Recent Advances in Surface Plasmon Resonance (SPR) Technology for Detecting Ovarian Cancer Biomarkers
Abstract
Simple Summary
Abstract
1. Introduction
2. Serum-Based EOC Biomarkers
3. Conventional Diagnostic EOC Biomarker in Immunoassay Technique
| Criteria | Value/ Target Cohort | Biomarkers | |||
|---|---|---|---|---|---|
| CA125 U/mL | HE4 pM/L | CA19-9 U/mL | CEA ng/mL | ||
| Type | Mucinous Glycoprotein | Glycoprotein | Mucinous Glycoprotein | Glycoprotein | |
| Diagnostic | ✓ | ✓ | ✓ | ✓ | |
| Prognosis | ✓ | ✓ | ✓ | ||
| Monitoring | ✓ | ✓ | |||
| FDA Approval (Year) | 1981 | 2008 | 2002 | 1985 | |
| Cut-off | 35 | >70 or 140 | <37 or 27 | 2.5–5 | |
| MW (kDa) | >200 | 25 | 1000 | 180 | |
| Immunoassay Technique: ECLIA/Magnetic bead assay | SP% | >60–80 | >96–100 | 79.01 | >88–100 |
| SN% | >60–96 | >63–83 | 35.71 | >38–66.3 | |
| Benign | 9.02–54.92 | 49.01–54.49 | 19.15 | 2.71 | |
| Malignant | 368 | 245.9 | |||
| Late-stage | 184.62 | 234.77 | 45.61 | 9.27 | |
| Ref. | [60,61] | [60,61,62,63] | [36,54,64] | [52,64,65,66] | |
| ROMA Test | RMI | OVA1 Test | OVA2 (Overa) | |
|---|---|---|---|---|
| FDA Approval (Year) | 2011 | N/A | 2009 | 2016 |
| Biomarker | ||||
| • CA 125 | x | x | x | x |
| • HE4 | x | x | ||
| • B2M | x | |||
| • ApoA-I | x | x | ||
| • FSH | x | |||
| • TRF | x | |||
| • Transferrin | x | x | ||
| Immunoassay technique | ECLIA [Cobas 8000, Roche Diagnostics Scandinavia AB, Sweden]// Magnetic bead assay [xMAP bead-based technology (Luminex, Austin, Texas)] | ECLIA Cobas 6000 (Roche, Germany) /IVDMIA [Quest Diagnostics Incorporated; Vermillion, (Austin, Texas)] | IVDMIA [Quest Diagnostics Incorporated; Vermillion, (Austin, Texas)] | |
| SP (%) | >80 to 91 | >80 to 90 | <75 to 26 | 28–35 |
| SN (%) | >85 to 97 | >87 to 97 | >78 to 98 | 77–96 |
| Ref. | [40,52,60,65,67] | [40,52,60,65,67] | [36,44,52,54,60] | [52,60,66] |
4. Advanced Diagnostic Multiplex EOC Biomarker in Immunoassay Technique
4.1. Significance of Multiplexed EOC Biomarker
4.2. Multiplex Panel Analysis via Current Labelled Immunoassay
| Paper | Biomarker | Target Cohort | SN% | AUC | Ref. |
|---|---|---|---|---|---|
| Chen et al. (2018) | CA125, HE4, CEA | Benign, Malignant | 88.52 | 0.972 | [64] |
| Guo et al. (2019) | CA125, MIF, OPN, IL-8 AAb | Early-stage | 82 | 0.974 | [72] |
| Kampan et al. (2020) | CA125, HE4, IL-6, IL-8, IL-6 + CA125, Il-6 + RMI score, IL-6 +HE4, IL6 + ROMA | Benign, Malignant | - | >0.9 | [40] |
| Yang et al. (2020) | CA125 + HE4 Ag-AAb | Early-stage | - | 0.986 | [73] |
| CA125 + HE4 Ag-AAb | Late stage | - | 0.985 | ||
| Boylan et al. (2017) | CA125, HE4, MK, KLK6, hK11, CXCL13, FR-alpha, IL 6, TNFSF14, FADD, PRSS8, FUR | Early-stage, Healthy | 93 | 0.99 | [42] |
| Leandersson et al. (2020) | CA125, HE4, CXCL6, CTSV, CEACAM1, S100A4, FOLR1, | Benign, Malignant | - | 0.921 | [39] |
| HE4, CA125, ITGAV, CXCL1, CEACAM1, IL-10RB | Benign, Malignant, Borderline | - | 0868 | ||
| Mukama et al. (2022) | CA125, HE4, KLK11, CXCL13, FOLR1, WISP1, MDK, MSLN, ADAM8 | Malignant, Control | - | ≥0.70 | [74] |
4.2.1. Luminex Technology
4.2.2. Proximity Extension Assay (PEA) Technology
4.3. Challenges of Multiplex Technology Using Advance Immunoassay
4.3.1. Target Cohort and Sample Size
4.3.2. Dependency-Appropriate Biomarker Combinations
4.3.3. The Technologies
4.3.4. The EOC Multiplex Signatures
| Multiplex Technology | Luminex Technology | PEA Technology | |
|---|---|---|---|
| Key Points | |||
| Principle | Bead-based Immunoassay | Proximity ligation and Amplification | |
| Number of analytes | 100 | 92 | |
| Sensitivity | Variable, Depending on the assay design, The dilution factor of an analyte | High sensitivity | |
| Application | Protein biomarker analysis, cytokine profiling | Protein biomarker analysis, New biomarker discovery, Drug development | |
| Commercial availability | Widely | Available from specific vendors | |
| Limitation |
| ||
5. Point-of-Care (POC) Multiplex Sensors for Detecting EOC Biomarkers
Label-Free Optical Biosensor
6. Surface Plasmon Resonance (SPR) Optical Biosensing
6.1. SPR in Multiplexed Detection of EOC Biomarkers
6.2. Detection of CA125 and HE4 Biomarkers Using SPR
6.2.1. SPR-Based Biosensors
6.2.2. SPRi
6.2.3. LSPR Co-Enhanced Raman Scattering
6.3. Challenges and Future Preceptive in Developing New SPR-Based Biosensor
7. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Glossary
| Diagnostic | To detect or confirm the presence of an early disease state or condition of interest or Classification into disease subtypes. |
| Progression | Prediction of disease progression or recurrence in patients who have the disease or medical condition of interest. |
| Monitoring | Identification status of a disease or medical condition or for evidence of exposure to a blood test to monitor changes in the levels of specific biomarkers. |
| Benign | Benign tumors may grow larger but the absence of cell proliferation and invasion to other parts of the body. 60% of these tumors occur in females aged <40 years. |
| Borderline | A tumor is characterized by cell proliferation, a minor degree of nuclear atypia, and without stromal invasion. Occur at a younger age than carcinoma, age 45 years. |
| Malignant | Malignant cells proliferate nuclear atypia and stromal invasion to other parts of the body. Primarily found in elderly patients, median age of 60 years. |
| Early-stage | Biomarker detection focuses on identifying biomarkers that can indicate the presence of a disease or condition at its initial or early stages. |
| Late-stage | Biomarker detection refers to the identification and measurement of biomarkers that are associated with advanced stages of a disease or condition |
| Sensitivity (SN) | Identify individuals who have the disease (true positive). |
| Specificity (SP) | Identify individuals who do not have the disease (true negative). |
| Cut-off | Biomarker-oriented (or reference intervals) approach the mean of biomarker biomarkers are sets of values. |
| AUC | To estimate the accuracy of a diagnostic test or predictive model use the standard method to receiver operating characteristic area under the curve (ROC AUC). |
| Sensitivity (SSPR) | Sensitivity in terms of detecting molecular binding in SPR biosensors (referred to as surface sensitivity). |
References
- Institut Kanser Negara. Summary of Malaysia National Cancer-Registry Report. 2012–2016; Institut Kanser Negara: Putrajaya, Malaysia, 2019.
- Takemura, K. Surface Plasmon Resonance (SPR)- and Localized SPR (LSPR)-Based Virus Sensing Systems: Optical Vibration of Nano- and Micro-Metallic Materials for the Development of Next-Generation Virus Detection Technology. Biosensors 2021, 11, 250. [Google Scholar] [CrossRef] [PubMed]
- Colombo, N.; Sessa, C.; du Bois, A.; Ledermann, J.; McCluggage, W.G.; McNeish, I.; Morice, P.; Pignata, S.; Ray-Coquard, I.; Vergote, I.; et al. ESMO-ESGO consensus conference recommendations on ovarian cancer: Pathology and molecular biology, early and advanced stages, borderline tumors and recurrent diseasedagger. Ann. Oncol. 2019, 30, 672–705. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Gupta, S.; Sachan, M. Epigenetic Biomarkers in the Management of Ovarian Cancer: Current Prospectives. Front. Cell Dev. Biol. 2019, 7, 182. [Google Scholar] [CrossRef] [PubMed]
- Atallah, G.A.; Aziz, N.A.; Teik, C.K.; Shafiee, M.N.; Kampan, N.C. New Predictive Biomarkers for Ovarian Cancer. Diagnostics 2021, 11, 465. [Google Scholar] [CrossRef]
- Atallah, G.A.; Kampan, N.C.; Teik, C.K.; Mokhtar, M.N.; Zin, R.R.M.; Shafiee, M.N.; Aziz, N.A. Predicting Prognosis and Platinum Resistance in Ovarian Cancer: Role of Immunohistochemistry Biomarkers. Int. J. Mol. Sci. 2023, 24, 1973. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef]
- Ovarian Cancer Research Alliance. Stages of Ovarian Cancer; Ovarian Cancer Research Alliance: New York, NY, USA, 2023. [Google Scholar]
- Centre of Cancer Research. Studies of an Experimental Ovarian Cancer Therapy Identify a Biomarker of Treatment Response; Center for Cancer Research: Nanchang, China, 2023. [Google Scholar]
- National Cancer Institute. Cancer Stat Facts: Ovarian Cancer, E. Surveillance, and End Results (SEER) Cancer Statistics Factsheets: Ovarian Cancer; National Cancer Institute, Ed.; National Cancer Institute: Bethesda, MD, USA, 2023.
- Ovarian Cancer Research Alliance. Ovarian Cancer Statistics; Ovarian Cancer Research Alliance: New York, NY, USA, 2023. [Google Scholar]
- American Cancer Society. Key Statistics for Ovarian Cancer; American Cancer Society: Atlanta, GA, USA, 2023. [Google Scholar]
- Rizzo, F. Optical Immunoassays Methods in Protein Analysis: An Overview. Chemosensors 2022, 10, 326. [Google Scholar] [CrossRef]
- Charkhchi, P.; Cybulski, C.; Gronwald, J.; Wong, F.O.; Narod, S.A.; Akbari, M.R. CA125 and Ovarian Cancer: A Comprehensive Review. Cancers 2020, 12, 3730. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Z.; Wei, Q. Comparison of CA125, HE4, and ROMA index for ovarian cancer diagnosis. Curr. Probl. Cancer 2018, 43, 135–144. [Google Scholar] [CrossRef]
- Lycke, M. Clinical Implementation of Novel Diagnostic Biomarkers for Epithelial Ovarian Cancer-Can We Improve Diagnosis? 2020. Available online: https://gupea.ub.gu.se/bitstream/handle/2077/63236/gupea_2077_63236_2.pdf?sequence=2&isAllowed=y.2020 (accessed on 1 April 2020).
- Jacobs, I.J.; Menon, U. Progress and challenges in screening for early detection of ovarian cancer. Mol. Cell Proteom. 2004, 3, 355–366. [Google Scholar] [CrossRef]
- Manole, E.; Bastian, A.E.; Popescu, I.D.; Constantin, C.; Mihai, S.; Gaina, G.F.; Codrici, E.; Neagu, M.T. Immunoassay Techniques Highlighting Biomarkers in Immunogenetic Diseases. In Immunogenetics; IntechOpen: Bucharest, Romania, 2018. [Google Scholar]
- Platchek, M.; Lu, Q.; Tran, H.; Xie, W. Comparative Analysis of Multiple Immunoassays for Cytokine Profiling in Drug Discovery. SLAS Discov. 2020, 25, 1197–1213. [Google Scholar] [CrossRef] [PubMed]
- Fadlalla, M.H.; Ling, S.; Wang, R.; Li, X.; Yuan, J.; Xiao, S.; Wang, K.; Tang, S.; Elsir, H.; Wang, S. Development of ELISA and Lateral Flow Immunoassays for Ochratoxins (OTA and OTB) Detection Based on Monoclonal Antibody. Front. Cell Infect. Microbiol. 2020, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Patel, P. Biosensors and Biomarkers: Promising Tools for Cancer Diagnosis. Int. J. Biosens. Bioelectron. 2017, 3, 72. [Google Scholar] [CrossRef][Green Version]
- Ahn, K.C.; Kim, H.J.; McCoy, M.R.; Gee, S.J.; Hammock, B.D. Immunoassays and biosensors for monitoring environmental and human exposure to pyrethroid insecticides. J. Agric. Food Chem. 2011, 59, 2792–2802. [Google Scholar] [CrossRef]
- Reuterswärd, P. Development of Array Systems for Molecular Diagnostic Assays. 2018. Available online: https://www.diva-portal.org/smash/get/diva2:1203278/FULLTEXT01.pdf (accessed on 1 April 2020).
- Hossain, K.R.; Escobar Bermeo, J.D.; Warton, K.; Valenzuela, S.M. New Approaches and Biomarker Candidates for the Early Detection of Ovarian Cancer. Front. Bioeng. Biotechnol. 2022, 10, 819183. [Google Scholar] [CrossRef] [PubMed]
- Bellassai, N.; D’Agata, R.; Jungbluth, V.; Spoto, G. Surface Plasmon Resonance for Biomarker Detection: Advances in Non-invasive Cancer Diagnosis. Front. Chem. 2019, 7, 570. [Google Scholar] [CrossRef]
- Zhao, X.; Chu-Su, Y.; Tsai, W.-H.; Wang, C.-H.; Chuang, T.-L.; Lin, C.-W.; Tsao, Y.-C.; Wu, M.-S. Improvement of the sensitivity of the surface plasmon resonance sensors based on multi-layer modulation techniques. Opt. Commun. 2015, 335, 32–36. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef]
- Falkowski, P.; Mrozek, P.; Lukaszewski, Z.; Oldak, L.; Gorodkiewi, E. An Immunosensor for the Determination of Cathepsin S in Blood Plasma by Array SPRi-A Comparison of Analytical Properties of Silver-Gold and Pure Gold Chips. Biosensors 2021, 11, 298. [Google Scholar] [CrossRef]
- Zilovic, D.; Čiurlienė, R.; Sabaliauskaite, R.; Jarmalaite, S. Future Screening Prospects for Ovarian Cancer. Cancers 2021, 13, 3840. [Google Scholar] [CrossRef]
- Cui, M.; Liu, Y.H.; Cheng, L.; Li, T.; Deng, Y.; Liu, D. Research progress on anti-ovarian cancer mechanism of miRNA regulating tumor microenvironment. Front. Immunol. 2022, 13, 1050917. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Fu, F.; Yu, F.; He, X. Advances of exosomal miRNAs in the diagnosis and treatment of ovarian cancer. Discov. Oncol. 2023, 14, 65. [Google Scholar] [CrossRef]
- Frisk, N.L.S.; Pedersen, O.B.V.; Dalgaard, L.T. Circulating microRNAs for Early Diagnosis of Ovarian Cancer: A Systematic Review and Meta-Analysis. Biomolecules 2023, 13, 871. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Hellstrom, I.; Heagerty, P.J.; Swisher, E.M.; Liu, P.; Jaffar, J.; Agnew, K.; Hellstromet, K.E. Detection of the HE4 protein in urine as a biomarker for ovarian neoplasms. Cancer Lett. 2010, 296, 43–48. [Google Scholar] [CrossRef]
- Trinidad, C.V.; Tetlow, A.L.; Bantis, L.E.; Godwin, A.K. Reducing Ovarian Cancer Mortality Through Early Detection: Approaches Using Circulating Biomarkers. Cancer Prev. Res. 2020, 13, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Muinao, T.; Boruah, H.P.D.; Pal, M. Multi-biomarker panel signature as the key to diagnosis of ovarian cancer. Heliyon 2019, 5, e02826. [Google Scholar] [CrossRef]
- Sölétormos, G.; Duffy, M.J.; Abu Hassan, S.O.; Verheijen, R.H.; Tholander, B.; Bast, R.C.; Gaarenstroom, K.N.; Sturgeon, C.M.; Bonfrer, J.M.; Petersen, P.H.; et al. Clinical Use of Cancer Biomarkers in Epithelial Ovarian Cancer: Updated Guidelines From the European Group on Tumor Markers. Int. J. Gynecol. Cancer 2016, 26, 43–51. [Google Scholar] [CrossRef]
- Dochez, V.; Caillon, H.; Vaucel, E.; Dimet, J.; Winer, N.; Ducarme, G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. J. Ovarian Res. 2019, 12, 28. [Google Scholar] [CrossRef]
- Leandersson, P.; Malander, S.; Borgfeldt, C. A multiplex biomarker assay improves the diagnostic performance of HE4 and CA125 in ovarian tumor patients. PLoS ONE 2020, 15, e0240418. [Google Scholar] [CrossRef]
- Kampan, N.C.; Madondo, M.T.; Reynolds, J.; Hallo, J.; McNally, O.M.; Jobling, T.W.; Stephens, A.N.; Quinn, M.A.; Plebanski, M. Pre-operative sera interleukin-6 in the diagnosis of high-grade serous ovarian cancer. Sci. Rep. 2020, 10, 2213. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.Y.; Kim, S. Determination of cutoff values for biomarkers in clinical studies. Precis. Future Med. 2020, 4, 2–8. [Google Scholar] [CrossRef]
- Boylan, K.L.M.; Geschwind, K.; Koopmeiners, J.S.; Geller, M.A.; Starr, T.K.; Skubitz, A.P.N. A multiplex platform for the identification of ovarian cancer biomarkers. Clin. Proteom. 2017, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Szymanska, B.; Lukaszewski, Z.; Zelazowska-Rutkowska, B.; Hermanowicz-Szamatowicz, K.; Gorodkiewicz, E. An SPRi Biosensor for Determination of the Ovarian Cancer Marker HE4 in Human Plasma. Sensors 2021, 21, 3567. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, A.; Utratna, M.; O’Dwyer, M.E.; Joshi, L.; Kilcoyne, M. Glycosylation-Based Serum Biomarkers for Cancer Diagnostics and Prognostics. Biomed. Res. Int. 2015, 2015, 490531. [Google Scholar] [CrossRef]
- McDermott, J.E.; Mitchell, H.; Webb-Robertson, B.J.; Hafen, R.; Ramey, J.; Rodland, K.D. Challenges in Biomarker Discovery: Combining Expert Insights with Statistical Analysis of Complex Omics Data. Expert. Opin. Med. Diagn. 2013, 7, 37–51. [Google Scholar] [CrossRef]
- Matsas, A.; Stefanoudakis, D.; Troupis, T.; Kontzoglou, K.; Eleftheriades, M.; Christopoulos, P.; Panoskaltsis, T.; Stamoula, E.; Iliopoulos, D.C. Tumor Markers and Their Diagnostic Significance in Ovarian Cancer. Life 2023, 13, 1689. [Google Scholar] [CrossRef]
- Wang, Q.; Feng, X.; Liu, X.; Zhu, S. Prognostic Value of Elevated Pre-treatment Serum CA-125 in Epithelial Ovarian Cancer: A Meta-Analysis. Front. Oncol. 2022, 12, 868061. [Google Scholar] [CrossRef]
- Furrer, D.; Grégoire, J.; Turcotte, S.; Plante, M.; Bachvarov, D.; Trudel, D.; Têtu, B.; Douville, P.; Bairati, I. Performance of preoperative plasma tumor markers HE4 and CA125 in predicting ovarian cancer mortality in women with epithelial ovarian cancer. PLoS ONE 2019, 14, e0218621. [Google Scholar] [CrossRef]
- Rong, Y.; Li, L. Early clearance of serum HE4 and CA125 in predicting platinum sensitivity and prognosis in epithelial ovarian cancer. J. Ovarian Res. 2021, 14, 2. [Google Scholar] [CrossRef]
- Zhu, J.; Jiang, L.; Wen, H.; Bi, R.; Wu, X.; Ju, X. Prognostic Value of Serum CA19-9 and Perioperative CA-125 Levels in Ovarian Clear Cell Carcinoma. Int. J. Gynecol. Cancer 2018, 28, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wu, M.; Wang, F. Research Progress in Prognostic Factors and Biomarkers of Ovarian Cancer. J. Cancer 2021, 12, 3976–3996. [Google Scholar] [CrossRef] [PubMed]
- Premera. Multimarker Serum Testing Related to Ovarian Cancer. 2023. Available online: https://www.premera.com/medicalpolicies/2.04.62.pdf (accessed on 1 February 2023).
- Chen, P.; Chung, M.T.; McHugh, W.; Nidetz, R.; Li, Y.; Fu, J.; Cornell, T.T.; Shanley, T.P.; Kurabayashi, K. Multiplex serum cytokine immunoassay using nanoplasmonic biosensor microarrays. ACS Nano 2015, 9, 4173–4781. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Herzog, T.J.; Chan, D.W.; Munroe, D.G.; Pappas, T.C.; Smith, A.; Zhang, Z.; Wolf, J. Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses. Am. J. Obstet. Gynecol. 2016, 215, e1–e82. [Google Scholar] [CrossRef]
- Davar, R.; Yalamanchili, M. Identification of a Panel of Biomarkers for the Early Detection of Ovarian Cancer; Journal of Student Research: Binghamton, NY, USA, 2022. [Google Scholar]
- Zhang, R.; Siu, M.K.Y.; Ngan, H.Y.S.; Chan, K.K.L. Molecular Biomarkers for the Early Detection of Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 12041. [Google Scholar] [CrossRef]
- Cinquanta, L.; Fontana, D.E.; Bizzaro, N. Chemiluminescent immunoassay technology: What does it change in autoantibody detection? Auto Immun. Highlights 2017, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Muthelo, P.M. Proximity Extension Assay; Diva portal: Uppsala, Sweden, 2022. [Google Scholar]
- Ahsan, H.; Ahmad, R. Multiplex Technology for Biomarker Immunoassays. In Innate Immunity in Health and Disease; IntechOpen: New Delhi, India, 2010. [Google Scholar]
- Wanyama, F.M.; Blanchard, V. Glycomic-Based Biomarkers for Ovarian Cancer: Advances and Challenges. Diagnostics 2021, 11, 643. [Google Scholar] [CrossRef] [PubMed]
- Hasenburg, A.; Eichkorn, D.; Vosshagen, F.; Obermayr, E.; Geroldinger, A.; Zeillinger, R.; Bossart, M. Biomarker-based early detection of epithelial ovarian cancer based on a five-protein signature in patient’s plasma—A prospective trial. BMC Cancer 2021, 21, 1037. [Google Scholar] [CrossRef]
- Hu, C.; Jiang, W.; Lv, M.; Fan, S.; Lu, Y.; Wu, Q.; Pi, J. Potentiality of Exosomal Proteinsas Novel Cancer Biomarkersfor Liquid Biopsy. Front. Immunol. 2022, 13, 792046. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Deng, H.; Xiong, X.; Zhang, H.; Liang, T.; Li, C. An aptamer biosensor for CA125 quantification in human serum based on upconversion luminescence resonance energy transfer. Microchem. J. 2021, 161, 105761. [Google Scholar] [CrossRef]
- Chen, F.; Shen, J.; Wang, J.; Cai, P.; Huang, Y. Clinical analysis of four serum tumor markers in 458 patients with ovarian tumors: Diagnostic value of the combined use of HE4, CA125, CA19-9, and CEA in ovarian tumors. Cancer Manag. Res. 2018, 10, 1313–1318. [Google Scholar] [CrossRef]
- Yurkovetsky, Z.; Skates, S.; Lomakin, A.; Nolen, B.; Pulsipher, T.; Modugno, F.; Marks, J.; Godwin, A.; Gorelik, E.; Jacobs, I.; et al. Development of a multimarker assay for early detection of ovarian cancer. J. Clin. Oncol. 2010, 28, 2159–2166. [Google Scholar] [CrossRef]
- Kumari, S. Serum Biomarker Based Algorithms in Diagnosis of Ovarian Cancer: A Review. Indian J. Clin. Biochem. 2018, 33, 382–386. [Google Scholar] [CrossRef]
- Lycke, M.; Kristjansdottir, B.; Sundfeldt, K. A multicenter clinical trial validating the performance of HE4, CA125, risk of ovarian malignancy algorithm and risk of malignancy index. Gynecol. Oncol. 2018, 151, 159–165. [Google Scholar] [CrossRef]
- Montagnana, M.; Benati, M.; Danese, E. Circulating biomarkers in epithelial ovarian cancer diagnosis: From present to future perspective. Ann. Transl. Med. 2017, 5, 276. [Google Scholar] [CrossRef]
- Nahm, F.S. Receiver operating characteristic curve: Overview and practical use for clinicians. Korean J. Anesthesiol. 2022, 75, 25–36. [Google Scholar] [CrossRef]
- Spindel, S.; Sapsford, K.E. Evaluation of optical detection platforms for multiplexed detection of proteins and the need for point-of-care biosensors for clinical use. Sensors 2014, 14, 22313–22341. [Google Scholar] [CrossRef]
- Gane, A. Cytiva, Magbeads 101: A Guide Tochoosing and Using Magneticbeads. 2019. Available online: https://www.cytivalifesciences.com/en/us/news-center/magnetic-beads-a-simple-guide-10001 (accessed on 29 May 2019).
- Guo, J.; Yang, W.-L.; Pak, D.; Celestino, J.; Lu, K.H.; Ning, J.; Lokshin, A.E.; Cheng, Z.; Lu, Z.; Bast, R.C. Osteopontin, Macrophage Migration Inhibitory Factor and Anti-Interleukin-8 Autoantibodies Complement CA125 for Detection of Early Stage Ovarian Cancer. Cancers 2019, 11, 596. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z.; Guo, J.; Fellman, B.M.; Ning, J.; Lu, K.H.; Menon, U.; Kobayashi, M.; Hanash, S.M.; Celestino, J.; et al. Human epididymis protein 4 antigen-autoantibody complexes complement cancer antigen 125 for detecting early-stage ovarian cancer. Cancer 2020, 126, 725–736. [Google Scholar] [CrossRef]
- Mukama, T.; Fortner, R.T.; Katzke, V.; Hynes, L.C.; Petrera, A.; Hauck, S.M.; Johnson, T.; Schulze, M.; Schiborn, C.; Rostgaard-Hansen, A.L.; et al. Prospective evaluation of 92 serum protein biomarkers for early detection of ovarian cancer. Br. J. Cancer 2022, 126, 1301–1309. [Google Scholar] [CrossRef]
- Tighe, P.; Negm, O.; Todd, I.; Fairclough, L. Utility, reliability and reproducibility of immunoassay multiplex kits. Methods 2013, 61, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.-X.; Qin, J.-J.; Ye, H.; Wang, P.; Wang, K.-J.; Zhang, J.Y. Tumor associated antigens or anti-TAA autoantibodies as biomarkers in the diagnosis of ovarian cancer: A systematic review with meta-analysis. Expert Rev. Mol. Diagn. 2015, 15, 829–852. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.M.; Martins, T.B.; Peterson, L.K.; Hill, H.R. Clinical significance of measuring serum cytokine levels as inflammatory biomarkers in adult and pediatric COVID-19 cases: A review. Cytokine 2021, 142, 155478. [Google Scholar] [CrossRef] [PubMed]
- Perricos, A.; Wenzl, R.; Husslein, H.; Eiwegger, T.; Gstoettner, M.; Weinhaeusel, A.; Beikircher, G.; Kuessel, L. Does the Use of the “Proseek((R)) Multiplex Oncology I Panel” on Peritoneal Fluid Allow a Better Insight in the Pathophysiology of Endometriosis, and in Particular Deep-Infiltrating Endometriosis? J. Clin. Med. 2020, 9, 2009. [Google Scholar] [CrossRef] [PubMed]
- Gyllensten, U.; Hedlund-Lindberg, J.; Svensson, J.; Manninen, J.; Öst, T.; Ramsell, J.; Åslin, M.; Ivansson, E.; Lomnytska, M.; Lycke, M.; et al. Next Generation Plasma Proteomics Identifies High-Precision Biomarker Candidates for Ovarian Cancer. Cancers 2022, 14, 1757. [Google Scholar] [CrossRef] [PubMed]
- Mekhlafi, A.A.; Becker, T.; Klawonn, F. Sample size and performance estimation for biomarker combinations based on pilot studies with small sample sizes. Commun. Stat.-Theory Methods 2020, 51, 5534–5548. [Google Scholar] [CrossRef]
- Mummareddy, S.; Pradhan, S.; Narasimhan, A.K.; Natarajan, A. On Demand Biosensors for Early Diagnosis of Cancer and Immune Checkpoints Blockade Therapy Monitoring from Liquid Biopsy. Biosensors 2021, 11, 500. [Google Scholar] [CrossRef]
- Japp, N.C.; Souchek, J.J.; Sasson, A.R.; Hollingsworth, M.A.; Batra, S.K.; Junker, W.M. Tumor Biomarker In-Solution Quantification, Standard Production, and Multiplex Detection. J. Immunol. Res. 2021, 2021, 9942605. [Google Scholar] [CrossRef]
- FitzGerald, E.A.; Vagrys, D.; Opassi, G.; Klein, H.F.; Hamilton, D.J.; Boronat, P.; Cederfelt, D.; Talibov, V.O.; Abramsson, M.; Moberg, A.; et al. Multiplexed experimental strategies for fragment library screening using SPR biosensors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bellassai, N.; Spoto, G. Biosensors for liquid biopsy: Circulating nucleic acids to diagnose and treat cancer. Anal. Bioanal. Chem. 2016, 408, 7255–7264. [Google Scholar] [CrossRef]
- Andryukov, B.G.; Besednova, N.N.; Romashko, R.V.; Zaporozhets, T.S.; Efimov, T.A. Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends. Biosensors 2020, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Wik, L.; Nordberg, N.; Broberg, J.; Björkesten, J.; Assarsson, E.; Henriksson, S.; Grundberg, I.; Pettersson, E.; Westerberg, C.; Liljeroth, E.; et al. Proximity Extension Assay in Combination with Next-Generation Sequencing for High-throughput Proteome-wide Analysis. Mol. Cell Proteom. 2021, 20, 100168. [Google Scholar] [CrossRef] [PubMed]
- Pourmadadi, M.; Moammeri, A.; Shamsabadipour, A.; Moghaddam, Y.F.; Rahdar, A.; Pandey, S. Application of Various Optical and Electrochemical Nanobiosensors for Detecting Cancer Antigen 125 (CA-125): A Review. Biosensors 2023, 13, 99. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Sun, H.; Li, D.; Yang, W.; Fan, K.; Liu, C.; Dong, L.; Wang, G. A Review of Biosensors for Detecting Tumor Markers in Breast Cancer. Life 2022, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Thorsen, S.B.; Ekman, D.; Eriksson, A.; Dickens, E.R.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef]
- Gaudreault, J.; Forest-Nault, C.; Crescenzo, G.D.; Durocher, Y.; Henry, O. On the Use of Surface Plasmon Resonance-Based Biosensors for Advanced Bioprocess Monitoring. Processes 2021, 9, 1996. [Google Scholar] [CrossRef]
- Breault-Turcot, J.; Poirier-Richard, H.P.; Couture, M.; Pelechacz, D.; Masson, J.F. Single chip SPR and fluorescent ELISA assay of prostate specific antigen. Lab Chip 2015, 15, 4433–4440. [Google Scholar] [CrossRef]
- Wang, X.; Hou, T.; Lin, H.; Lv, W.; Li, H.; Li, F. In situ template generation of silver nanoparticles as amplification tags for ultrasensitive surface plasmon resonance biosensing of microRNA. Biosens. Bioelectron. 2019, 137, 82–87. [Google Scholar] [CrossRef]
- Menon, P.S.; Mulyanti, B.; Jamil, N.A.; Wulandari, C.; Nugroho, H.S.; Mei, G.S.; Abidin, N.F.Z.; Hasanah, L.; Pawinanto, R.E.; Berhanuddin, D.D. Refractive Index and Sensing of Glucose Molarities determined using Au-Cr K-SPR at 670/785 nm Wavelength. Sains Malays. 2019, 48, 1259–1265. [Google Scholar] [CrossRef]
- Skubitz, A.P.; Boylan, K.L.; Geschwind, K.; Cao, Q.; Starr, T.K.; Geller, M.A.; Celestino, J.; Bast, R.C.; Lu, K.H.; Koopmeiners, J.S. Simultaneous Measurement of 92 Serum Protein Biomarkers for the Development of a Multiprotein Classifier for Ovarian Cancer Detection. Cancer Prev. Res. 2019, 12, 171–184. [Google Scholar] [CrossRef]
- Song, Y. Ultrafast Microfluidic Immunoassays towards Real-Time Intervention of Cytokine Storms. Ph.D. Thesis, University of Michigan, Ann Arbor, MI, USA, 2020. [Google Scholar]
- Wang, D.; Loo, J.F.C.; Chen, J.; Yam, Y.; Chen, S.-C.; He, H.; Kong, S.K.; Ho, H.P. Recent Advances in Surface Plasmon Resonance Imaging Sensors. Sensors 2019, 19, 1266. [Google Scholar] [CrossRef] [PubMed]
- Szymanska, B.; Lukaszewski, Z.; Hermanowicz-Szamatowicz, K.; Gorodkiewicz, E. A biosensor for determination of the circulating biomarker CA125/MUC16 by Surface Plasmon Resonance Imaging. Talanta 2020, 206, 120187. [Google Scholar] [CrossRef] [PubMed]
- Peltomaa, R.; Glahn-Martínez, B.; Benito-Peña, E.; Moreno-Bondi, M.C. Optical Biosensors for Label-Free Detection of Small Molecules. Sensors 2018, 18, 4126. [Google Scholar] [CrossRef] [PubMed]
- Puiu, M.; Bala, C. SPR and SPR Imaging: Recent Trends in Developing Nanodevices for Detection and Real-Time Monitoring of Biomolecular Events. Sensors 2016, 16, 870. [Google Scholar] [CrossRef]
- Damborsky, P.; Svitel, J.; Katrlik, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar]
- Nurrohman, D.T.; Wang, Y.H.; Chiu, N.F. Exploring Graphene and MoS(2) Chips Based Surface Plasmon Resonance Biosensors for Diagnostic Applications. Front. Chem. 2020, 8, 728. [Google Scholar] [CrossRef] [PubMed]
- Eddin, F.B.K.; Fen, Y.W. The Principle of Nanomaterials Based Surface Plasmon Resonance Biosensors and Its Potential for Dopamine Detection. Molecules 2020, 25, 2769. [Google Scholar] [CrossRef] [PubMed]
- Sameer, Q. The potential implementation of biosensors for the diagnosis of biomarkers of various cancer. Preprints 2022. [Google Scholar] [CrossRef]
- Said, F.A.; Menon, P.S.; Rajendran, V.; Shaari, S.; Majlis, B.Y. Investigation of graphene-on-metal substrates for SPR-based sensor using finite-difference time domain. IET Nanobiotechnol. 2017, 11, 981–986. [Google Scholar] [CrossRef]
- Sharma, S.; Raghav, R.; Kennedy, R. O’.; Srivastava, S. Advances in ovarian cancer diagnosis: A journey from immunoassays to immunosensors. Enzym. Microb. Technol. 2016, 89, 15–30. [Google Scholar] [CrossRef]
- Justino, C.I.L.; Duarte, A.C.; Rocha-Santos, T.A.P. Critical overview on the application of sensors and biosensors for clinical analysis. Trends Anal. Chem. 2016, 85, 36–60. [Google Scholar] [CrossRef]
- Szymanska, B.; Lukaszewski, Z.; Hermanowicz-Szamatowicz, K.; Gorodkiewicz, E. A Multiple-Array SPRi Biosensor as a Tool for Detection of Gynecological-Oncological Diseases. Biosensors 2023, 13, 279. [Google Scholar] [CrossRef]
- Szymanska, B.; Lukaszewski, Z.; Oldak, L.; Zelazowska-Rutkowska, B.; Hermanowicz-Szamatowicz, K.; Gorodkiewicz, E. Two Biosensors for the Determination of Interleukin-6 in Blood Plasma by Array SPRi. Biosensors 2022, 12, 412. [Google Scholar] [CrossRef]
- Çimen, D.; Bereli, N.; Denizli, A. Molecularly Imprinted Surface Plasmon Resonance Sensor-Based Devices for Clinical Applications. Biomed. Mater. Devices 2022, 11. [Google Scholar] [CrossRef]
- Ryu, J.-H.; Lee, H.Y.; Lee, J.-Y.; Kim, H.-S.; Kim, S.-H.; Ahn, H.S.; Ha, D.H.; Yi, S.N. Enhancing SERS Intensity by Coupling PSPR and LSPR in a Crater Structure with Ag Nanowires. Appl. Sci. 2021, 11, 11855. [Google Scholar] [CrossRef]
- Rebelo, T.; Costa, R.; Brandao, A.; Silva, A.F.; Sales, M.G.F.; Pereira, C.M. Molecularly imprinted polymer SPE sensor for analysis of CA-125 on serum. Anal. Chim. Acta 2019, 1082, 126–135. [Google Scholar] [CrossRef]
- Islam, M.S.; Kouzani, A.Z.; Dai, X.J.; Michalski, W.P. Parameter Sensitivity Analysis of Surface Plasmon Resonance Biosensor through Numerical Simulation. In Proceedings of the IEEE/IICME International Conference on Complex Medical Engineering, Gold Coast, QLD, Australia, 13–15 July 2010. [Google Scholar]
- Yi, R.; Zhang, Z.; Liu, C.; Qi, Z. Gold-silver alloy film based surface plasmon resonance sensor for biomarker detection. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 116, 111126. [Google Scholar] [CrossRef]
- Eom, G.; Hwang, A.; Kim, H.; Moon, J.; Kang, H.; Jung, J.; Lim, E.-K.; Jeong, J.; Park, H.G.; Kang, T. Ultrasensitive detection of ovarian cancer biomarker using Au nanoplate SERS immunoassay. BioChip J. 2021, 15, 348–355. [Google Scholar] [CrossRef]
- Meyer, S.A.; Le, E.C.R.; Etchegoin, P.G. Combining surface plasmon resonance (SPR) spectroscopy with surface-enhanced Raman scattering (SERS). Anal. Chem. 2011, 83, 2337–2344. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Ashton, K.M.; Stringfellow, H.F.; Wood, N.J.; Keating, P.J.; Rowbottom, A.W.; Martin-Hirsch, P.L.; Martin, F.L. Raman spectroscopic techniques to detect ovarian cancer biomarkers in blood plasma. Talanta 2018, 189, 281–288. [Google Scholar] [CrossRef]
- Farzin, L.; Shamsipur, M.; Samandari, L.; Sheibani, S. HIV biosensors for early diagnosis of infection: The intertwine of nanotechnology with sensing strategies. Talanta 2020, 206, 120201. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.K.; Kitahama, Y.; Huang, G.G.; Han, X.; Ozaki, Y. Surface-enhanced Raman scattering: Realization of localized surface plasmon resonance using unique substrates and methods. Anal. Bioanal. Chem. 2009, 394, 1747–1760. [Google Scholar] [CrossRef] [PubMed]
- Geka, G.; Kanioura, A.; Likodimos, V.; Gardelis, S.; Papanikolaou, N.; Kakabakos, S.; Petrou, P. SERS Immunosensors for Cancer Markers Detection. Materials 2023, 16, 3733. [Google Scholar] [CrossRef] [PubMed]
- TunC, I.; Susapto, H.H. Label-Free Detection of Ovarian Cancer Antigen CA125 by Surface Enhanced Raman Scattering. J. Nanosci. Nanotechnol. 2020, 20, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Cheng, L.; Wang, W. Surface Plasmon Resonance of Large-Size Ag Nanobars. Micromachines 2022, 13, 638. [Google Scholar] [CrossRef] [PubMed]
- Gur, S.D.; Sinem, D.G.; Bakhshpour, M.; Denizli, A. Nanoscale SPR sensor for the ultrasensitive detection of the ovarian cancer marker carbohydrate antigen 125. New J. Chem. 2022, 46, 7263–7270. [Google Scholar]
- Ruscito, A.; DeRosa, M.C. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Kim, T. Review of microfluidic approaches for surface-enhanced Raman scattering. Sens. Actuators B Chem. 2015, 227, 504–514. [Google Scholar] [CrossRef]
- Divya, J.; Selvendran, S.; Raja, A.S.; Sivasubramanian, A. Surface plasmon based plasmonic sensors: A review on their past, present and future. Biosens. Bioelectron. X 2022, 11, 100175. [Google Scholar] [CrossRef]
- Menon, P.S.; Jamil, N.A.; Mei, G.S.; Zain, A.R.M.; Hewak, D.W.; Huang, C.-C.; Mohamed, M.A.; Majlis, B.Y.; Mishra, R.K.; Raghavan, S.; et al. Multilayer CVD-Graphene and MoS₂ Ethanol Sensing and Characterization Using Kretschmann-Based SPR. IEEE J. Electron. Devices Soc. 2020, 8, 1227–1235. [Google Scholar] [CrossRef]

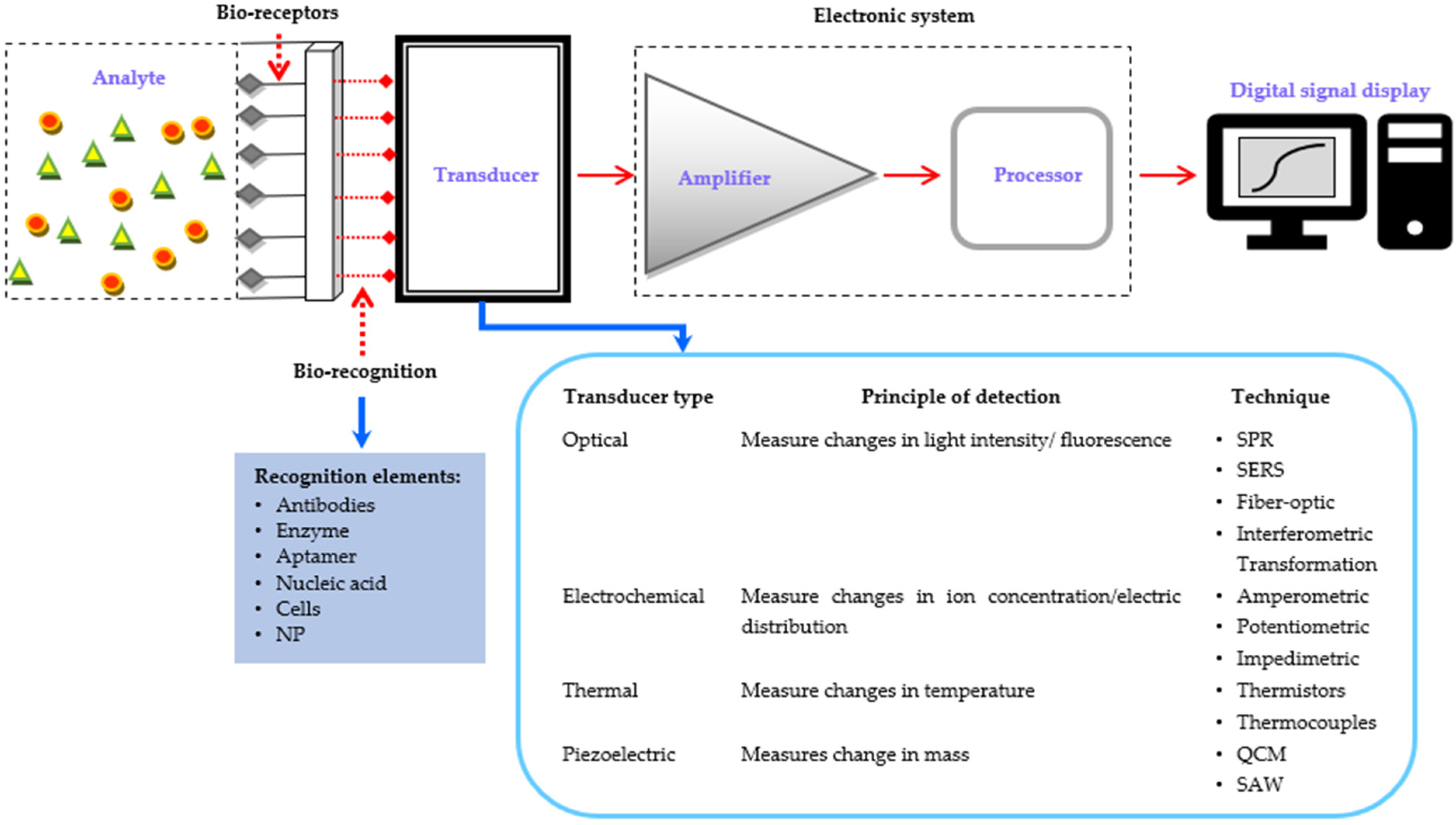

| Biosensor Types | Advantages | Disadvantages | Type of Signal | Surface Substrate | Ref. |
|---|---|---|---|---|---|
| SPR |
|
| RI | Metal sensor thickness | [2,81,107] |
| LSPR |
|
| Absorption peak shift | Particle size and shape | [28,81,108] |
| SPRi |
|
| Intensity contrast image | Control the intensity of light with a fixed angle | [96,99] |
| Platform | Biomarker | LOD | Dynamic Range | Wavelength (nm) | Ref. |
|---|---|---|---|---|---|
| SPR | CA125 | 0.1 U/mL−1 | 0.1–300 U/mL−1 | 670 | [111] |
| 0.1 U/mL−1 | 0.1–1 U/mL−1 | 600–700 | [113] | ||
| SPRi | CA125 | 0.01 U/mL−1 | 0.1–10 U/mL−1 | 670 | [122] |
| 2.2 U/mL−1 | 2.2–150 U/mL−1 | 633 | [97] | ||
| HE4 | 2pM | 2–110 pM | - | [43] | |
| LSPR-SERS | CA125 | 20 ug/mL | - | 532 | [120] |
| HE4 | 10−17 M | 10−17–10−9 M | 633 | [114] |
| Paper | Recognition Element | Metal Flim/Fabrication Strategy | Stability | Advantages | Limitation |
|---|---|---|---|---|---|
| Rebelo et al. (2019) [111] | MIP | Au/Py/SPE | - | Good selectivity. Reusability. | SSPR to pH, and temperature, which can affect its conductivity and stability |
| Gür et al. (2020) [122] | Imprinted polymeric NPs | Au/Poly(HEMA-MATrp)/NS | 5 months | High stability. Reusability. | Susceptibility to hydrolysis, which can lead to its degradation over time |
| Szymańska et al. (2020) [97] & (2021) [43] | pcAb | Au-CysA | - | Good recovery. High specificity. | Low absorptivity. Instability. |
| Yi et al. (2020) [113] | pcAb | AuAg | - | Excellent biocompatibility. | Instability. |
| TunC et al. (2020) [120] | mAbs/Ag | AuNPs/SERS | - | SERS signal remains consistent even at low concentrations. | Limit the extent of Ag-NP interactions |
| Eom et al. (2021) [114] | mAbs | Au/NPl/(Cys)3-protein G/SERS | - | High specificity. | Complexity of the substrate preparation. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, V.R.; Kampan, N.C.; Abd Aziz, N.H.; Teik, C.K.; Shafiee, M.N.; Menon, P.S. Recent Advances in Surface Plasmon Resonance (SPR) Technology for Detecting Ovarian Cancer Biomarkers. Cancers 2023, 15, 5607. https://doi.org/10.3390/cancers15235607
Kumar VR, Kampan NC, Abd Aziz NH, Teik CK, Shafiee MN, Menon PS. Recent Advances in Surface Plasmon Resonance (SPR) Technology for Detecting Ovarian Cancer Biomarkers. Cancers. 2023; 15(23):5607. https://doi.org/10.3390/cancers15235607
Chicago/Turabian StyleKumar, Vikneswary Ravi, Nirmala Chandralega Kampan, Nor Haslinda Abd Aziz, Chew Kah Teik, Mohamad Nasir Shafiee, and P. Susthitha Menon. 2023. "Recent Advances in Surface Plasmon Resonance (SPR) Technology for Detecting Ovarian Cancer Biomarkers" Cancers 15, no. 23: 5607. https://doi.org/10.3390/cancers15235607
APA StyleKumar, V. R., Kampan, N. C., Abd Aziz, N. H., Teik, C. K., Shafiee, M. N., & Menon, P. S. (2023). Recent Advances in Surface Plasmon Resonance (SPR) Technology for Detecting Ovarian Cancer Biomarkers. Cancers, 15(23), 5607. https://doi.org/10.3390/cancers15235607











