.H.; Ignatz-Hoover, J.J.; Driscoll, J.J. Novel Therapies Emerging in Oncology to Target the TGF-β Pathway. J. Hematol. Oncol. 2021, 14, 55. [Google Scholar] [CrossRef]
- Reustle, A.; Di Marco, M.; Meyerhoff, C.; Nelde, A.; Walz, J.S.; Winter, S.; Kandabarau, S.; Büttner, F.; Haag, M.; Backert, L. Integrative-Omics and HLA-ligandomics Analysis to Identify Novel Drug Targets for ccRCC Immunotherapy. Genome Med. 2020, 12, 32. [Google Scholar] [CrossRef]
- Wang, C.; Xiong, C.; Hsu, Y.-C.; Wang, X.; Chen, L. Human Leukocyte Antigen (HLA) and Cancer Immunotherapy: HLA-Dependent and-Independent Adoptive Immunotherapies. Ann. Blood 2020, 5, 10.21037. [Google Scholar] [CrossRef]
- Maggs, L.; Sadagopan, A.; Moghaddam, A.S.; Ferrone, S. HLA Class I Antigen Processing Machinery Defects in Antitumor Immunity and Immunotherapy. Trends Cancer 2021, 7, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Serzan, M.T.; Atkins, M.B. Current and Emerging Therapies for First Line Treatment of Metastatic Clear Cell Renal Cell Carcinoma. J. Cancer Metastasis Treat. 2021, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Wang, T.; Zhang, J.; Chen, S.; Wang, X. HIF1A Predicts the Efficacy of Anti-PD-1 Therapy in Advanced Clear Cell Renal Cell Carcinoma. Transl. Oncol. 2022, 26, 101554. [Google Scholar] [CrossRef]
- Shen, C.; Beroukhim, R.; Schumacher, S.E.; Zhou, J.; Chang, M.; Signoretti, S.; Kaelin, W.G., Jr. Genetic and Functional Studies Implicate HIF1α as a 14q Kidney Cancer Suppressor Gene. Cancer Discov. 2011, 1, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Diesing, K.; Ribback, S.; Winter, S.; Gellert, M.; Oster, A.M.; Stühler, V.; Gläser, E.; Adler, F.; Hartwig, C.; Scharpf, M.; et al. p53 Is Functionally Inhibited in Clear Cell Renal Cell Carcinoma (ccRCC): A Mechanistic and Correlative Investigation into Genetic and Molecular Characteristics. J. Cancer Res. Clin. Oncol. 2021, 147, 3565–3576. [Google Scholar] [CrossRef]
- Godlewski, J.; Krazinski, B.E.; Kowalczyk, A.E.; Kiewisz, J.; Kiezun, J.; Kwiatkowski, P.; Sliwińska-Jewsiewicka, A.; Wierzbicki, P.W.; Kmieć, Z. Expression and Prognostic Significance of EP300, TP53 and BAX in Clear Cell Renal Cell Carcinoma. Anticancer. Res. 2017, 37, 2927–2937. [Google Scholar]
- Chiu, J.W.; Binte Hanafi, Z.; Chew, L.C.Y.; Mei, Y.; Liu, H. IL-1α Processing, Signaling and Its Role in Cancer Progression. Cells 2021, 10, 92. [Google Scholar] [CrossRef]
- Aggen, D.H.; Ager, C.R. Blocking IL1 Beta Promotes Tumor Regression and Remodeling of the Myeloid Compartment in a Renal Cell Carcinoma Model: Multidimensional Analyses. Clin. Cancer Res. 2021, 27, 608–621. [Google Scholar] [CrossRef]
- Wang, S.; Yu, Z.H.; Chai, K.Q. Identification of EGFR as a Novel Key Gene in Clear Cell Renal Cell Carcinoma (ccRCC) through Bioinformatics Analysis and Meta-Analysis. BioMed Res. Int. 2019, 2019, 6480865. [Google Scholar] [CrossRef]
- Muroni, M.R.; Ribback, S.; Sotgiu, G.; Kroeger, N.; Saderi, L.; Angius, A.; Cossu-Rocca, P.; De Miglio, M.R. Prognostic Impact of Membranous/Nuclear Epidermal Growth Factor Receptor Localization in Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 8747. [Google Scholar] [CrossRef] [PubMed]
- Stühler, V.; Maas, J.M.; Rausch, S.; Stenzl, A.; Bedke, J. Immune Checkpoint Inhibition for the Treatment of Renal Cell Carcinoma. Expert. Opin. Biol. Ther. 2020, 20, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Rebuzzi, S.E.; Perrone, F.; Bersanelli, M.; Bregni, G.; Milella, M.; Buti, S. Prognostic and Predictive Molecular Biomarkers in Metastatic Renal Cell Carcinoma Patients Treated with Immune Checkpoint Inhibitors: A Systematic Review. Expert. Rev. Mol. Diagn. 2020, 20, 169–185. [Google Scholar] [CrossRef] [PubMed]
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 11, 79. [Google Scholar] [CrossRef]
- Maqbool, M.; Khan, A.; Shahzad, A.; Sarfraz, Z.; Sarfraz, A.; Aftab, H.; Jaan, A. Predictive Biomarkers for Colorectal Cancer: A State-of-the-Art Systematic Review. Biomarkers 2023, 28, 562–598. [Google Scholar] [CrossRef]
- Gazouli, M.; Souliotis, K. The Economic Considerations and Implications of the Stratification of Future Oncology Therapeutics. Mol. Diagn. Ther. 2014, 18, 403–408. [Google Scholar] [CrossRef]

| Characteristic | Number of Patients (%) | |
|---|---|---|
| Gender | Male | 21 (77.78%) |
| Female | 6 (22.22%) | |
| Median age (Range) | 66.4 ± 10.7 | |
| Treatment | Nivolumab + Ipilimumab | 11 (40.74%) |
| Nivolumab + Cabozantinib | 8 (29.63%) | |
| Pemprolizumab + Axitinib | 8 (29.63%) | |
| Response Status | Clinical Benefit (CB) | 14 (54.85%) |
| Progressive Disease (PD) | 13 (48.15%) |
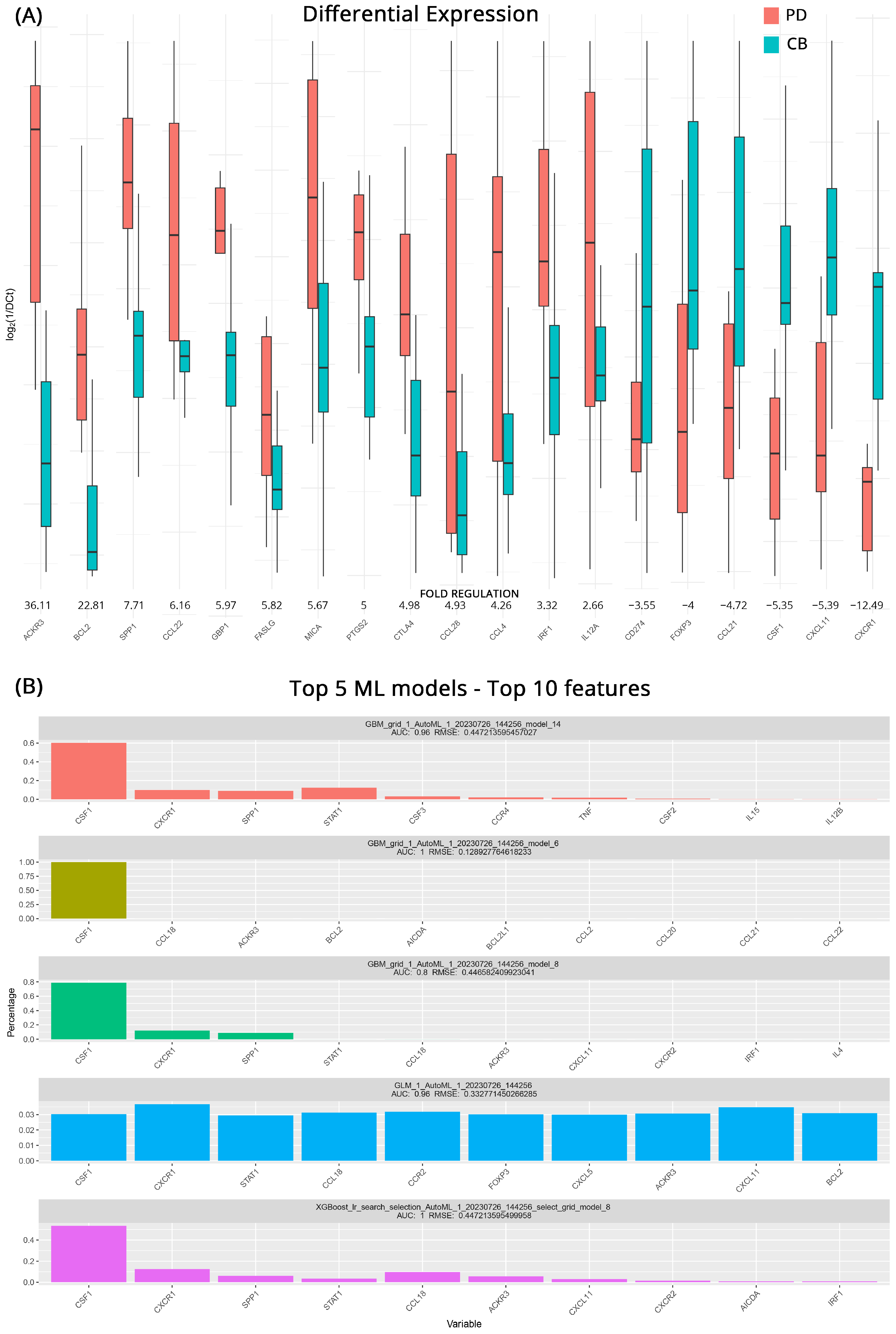
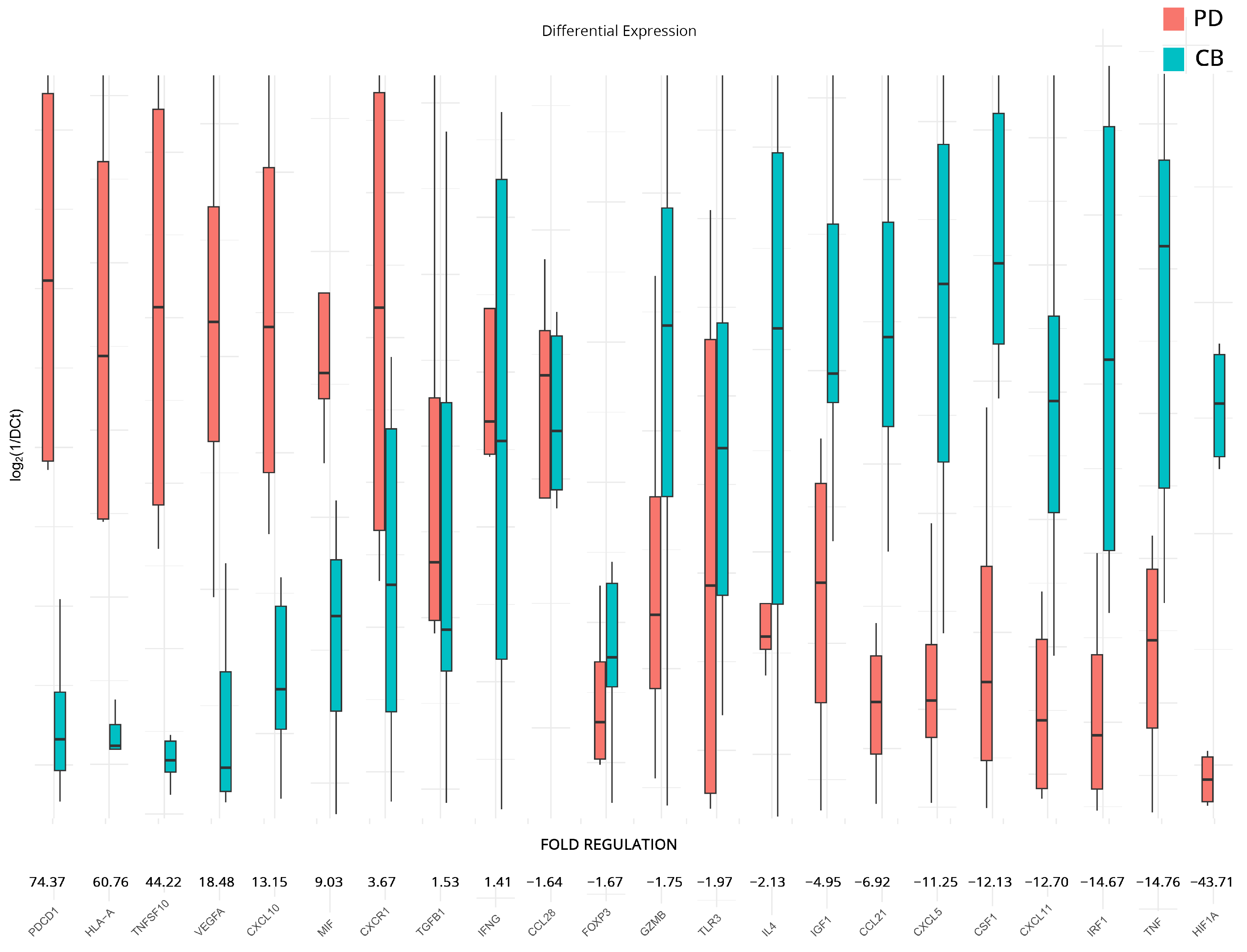
| PD vs. CB at Baseline | PD vs. CB after Therapy | ICI/ICI vs. ICI/TKI after Therapy | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Fold Regulation | p-Value | Gene Symbol | Fold Regulation | p-Value | Gene Symbol | Fold Regulation | p-Value |
| ACKR3 | 36.1 | >0.001 | PDCD1 | 74.4 | 0.030 | TP53 | 9.3 | 0.310 |
| BCL2 | 22.8 | 0.026 | HLA-A | 60.8 | 0.050 | IL1A | 3.7 | 0.884 |
| SPP1 | 7.7 | >0.001 | TNFSF10 | 44.2 | 0.036 | ACKR3 | 3.3 | 0.377 |
| CCL22 | 6.2 | 0.016 | VEGFA | 18.5 | 0.041 | HLA-A | 3.3 | 0.783 |
| GBP1 | 6.0 | 0.013 | CXCL10 | 13.2 | 0.045 | BCL2L1 | 2.8 | 0.183 |
| FASLG | 5.8 | 0.031 | MIF | 9.0 | 0.055 | CCR9 | 2.8 | 0.287 |
| MICA | 5.7 | 0.019 | CXCR1 | 3.7 | 0.053 | KITLG | 2.8 | 0.553 |
| PTGS2 | 5.0 | 0.034 | TGFB1 | 1.5 | 0.678 | NOS2 | 2.8 | 0.901 |
| CTLA4 | 5.0 | 0.050 | IFNG | 1.4 | 0.747 | MIF | 2.5 | 0.242 |
| CCL28 | 4.9 | 0.023 | CCL28 | −1.6 | 0.676 | CXCL8 | 2.5 | 0.966 |
| CCL4 | 4.3 | 0.017 | FOXP3 | −1.7 | 0.376 | CXCL5 | −2.4 | 0.942 |
| IRF1 | 3.3 | 0.023 | GZMB | −1.8 | 0.227 | TNF | −2.4 | 0.905 |
| IL12A | 2.7 | 0.017 | TLR3 | −2.0 | 0.659 | CSF1 | −2.5 | 0.747 |
| CD274 | −3.6 | 0.026 | IL4 | −2.1 | 0.121 | EGFR | −2.5 | 0.751 |
| FOXP3 | −4.0 | 0.025 | IGF1 | −5.0 | 0.097 | TGFB1 | −2.5 | 0.110 |
| CCL21 | −4.7 | 0.005 | CCL21 | −6.9 | 0.034 | CD274 | −2.8 | 0.952 |
| CSF1 | −5.4 | 0.004 | CXCL5 | −11.3 | 0.046 | CSF2 | −2.9 | 0.487 |
| CXCL11 | −5.4 | 0.005 | CSF1 | −12.1 | 0.026 | HIF1A | −3.0 | 0.782 |
| CXCR1 | −12.5 | 0.008 | CXCL11 | −12.7 | 0.049 | IL15 | −4.0 | 0.052 |
| IRF1 | −14.7 | 0.043 | EGF | −5.1 | 0.244 | |||
| TNF | −14.8 | 0.050 | ||||||
| HIF1A | −43.7 | 0.030 | ||||||
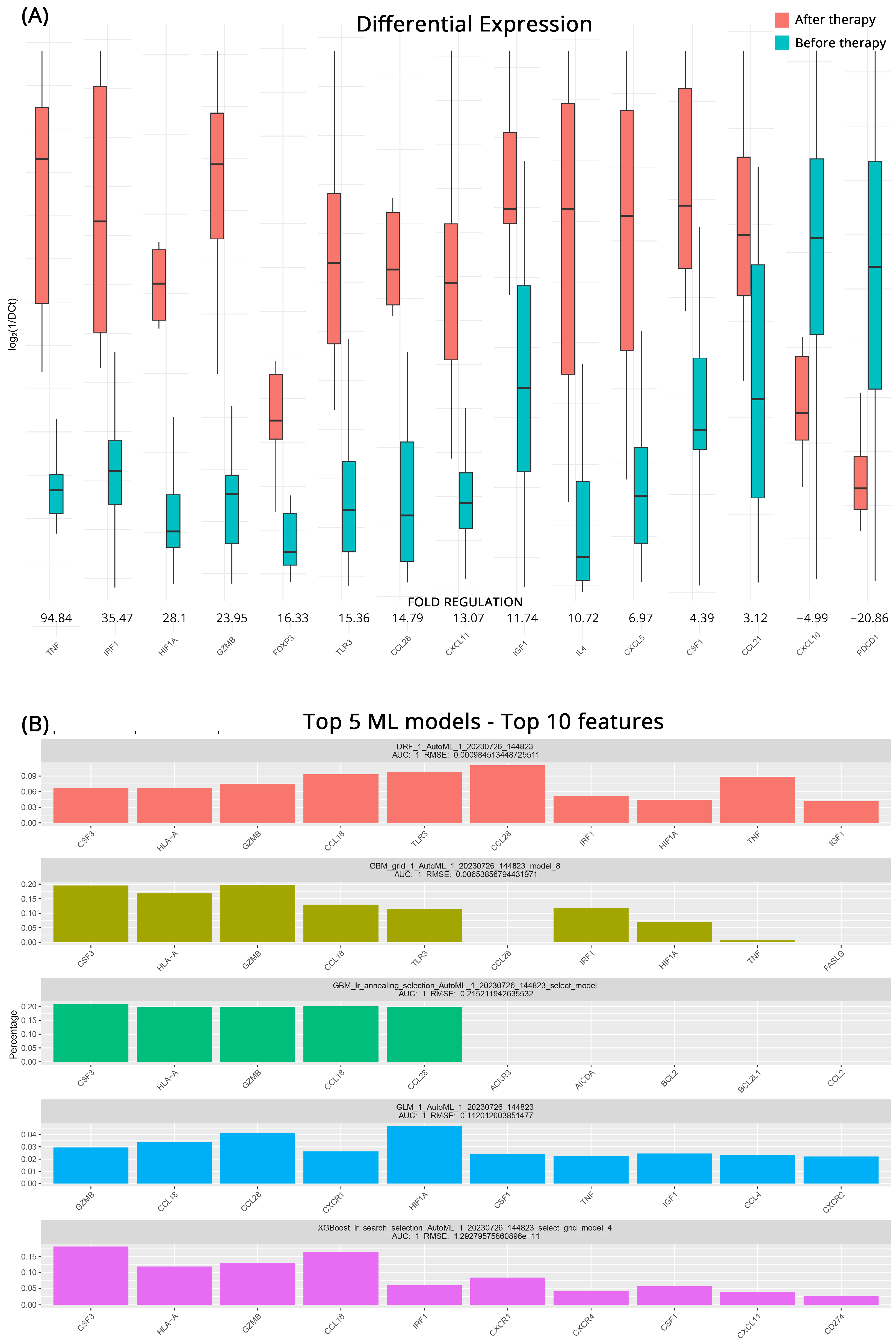
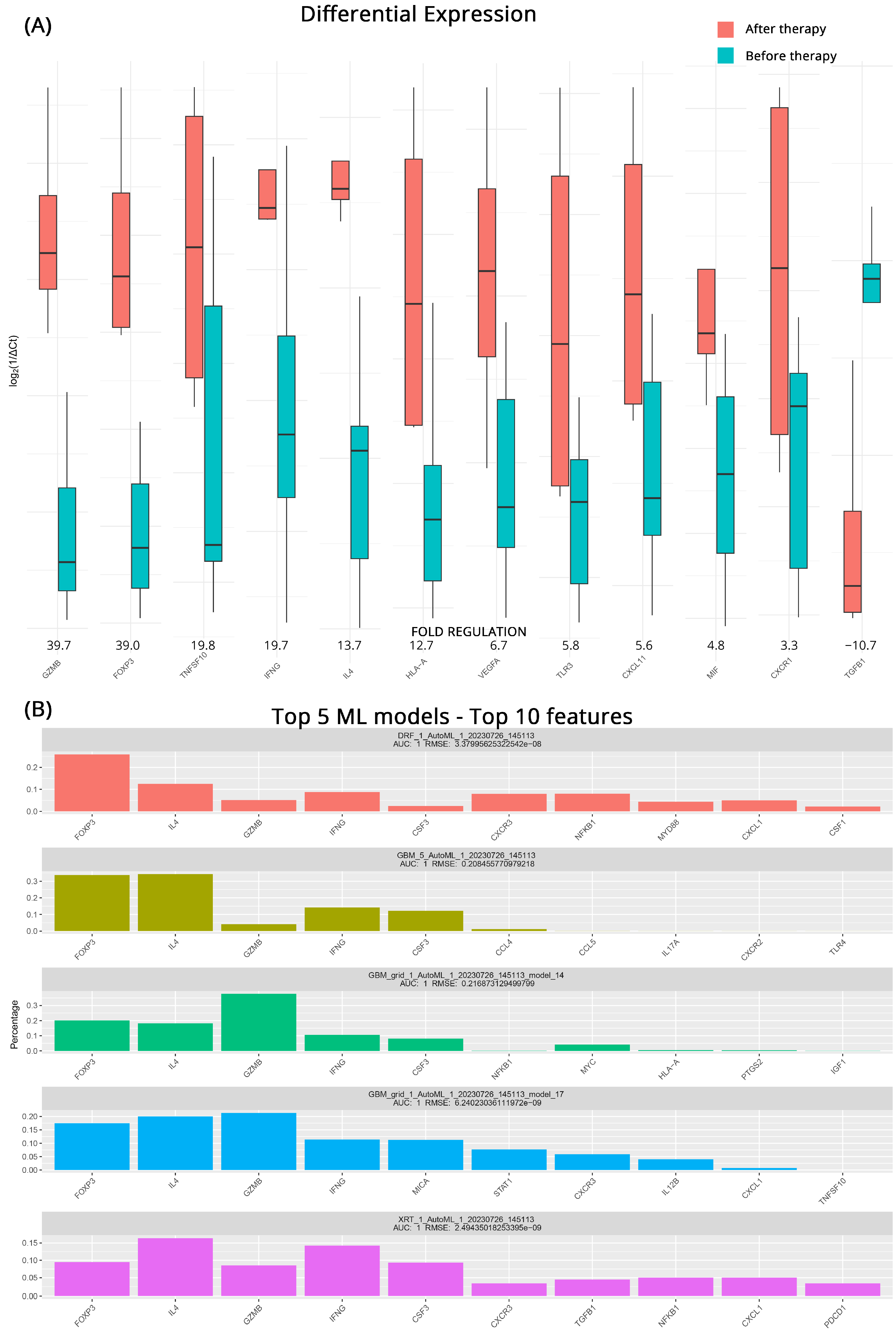
| CB after vs. before Therapy | PD after vs. before Therapy | |||||||
| Gene Symbol | Fold Regulation | p-Value | Gene Symbol | Fold Regulation | p-Value | |||
| TNF | 94.8 | >0.001 | GZMB | 39.7 | >0.001 | |||
| IRF1 | 35.5 | 0.001 | FOXP3 | 39.0 | >0.001 | |||
| HIF1A | 28.1 | >0.001 | TNFSF10 | 19.8 | 0.045 | |||
| GZMB | 24.0 | >0.001 | IFNG | 19.7 | 0.004 | |||
| FOXP3 | 16.3 | 0.003 | IL4 | 13.7 | >0.001 | |||
| TLR3 | 15.4 | 0.003 | HLA-A | 12.7 | 0.008 | |||
| CCL28 | 14.8 | 0.003 | VEGFA | 6.7 | 0.031 | |||
| CXCL11 | 13.1 | 0.001 | TLR3 | 5.8 | 0.008 | |||
| IGF1 | 11.7 | 0.003 | CXCL11 | 5.6 | 0.008 | |||
| IL4 | 10.7 | >0.001 | MIF | 4.8 | 0.014 | |||
| CXCL5 | 7.0 | 0.001 | CXCR1 | 3.3 | 0.010 | |||
| CSF1 | 4.4 | 0.001 | TGFB1 | −10.7 | 0.028 | |||
| CCL21 | 3.1 | 0.031 | ||||||
| CXCL10 | −5.0 | 0.039 | ||||||
| PDCD1 | −20.9 | 0.039 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dovrolis, N.; Katifelis, H.; Grammatikaki, S.; Zakopoulou, R.; Bamias, A.; Karamouzis, M.V.; Souliotis, K.; Gazouli, M. Inflammation and Immunity Gene Expression Patterns and Machine Learning Approaches in Association with Response to Immune-Checkpoint Inhibitors-Based Treatments in Clear-Cell Renal Carcinoma. Cancers 2023, 15, 5637. https://doi.org/10.3390/cancers15235637
Dovrolis N, Katifelis H, Grammatikaki S, Zakopoulou R, Bamias A, Karamouzis MV, Souliotis K, Gazouli M. Inflammation and Immunity Gene Expression Patterns and Machine Learning Approaches in Association with Response to Immune-Checkpoint Inhibitors-Based Treatments in Clear-Cell Renal Carcinoma. Cancers. 2023; 15(23):5637. https://doi.org/10.3390/cancers15235637
Chicago/Turabian StyleDovrolis, Nikolas, Hector Katifelis, Stamatiki Grammatikaki, Roubini Zakopoulou, Aristotelis Bamias, Michalis V. Karamouzis, Kyriakos Souliotis, and Maria Gazouli. 2023. "Inflammation and Immunity Gene Expression Patterns and Machine Learning Approaches in Association with Response to Immune-Checkpoint Inhibitors-Based Treatments in Clear-Cell Renal Carcinoma" Cancers 15, no. 23: 5637. https://doi.org/10.3390/cancers15235637
APA StyleDovrolis, N., Katifelis, H., Grammatikaki, S., Zakopoulou, R., Bamias, A., Karamouzis, M. V., Souliotis, K., & Gazouli, M. (2023). Inflammation and Immunity Gene Expression Patterns and Machine Learning Approaches in Association with Response to Immune-Checkpoint Inhibitors-Based Treatments in Clear-Cell Renal Carcinoma. Cancers, 15(23), 5637. https://doi.org/10.3390/cancers15235637










